Resistance to Antimicrobials Mediated by Efflux Pumps in Staphylococcus aureus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Evaluation of Efflux Activity
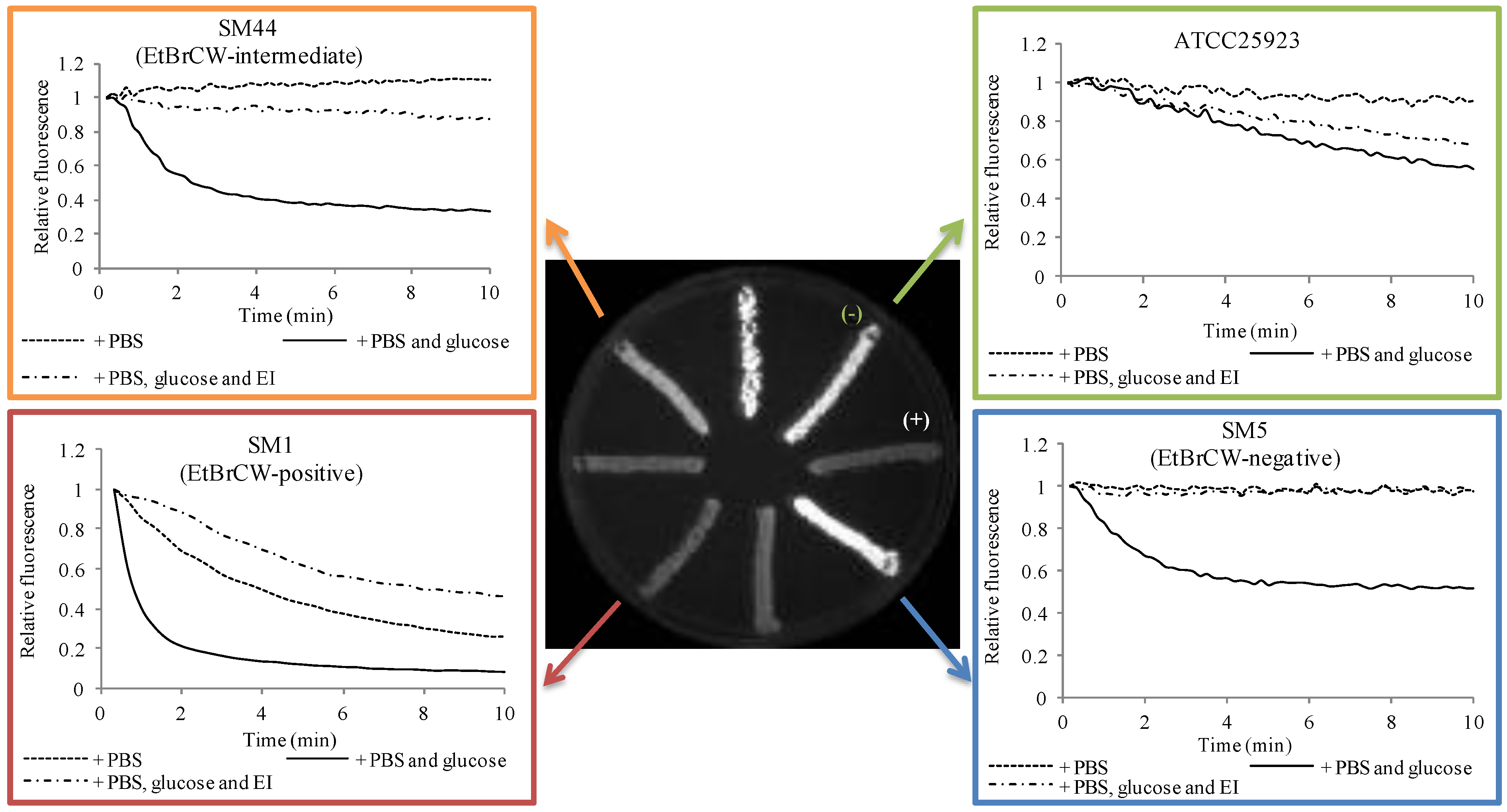
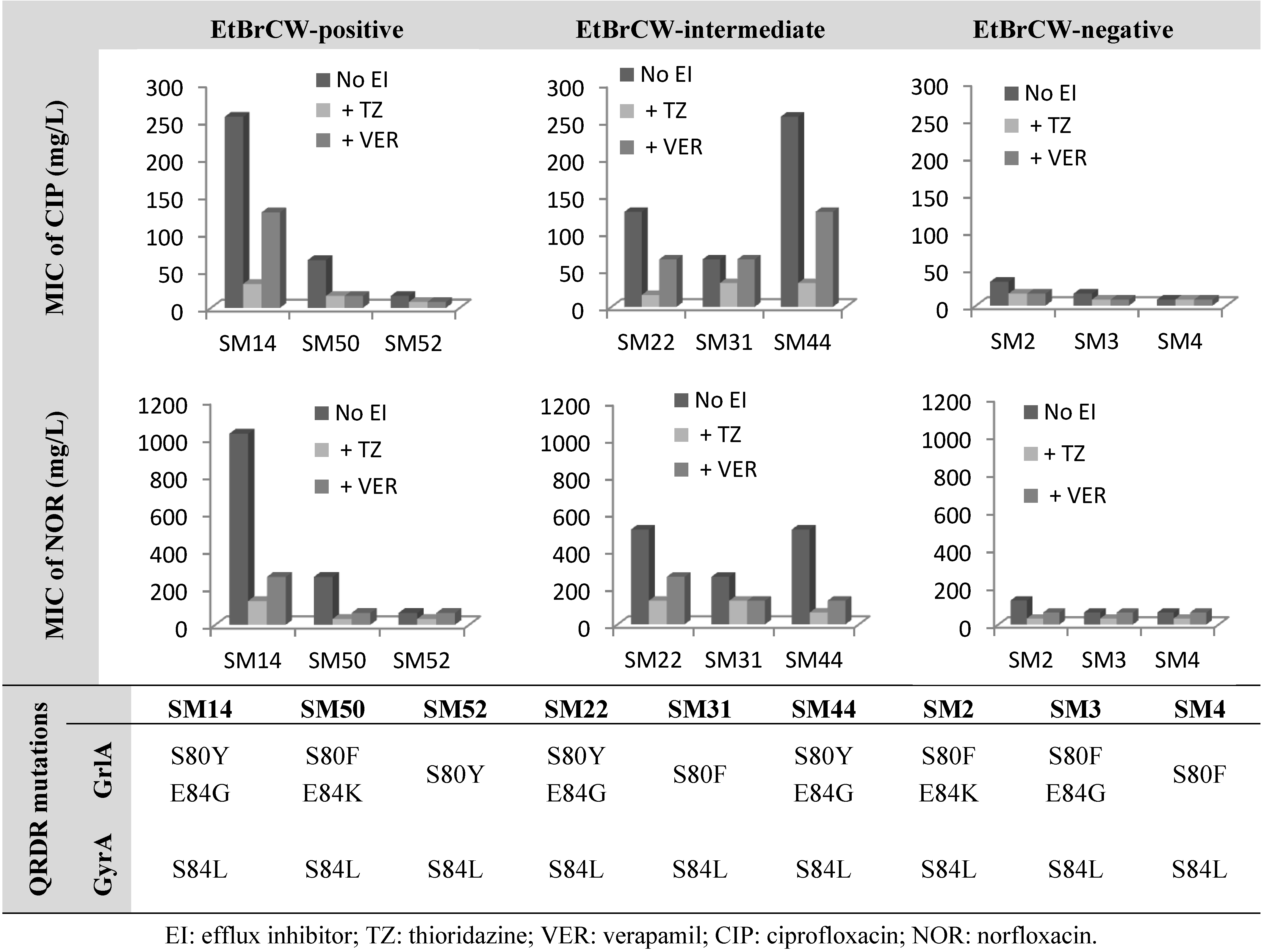
2.2. Contribution of Efflux to Fluoroquinolone Resistance

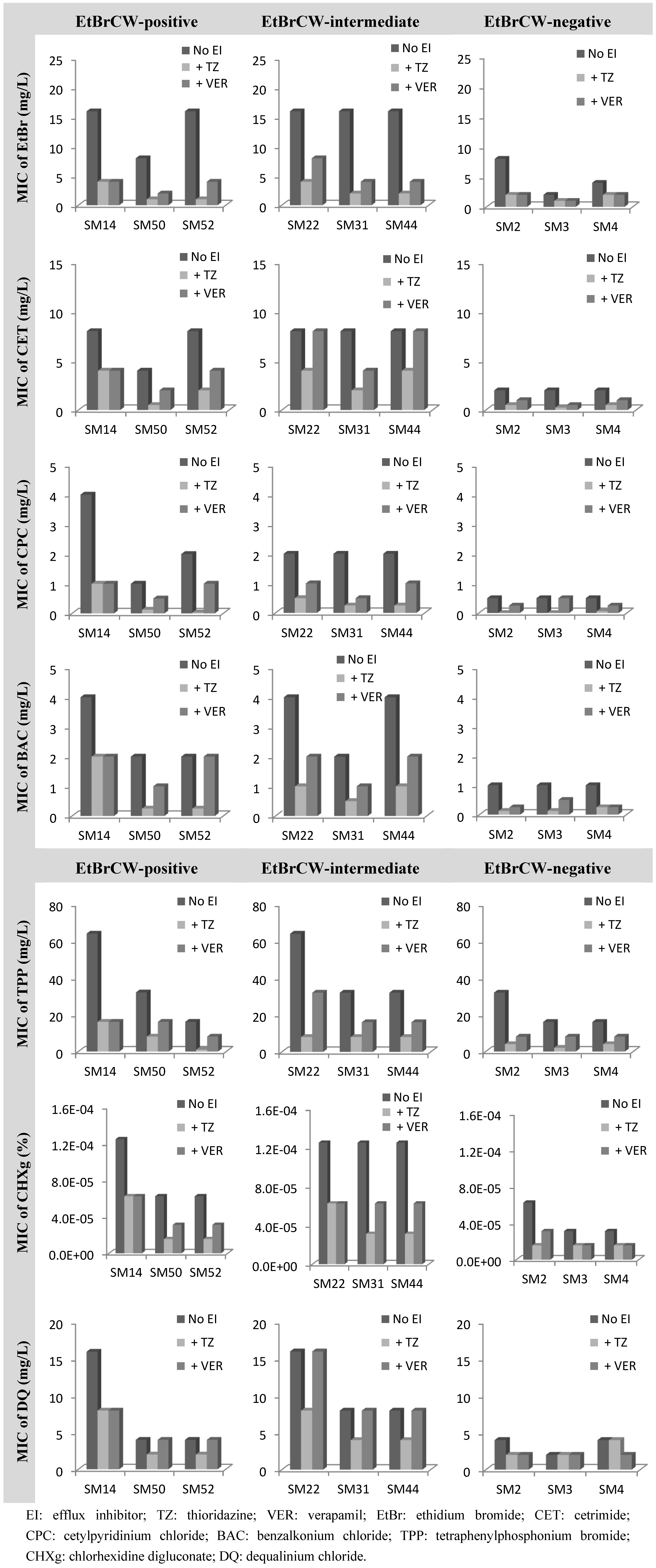
2.3. Contribution of Efflux to Biocide Reduced Susceptibility
3. Experimental
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Conflict of Interest
References and Notes
- Paulsen, I.T.; Brown, M.H.; Skurray, R.A. Proton-dependent multidrug efflux systems. Microbiol. Rev. 1996, 60, 575–608. [Google Scholar]
- Poole, K. Bacterial multidrug efflux pumps serve other functions. Microbe 2008, 3, 179–185. [Google Scholar]
- Piddock, L.J.V. Multidrug-resistance efflux pumps? not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Piddock, L.J.V. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- DeMarco, C.E.; Cushing, L.A.; Frempong-Manso, E.; Seo, S.M.; Jaravasa, T.A.A.; Kaatz, G.W. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3235–3239. [Google Scholar]
- Costa, S.S.; Falcão, C.; Viveiros, M.; Machado, D.; Martins, M.; Melo-Cristino, J.; Amaral, L.; Couto, I. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Staphylococcus aureus. BMC Microbiol. 2011, 11, e241. [Google Scholar] [CrossRef]
- Kosmidis, C.; Schindler, B.D.; Jacinto, P.L.; Patel, D.; Bains, K.; Kaatz, G.W. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 2012, 40, 204–209. [Google Scholar] [CrossRef]
- Yoshida, H.; Bogaki, M.; Nakamura, S.; Ubukata, K.; Konno, M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 1990, 172, 6942–6949. [Google Scholar]
- Kaatz, G.W.; Seo, S.M.; O'Brien, L.; Wahiduzzaman, M.; Foster, T.J. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 44, 1404–1406. [Google Scholar] [CrossRef]
- Viveiros, M.; Martins, M.; Couto, I.; Rodrigues, L.; Spengler, G.; Martins, A.; Kristiansen, J.E.; Molnar, J.; Amaral, L. New methods for the identification of efflux mediated MDR bacteria, genetic assessment of regulators and efflux pump constituents, characterization of efflux systems and screening of inhibitors of efflux pumps. Curr. Drug Targets 2008, 9, 760–768. [Google Scholar] [CrossRef]
- Martins, M.; Viveiros, M.; Couto, I.; Costa, S.S.; Pacheco, T.; Fanning, S.; Pagès, J.M.; Amaral, L. Identification of efflux pump-mediated multidrug-resistant bacteria by the Ethidium Bromide-Agar Cartwheel Method. In Vivo 2011, 25, 171–178. [Google Scholar]
- Viveiros, M.; Rodrigues, L.; Martins, M.; Couto, I.; Spengler, G.; Martins, A.; Amaral, L. Evaluation of Efflux Activity of Bacteria by A Semi-automated Fluorometric System. In Antibiotic Resistance Protocols; Gillespie, S.H., McHugh, T.D., Eds.; Humana Press: New York, NY, USA, 2010; Methods in Molecular Biology Series; Volume 642, pp. 159–172. [Google Scholar]
- Patel, D.; Kosmidis, C.; Seo, S.M.; Kaatz, G.W. Ethidium bromide MIC screening for enhanced efflux pump gene expression or efflux activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5070–5073. [Google Scholar] [CrossRef]
- Wolfson, J.S.; Hooper, D.C. The fluoroquinolones: Structures, mechanisms of action and resistance, and spectra of activity in vitro. Antimicrob. Agents Chemother. 1985, 28, 581–586. [Google Scholar] [CrossRef]
- Hooper, D.C. Mechanisms of fluoroquinolone resistance. Drug Res. Upd. 1999, 2, 38–55. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC), Antimicrobial resistance surveillance in Europe 2010. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). ECDC: Stockholm, Sweden, 2011.
- Ferrero, L.; Cameron, B.; Crouzet, J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 1995, 39, 1554–1558. [Google Scholar] [CrossRef]
- Tanaka, M.; Wang, T.; Onodera, Y.; Uchida, Y.; Sato, K. Mechanism of quinolone resistance in Staphylococcus aureus. J. Infect. Chemother. 2000, 6, 131–139. [Google Scholar] [CrossRef]
- Noguchi, N.; Okihara, T.; Namiki, Y.; Kumaki, Y.; Yamanaka, Y.; Koyama, M.; Wakasugi, K.; Sasatsu, M. Susceptibility and resistance genes to fluoroquinolones in methicillin-resistant Staphylococcus aureus isolated in 2002. Int. J. Antimicrob. Agents 2005, 25, 374–379. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Seo, S.M.; Ruble, C.A. Mechanisms of fluoroquinolone resistance in Staphylococcus aureus. J. Infect. Dis. 1991, 163, 1080–1086. [Google Scholar] [CrossRef]
- Schmitz, F.-J.; Fluit, A.C.; Lückefahr, M.; Engler, B.; Hofmann, B.; Verhoef, J.; Heinz, H.-P.; Hadding, U.; Jones, M.E. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 42, 807–810. [Google Scholar] [CrossRef]
- Muñoz-Bellido, J.L.; Manzanares, M.A.A.; Martínez, J.Á.; Guttiérrez Zufiaurre, M.N.; Ortiz, G.; Segovia Hernández, M.; García-Rodríguez, J.A. Efflux pump-mediated quinolones resistance in Staphylococcus aureus strains wild-type for gyrA, gyrB, grlA, and norA. Antimicrob. Agents Chemother. 1999, 43, 354–356. [Google Scholar]
- Tanaka, M.; Zhang, Y.X.; Ishida, H.; Akasaka, T.; Sato, K.; Hayakawa, I. Mechanisms of 4-quinolone resistance in quinolones-resistant and methicillin-resistant Staphylococcus aureus isolates from Japan and China. J. Med. Microbiol. 1995, 42, 214–219. [Google Scholar] [CrossRef]
- Schmitz, F.-J.; Köhrer, K.; Scheuring, S.; Verhoef, J.; Fluit, A.; Heinz, H.P.; Jones, M.E. The stability of grlA, grlB, gyrA, gyrB and norA mutations and MIC values of five fluoroquinolones in three different clonal populations of methicillin-resistant Staphylococcus aure. Clin. Microbiol. Infect. 1999, 5, 287–290. [Google Scholar] [CrossRef]
- Schmitz, F.-J.; Fluit, A.C.; Brisse, S.; Verhoef, J.; Köhrer, K.; Milatovic, D. Molecular epidemiology of quinolones resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol. Med. Microbiol. 1999, 26, 281–287. [Google Scholar] [CrossRef]
- Guirao, G.Y.; Martínez Toldos, M.C.; Mora Peris, B.; Alonso Manzanares, M.A.; Gutiérrez Zufiaurre, M.N.; Martínez Andrés, J.A.; Muñoz Bellido, J.L.; García-Rodríguez, J.A.; Segovia Hernández, M. Molecular diversity of quinolones resistance in genetically related clinical isolates of Staphylococcus aureus and susceptibility to newer quinolones. J. Antimicrob. Chemother. 2001, 47, 157–161. [Google Scholar] [CrossRef]
- Horii, T.; Suzuki, Y.; Monji, A.; Morita, M.; Muramatsu, H.; Kondo, Y.; Doi, M.; Takeshita, A.; Kanno, T.; Maekawa, M. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: Effects of the mutations on fluoroquinolone MICs. Diagn. Microbiol. Infect. Dis. 2003, 46, 139–145. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Dunman, P.M.; Strahilevitz, J.; Projan, S.J.; Hooper, D.C. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 2005, 187, 2395–2405. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Strahilevitz, J.; Hooper, D.C. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 1104–1107. [Google Scholar] [CrossRef]
- Kaatz, G.W.; McAleese, F.; Seo, S.M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005, 49, 1857–1864. [Google Scholar] [CrossRef]
- Wang, T.; Tanaka, M.; Sato, K. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob. Agents Chemother. 1998, 42, 236–240. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI), Performance standards for antimicrobial susceptibility testing; twenty-second Informational Supplement M100-S22. CLSI: Wayne, PA, USA, 2012.
- Singh, R.; Swick, M.C.; Ledesma, K.R.; Yang, Z.; Hu, M.; Zechiedrich, L.; Tam, V.H. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 2012, 56, 1680–1685. [Google Scholar] [CrossRef]
- Machado, D.; Couto, I.; Perdigão, J.; Rodrigues, L.; Portugal, I.; Baptista, P.; Veigas, B.; Amaral, L.; Viveiros, M. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One 2012, 7, e34538. [Google Scholar]
- Markham, P.N.; Neyfakh, A.A. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996, 40, 2673–2674. [Google Scholar]
- Sulavik, M.C.; Barg, N.L. Examination of methicillin-resistant and methicillin-susceptible Staphylococcus aureus mutants with low-level fluoroquinolone resistance. Antimicrob. Agents Chemother. 1998, 42, 3317–3319. [Google Scholar]
- Huet, A.A.; Raygada, J.L.; Mendiratta, K.; Seo, S.M.; Kaatz, G.W. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 2008, 154, 3144–3153. [Google Scholar] [CrossRef]
- McDonnell, G.; Russel, A.D. Antiseptics and disinfectants: Activity, action and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar]
- Gilbert, P.; Moore, L.E. Cationic antiseptics: Diversity of action under a common epithet. J. Appl. Microbiol. 2005, 99, 703–715. [Google Scholar] [CrossRef]
- Gilbert, P.; McBain, A.J. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 2003, 16, 189–208. [Google Scholar] [CrossRef]
- Maillard, J.Y. Bacterial resistance to biocides in the healthcare environment: Should it be of genuine concern? J. Hosp. Infect. 2007, 65, 60–72. [Google Scholar] [CrossRef]
- Grinius, L.L.; Goldberg, E.B. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 1994, 269, 29998–30004. [Google Scholar]
- Mayer, S.; Boos, M.; Beyer, A.; Fluit, A.C.; Schmitz, F.-J. Distribution of the antiseptic resistance genes qacA, qacB, and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 47, 893–895. [Google Scholar] [CrossRef]
- Zmantar, T.; Kouidhi, B.; Miladi, H.; Bakhrouf, A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res. Notes 2011, 4, e453. [Google Scholar] [CrossRef]
- Longtin, J.; Seah, C.; Siebert, K.; McGeer, A.; Simor, A.; Longtin, Y.; Low, D.E.; Melano, R.G. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob. Agents Chemother. 2011, 55, 2999–3001. [Google Scholar] [CrossRef]
- Weber, D.J.; Rutala, W.A.; Sickbert-Bennett, E.E. Outbreaks associated with contaminated antiseptics and disinfectants. Antimicrob. Agents Chemother. 2007, 51, 4217–4224. [Google Scholar] [CrossRef]
- Buffet-Bataillon, S.; Tattevin, P.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Emergence of resistance to antibacterial agents: The role of quaternary ammonium compounds—A critical review. Int. J. Antimicrob. Agents. 2012, 39, 381–389. [Google Scholar] [CrossRef]
- Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR), Assessment of the antibiotic resistance effects of biocides. 19 January 2009.
- Couto, I.; Costa, S.S.; Viveiros, M.; Martins, M.; Amaral, L. Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. J. Antimicrob. Chemother. 2008, 62, 504–513. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Costa, S.S.; Junqueira, E.; Palma, C.; Viveiros, M.; Melo-Cristino, J.; Amaral, L.; Couto, I. Resistance to Antimicrobials Mediated by Efflux Pumps in Staphylococcus aureus. Antibiotics 2013, 2, 83-99. https://doi.org/10.3390/antibiotics2010083
Costa SS, Junqueira E, Palma C, Viveiros M, Melo-Cristino J, Amaral L, Couto I. Resistance to Antimicrobials Mediated by Efflux Pumps in Staphylococcus aureus. Antibiotics. 2013; 2(1):83-99. https://doi.org/10.3390/antibiotics2010083
Chicago/Turabian StyleCosta, Sofia S., Elisabete Junqueira, Cláudia Palma, Miguel Viveiros, José Melo-Cristino, Leonard Amaral, and Isabel Couto. 2013. "Resistance to Antimicrobials Mediated by Efflux Pumps in Staphylococcus aureus" Antibiotics 2, no. 1: 83-99. https://doi.org/10.3390/antibiotics2010083




