Natural Products from Actinobacteria Associated with Fungus-Growing Termites
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Diversity
2.2. Antimicrobial Activities Against Test Strains
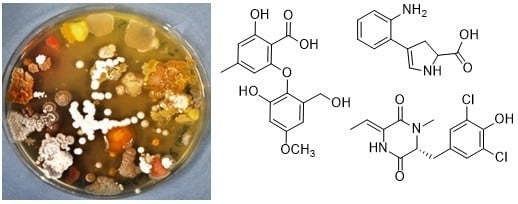
2.3. Chemical Analysis
3. Discussion
3.1. Phylogenetic and Ecological Relevance of the Actinobacteria
3.2. Bioactivities and Natural Products
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. Chem. 2011, 46, 4769–4807. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Kim, H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016, 5, 69–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molloy, E.M.; Hertweck, C. Antimicrobial discovery inspired by ecological interactions. Curr. Opin. Microbiol. 2017, 39, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.; Tuttle, R.N.; Jensen, P.R. Omics-based natural product discovery and the lexicon of genome mining. Curr. Opin. Microbiol. 2017, 39, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Guo, H.; Rischer, M.; Poulsen, M. Natural products from microbes associated with insects. Beilstein J. Org. Chem. 2016, 12, 314–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Salem, H.; Kreutzer, E.; Sudakaran, S.; Kaltenpoth, M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 2013, 15, 1956–1968. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Yildirim, E.; Gürbüz, M.F.; Herzner, G.; Strohm, E. Refining the roots of the beewolf-Streptomyces symbiosis: Antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl. Environ. Microbiol. 2012, 78, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Currie, C.R.; Mueller, U.G.; Malloch, D. The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. USA 1999, 96, 7998–8002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulsen, M.; Cafaro, M.J.; Erhardt, D.P.; Little, A.E.; Gerardo, N.M.; Tebbets, B.; Klein, B.S.; Currie, C.R. Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ. Microbiol. Rep. 2010, 2, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.C.; Poulsen, M.; Currie, C.R.; Clardy, J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 2009, 5, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Peleg, B.; Norris, D.M. Symbiotic interrelationships between microbes and ambrosia beetles. J. Invertebr. Pathol. 1972, 20, 59–65. [Google Scholar] [CrossRef]
- Shukla, S.P.; Sanders, J.G.; Byrne, M.J.; Pierce, N.E. Gut microbiota of dung beetles correspond to dietary specializations of adults and larvae. Mol. Ecol. 2016, 25, 6092–6106. [Google Scholar] [CrossRef] [PubMed]
- Um, S.; Park, S.H.; Kim, J.; Park, H.J.; Ko, K.; Bang, H.S.; Lee, S.K.; Shin, J.; Oh, D.C. Coprisamides A and B, new branched cyclic peptides from a gut bacterium of the Dung Beetle Copris tripartitus. Org. Lett. 2017, 5, 1272–1275. [Google Scholar] [CrossRef] [PubMed]
- Visser, A.A.; Nobre, T.; Currie, C.R.; Aanen, D.K.; Poulsen, M. Exploring the potential for actinobacteria as defensive symbionts in fungus-growing termites. Microb. Ecol. 2012, 63, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Kurtböke, D.I.; French, J.R.; Hayes, R.A.; Quinn, R.J. Eco-taxonomic insights into actinomycete symbionts of termites for discovery of novel bioactive compounds. Adv. Biochem. Eng. Biotechnol. 2015, 147, 111–135. [Google Scholar] [PubMed]
- Aanen, D.K.; Eggleton, P.; Rouland-Lefevre, C.; Guldberg-Froslev, T.; Rosendahl, S.; Boomsma, J.J. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl. Acad. Sci. USA 2002, 99, 14887–14892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobre, T.; Rouland-Lefevre, C.; Aanen, D.K. Comparative biology of fungus cultivation in termites and ants. In Biology of Termites: A Modern Sysnthesis; Bignell, D., Roisin, Y.L.N., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 193–210. [Google Scholar]
- Um, S.; Fraimout, A.; Sapountzis, P.; Oh, D.C.; Poulsen, M. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 2013, 3, 3250. [Google Scholar] [CrossRef] [PubMed]
- Visser, A.A.; Kooij, P.W.; Debets, A.J.M.; Kuyper, T.W.; Aanen, D.K. Pseudoxylaria as stowaway of the fungus-growing termite nest: Interaction asymmetry between Pseudoxylaria, Termitomyces and free-living relatives. Fungal Ecol. 2011, 4, 322–332. [Google Scholar] [CrossRef]
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin a, an ansamycin from a termite-associated Streptomyces sp. Chem. Sci. 2014, 5, 4333–4338. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org. Lett. 2017, 19, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Ruzzini, A.C.; Beemelmanns, C.; Kim, K.H.; Klassen, J.L.; Cao, S.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; Clardy, J. Linear peptides are the major products of a biosynthetic pathway that encodes for cyclic depsipeptides. Org. Lett. 2017, 19, 1772–1775. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.R.; Lee, D.; Benndorf, R.; Jung, W.H.; Beemelmanns, C.; Kang, K.S.; Kim, K.H. Termisoflavones A-C, isoflavonoid glycosides from termite-associated Streptomyces sp. RB1. J. Nat. Prod. 2016, 79, 3072–3078. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Benndorf, R.; Leichnitz, D.; Klassen, J.L.; Vollmers, J.; Görls, H.; Steinacker, M.; Weigel, C.; Dahse, H.-M.; Kaster, A.K.; et al. Isolation, biosynthesis and chemical modifications of rubterolones A-F: Rare tropolone alkaloids from Actinomadura sp. 5–2. Chem. Eur. J. 2017, 23, 9338–9345. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, R.R.; Hu, H.; Pilgaard, B.; Vreeburg, S.M.E.; Schückel, J.; Pedersen, K.S.K.; Kračun, S.K.; Busk, P.K.; Harholt, J.; Sapountzis, P.; et al. Enzyme activities at different stages of plant biomass decomposition in three species of fungus-growing termites. Appl. Environ. Microbiol. 2018, 14, e01815-17. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Gupta, R.S. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 66–112. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Goker, M.; Sproer, C.; Klenk, H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wang, S. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Sbaraini, N.; Guedes, R.L.; Andreis, F.C.; Junges, Â.; de Morais, G.L.; Vainstein, M.; de Vasconcelos, A.T.; Schrank, A. Secondary metabolite gene clusters in the entomopathogen fungus Metarhizium anisopliae: Genome identification and patterns of expression in a cuticle infection model. BMC Genom. 2016, 17, 736. [Google Scholar] [CrossRef] [PubMed]
- BLAST: Basic Local Alignment Search Tool—NCBI-NIH. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 26 July 2018).
- Jayasuriya, H.; Ball, R.G.; Zink, D.L.; Smith, J.L.; Goetz, M.A.; Jenkins, R.G.; Nallin-Omstead, M.; Silverman, K.C.; Bills, G.F.; Lingham, R.B.; et al. Barceloneic acid A, a new farnesyl-protein transferase inhibitor from a Phoma species. J. Nat. Prod. 1995, 58, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Aurora Screening Library; Order Number: K13.438.893; CAS Registry Number: 1798281-02-3. Available online: http://www.aurorafinechemicals.com (accessed on 3 July 2018).
- GNPS: Global Natural Products Social Molecular Networking. Available online: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp (accessed on 29 June 2018).
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Duran, H.G.S.; de los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. AntiSMASH4.0-improvments in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; van der Donk, W.A. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef] [PubMed]
- Ökesli, A.; Cooper, L.E.; Fogle, E.J.; van der Donk, W.A. Nine post-translational modifications during biosynthesis of cinnamycin. J. Am. Chem. Soc. 2011, 133, 13753–13760. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Songsumanus, A.; Kudo, T.; Ohkuma, M.; Phongsopitanun, W.; Tanasupawat, S. Actinomadura montaniterrae sp. nov., isolated from mountain soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 3310–3316. [Google Scholar] [CrossRef] [PubMed]
- Promnuan, Y.; Kudo, T.; Ohkuma, M.; Chantawannakul, P. Actinomadura apis sp. nov., isolated from a honey bee (Apis mellifera) hive, and the reclassification of Actinomadura cremea subsp. Rifamycini gauze et al. 1987 as Actinomadura rifamycini (gauze et al. 1987) sp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 2271–2277. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, C.; Zhao, J.; Guo, L.; Lu, C.; Li, J.; Jia, F.; Wang, X.; Xiang, W. Microbispora camponoti sp. nov., a novel actinomycete isolated from the cuticle of Camponotus japonicus mayr. Antonie Van Leeuwenhoek 2016, 109, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Otani, S.; Mikaelyan, A.; Nobre, T.; Hansen, L.H.; Kone, N.A.; Sorensen, S.J.; Aanen, D.K.; Boomsma, J.J.; Brune, A.; Poulsen, M. Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 2014, 23, 4631–4644. [Google Scholar] [CrossRef] [PubMed]
- Shinzato, N.; Muramatsu, M.; Matsui, T.; Watanabe, Y. Phylogenetic analysis of the gut bacterial microflora of the fungus-growing termite Odontotermes formosanus. Biosci. Biotechnol. Biochem. 2007, 71, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Mikaelyan, A.; Dietrich, C.; Köhler, T.; Poulsen, M.; Sillam-Dussès, D.; Brune, A. Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol. Ecol. 2015, 24, 5284–5295. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.H.; Xie, L.; Liu, N.; Yan, X.; Li, M.H.; Fan, M.Z.; Wang, Q. Comparison of gut-associated and nest-associated microbial communities of a fungus-growing termite (Odontotermes yunnanensis). Insect Sci. 2010, 17, 265–276. [Google Scholar] [CrossRef]
- Haofu, H. Gut Bacterial Contributions to Antimicrobial Defense and Biomass Decomposition in Fungus-Farming Termites. Ph.D. Thesis, University of Copenhagen, Copenhagen, Denmark, 2018. [Google Scholar]
- Otani, S.; Hansen, L.H.; Sorensen, S.J.; Poulsen, M. Bacterial communities in termite fungus combs are comprised of consistent gut deposits and contributions from the environment. Microb. Ecol. 2016, 71, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jiang, Y.; Wang, X.; Chen, D.; Chen, X.; Wang, L.; Han, L.; Huang, X.; Jiang, C. Diversity, antimicrobial activity, and biosynthetic potential of cultivable actinomycetes associated with lichen symbiosis. Microb. Ecol. 2017, 74, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Cain, C.C.; Lee, D.; Waldo Iii, R.H.; Henry, A.T.; Casida, E.J.; Wani, M.C.; Wall, M.E.; Oberlies, N.H.; Falkinham, J.O., III. Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 2003, 47, 2113–2117. [Google Scholar] [CrossRef] [PubMed]
- Grgurina, I.; Mariotti, F. Biosynthetic origin of syringomycin and syringopeptin 22, toxic secondary metabolites of the phytopathogenic bacterium Pseudomonas syringae pv. Syringae. FEBS Lett. 1999, 462, 151–154. [Google Scholar] [CrossRef]
- Cheng, Y.; Yang, M.; Matter, A.M. Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968. Appl. Environ. Microbiol. 2007, 73, 3460–3469. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; van der Donk, W.A. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 2016, 23, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Ogino, T.; Sunazuka, T.; Tanaka, H.; Omura, S. Chloropeptins, new anti-HIV antibiotics inhibiting gp120-CD4 binding from Streptomyces sp. II. Structure elucidation of chloropeptin I. J. Antibiot. 1997, 50, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.O. Chlorinated tyrosine derivatives in insect cuticle. Insect Biochem. Mol. Biol. 2004, 34, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Matselyukh, B.; Mohammadipanah, F.; Laatsch, H.; Rohr, J.; Efremenkova, O.; Khilya, V. N-methylphenylalanyl-dehydrobutyrine diketopiperazine, an A-factor mimic that restores antibiotic biosynthesis and morphogenesis in Streptomyces globisporus 1912-B2 and Streptomyces griseus 1439. J. Antibiot. 2015, 68, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequences alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Wright, E.S.; Yilmaz, L.S.; Noguera, D.R. Decipher, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 2012, 78, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC BioInform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- iTOL v3. Available online: https://itol.embl.de/ (accessed on 31 July 2018).
- Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. Available online: https://www.dsmz.de/ (accessed on 24 July 2018).
- Fdhila, F.; Vazquez, V.; Sanchez, J.L.; Riguera, R. dd-Diketopiperazines: Antibiotics active against Vibrio anguillarum isolated from marine bacteria associated with cultures of Pecten maximus. J. Nat. Prod. 2003, 66, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Stierle, A.C.; Cardellina, J.H., II; Strobel, G.A. Maculosin, a host-specific phytotoxin for spotted knapweed from Alternaria alternata. Proc. Natl. Acad. Sci. USA 1988, 85, 8008–8011. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Kersten, R.D.; Liu, W.T.; Wang, M.; Purvine, S.O.; Wu, S.; Brewer, H.M.; Pasa-Tolic, L.; Bandeira, N.; Moore, B.S.; et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014, 9, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591. [Google Scholar] [CrossRef]
- Kodani, S.; Komaki, H.; Ishimura, S.; Hemmi, H.; Ohnishi-Kameyama, M. Isolation and structure determination of a new lantibiotic cinnamycin B from Actinomadura atramentaria based on genome mining. J. Ind. Microbiol. Biotechnol. 2016, 43, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Goh, E.B.; Yim, G.; Tsui, W.; McClure, J.; Surette, M.G.; Davies, J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 2002, 99, 17025–31700. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Spiegelman, G.B.; Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006, 9, 445–453. [Google Scholar] [CrossRef] [PubMed]









| Genus | Family | This Study: Origin of Isolation (Number of Isolates) | Visser et al. [17] Origin of Isolation (Number of Isolates) | |||
|---|---|---|---|---|---|---|
| Termite Gut | Termite Exoskeleton | Fungus Comb | Termite Exoskeleton | Fungus Comb | ||
| Streptomyces | Streptomycetaceae | 46 | 14 | 13 | 13 | 2 |
| Kitasatospora | Streptomycetaceae | 0 | 0 | 0 | 2 | 0 |
| Actinomadura | Thermomonosporaceae | 4 | 1 | 0 | 1 | 0 |
| Leifsonia | Microbacteriaceae | 3 | 0 | 0 | 0 | 0 |
| Curtobacterium | Microbacteriaceae | 1 | 0 | 0 | 0 | 0 |
| Arthrobacter | Micrococcaceae | 0 | 1 | 0 | 0 | 0 |
| Micromonospora | Micromonosporaceae | 3 | 0 | 0 | 1 | 1 |
| Nocardia | Nocardiaceae | 3 | 0 | 0 | 0 | 0 |
| Aeromicrobium | Nocardioidaceae | 1 | 0 | 0 | 0 | 0 |
| Cellulosimicrobium | Promicromonosporaceae | 1 | 0 | 0 | 0 | 0 |
| Mycobacterium | Mycobacteriaceae | 3 | 0 | 0 | 0 | 0 |
| Sphaerisporangium | Streptosporangiaceae | 1 | 0 | 0 | 0 | 0 |
| Microbispora | Streptosporangiaceae | 1 | 0 | 0 | 0 | 0 |
| Luteimicrobium | family of order Micrococcales | 1 | 0 | 0 | 0 | 0 |
| Medium (Abbreviation) | Content Per L |
|---|---|
| Potato Dextrose Broth (PDB) | 26.5 g potato extract glucose (6.5 g potato extract, 20 g glucose) |
| Potato Dextrose Agar (PDA) | 26.5 g potato extract glucose, 20.0 g agar |
| ISP2 Broth | 4.0 g yeast extract, 10.0 g malt extract, 4.0 g glucose |
| ISP2 Agar | 4.0 g yeast extract, 10.0 g malt extract, 4.0 g glucose, 20.0 g agar |
| Chitin Agar | 4.0 g chitin, 0.7 g K2HPO4, 0.3 g KH2PO4, 0.57 g MgSO4·7H2O, 0.01 g FeSO4·7H2O, 0.0018 g ZnSO4·7H2O, 0.0016 g MnCl2·4H2O |
| Minimal Media | 2.0 g Na-acetate, 2.0 g NH4Cl 0.7 g K2HPO4, 0.3 g KH2PO4, 0.57 g MgSO4·7H2O, 0.01 g FeSO4·7H2O, 0.0018 g ZnSO4·7H2O, 0.0016 g MnCl2·4H2O |
| MC Agar | 5.0 g microcrystalline cellulose, 20.0 g agar |
| Soya Broth | 20.0 g soya flour, 20.0 g glucose, 5.0 g NaCl, 3.0 g CaCO3, 0.25 mL desmophen |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benndorf, R.; Guo, H.; Sommerwerk, E.; Weigel, C.; Garcia-Altares, M.; Martin, K.; Hu, H.; Küfner, M.; De Beer, Z.W.; Poulsen, M.; et al. Natural Products from Actinobacteria Associated with Fungus-Growing Termites. Antibiotics 2018, 7, 83. https://doi.org/10.3390/antibiotics7030083
Benndorf R, Guo H, Sommerwerk E, Weigel C, Garcia-Altares M, Martin K, Hu H, Küfner M, De Beer ZW, Poulsen M, et al. Natural Products from Actinobacteria Associated with Fungus-Growing Termites. Antibiotics. 2018; 7(3):83. https://doi.org/10.3390/antibiotics7030083
Chicago/Turabian StyleBenndorf, René, Huijuan Guo, Elisabeth Sommerwerk, Christiane Weigel, Maria Garcia-Altares, Karin Martin, Haofu Hu, Michelle Küfner, Z. Wilhelm De Beer, Michael Poulsen, and et al. 2018. "Natural Products from Actinobacteria Associated with Fungus-Growing Termites" Antibiotics 7, no. 3: 83. https://doi.org/10.3390/antibiotics7030083
APA StyleBenndorf, R., Guo, H., Sommerwerk, E., Weigel, C., Garcia-Altares, M., Martin, K., Hu, H., Küfner, M., De Beer, Z. W., Poulsen, M., & Beemelmanns, C. (2018). Natural Products from Actinobacteria Associated with Fungus-Growing Termites. Antibiotics, 7(3), 83. https://doi.org/10.3390/antibiotics7030083