Potential of a Bead-Based Multiplex Assay for SARS-CoV-2 Antibody Detection
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
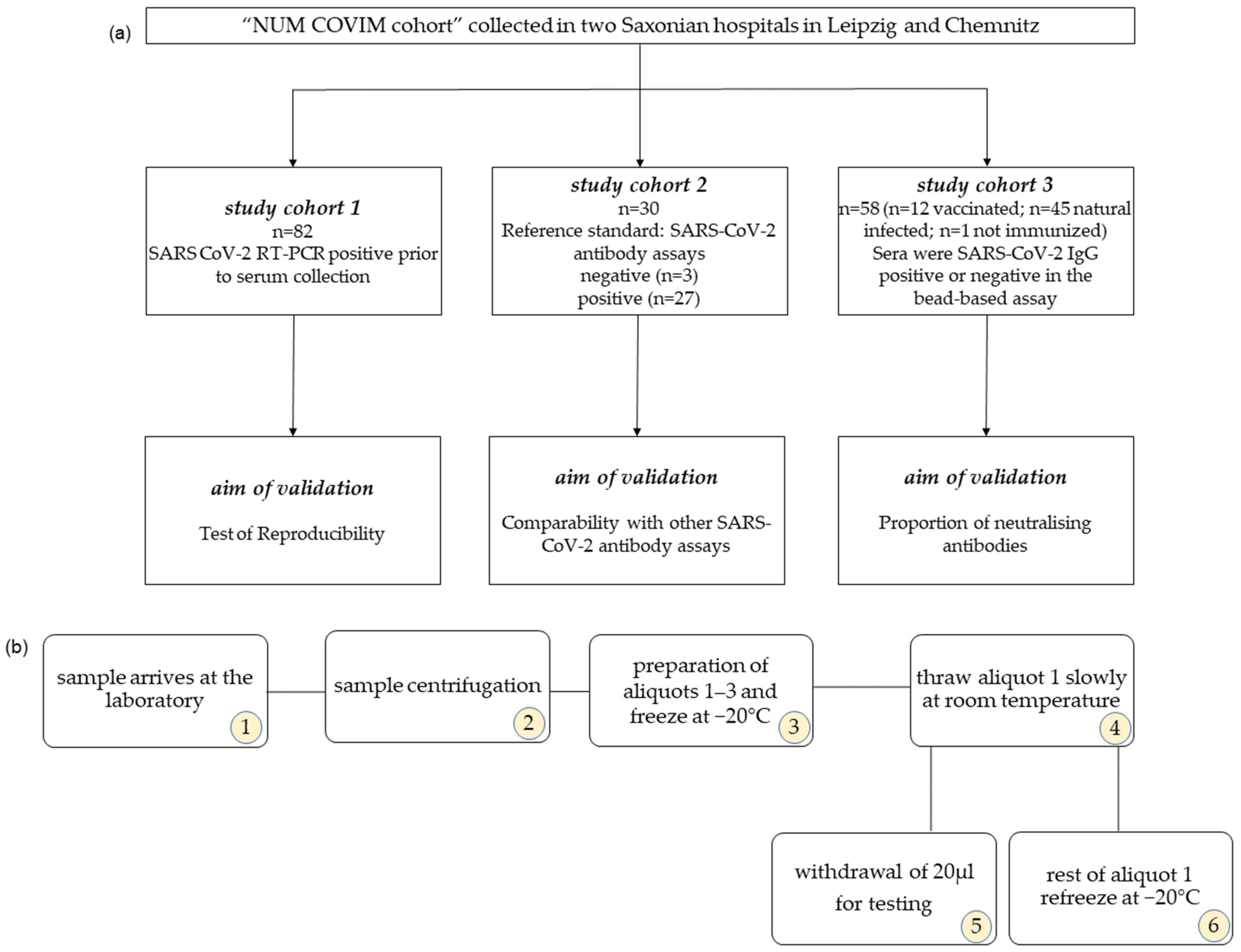
2.1. Study Cohort
2.2. Serum Storage
2.3. Bead-Based Immunoassay
2.4. Data Analysis of Bead-Based Immunoassay
2.5. IgG ELISA and ACE-2 Assay for the Detection of Neutralising Antibodies
2.6. Statistical Analysis
3. Results
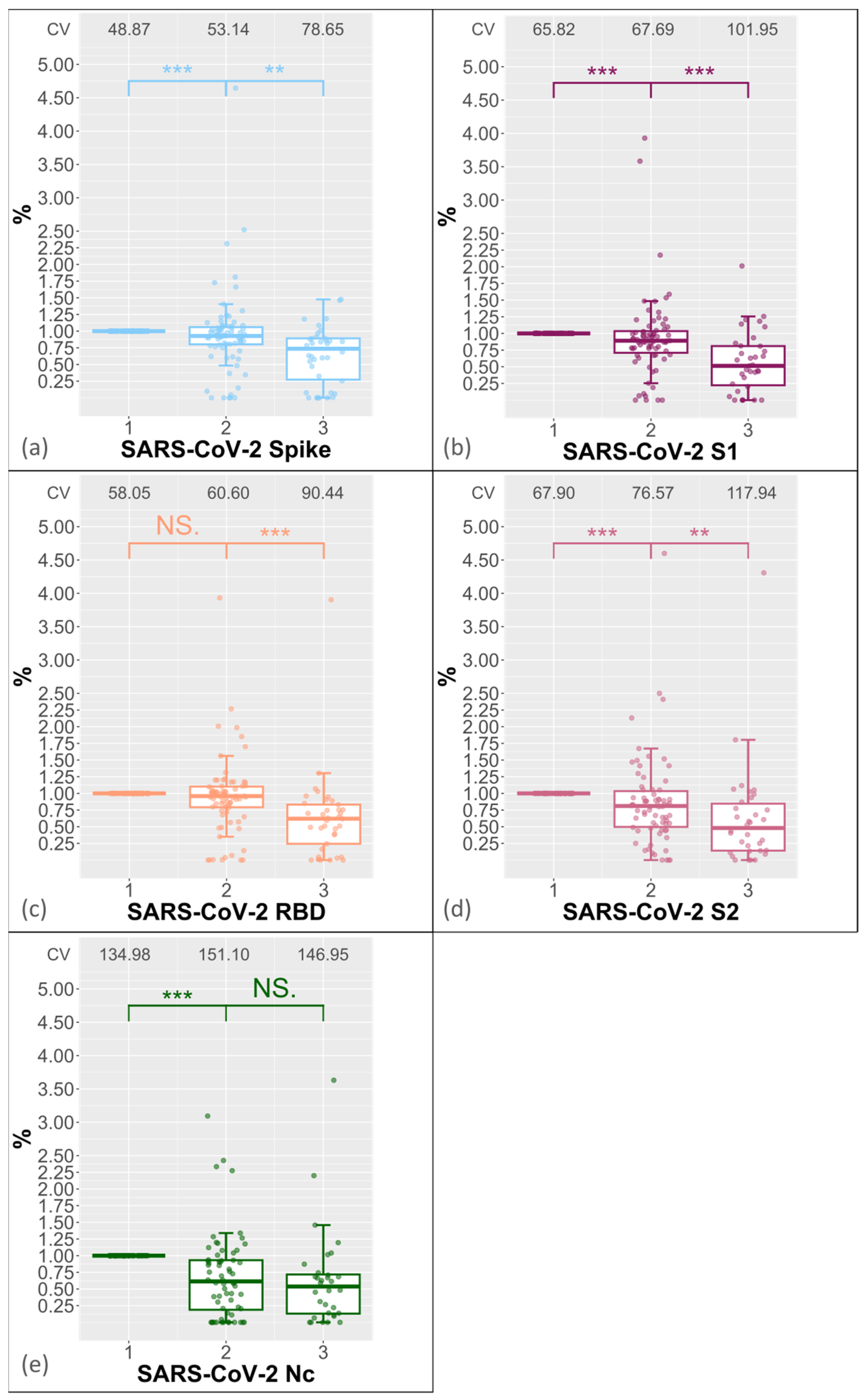
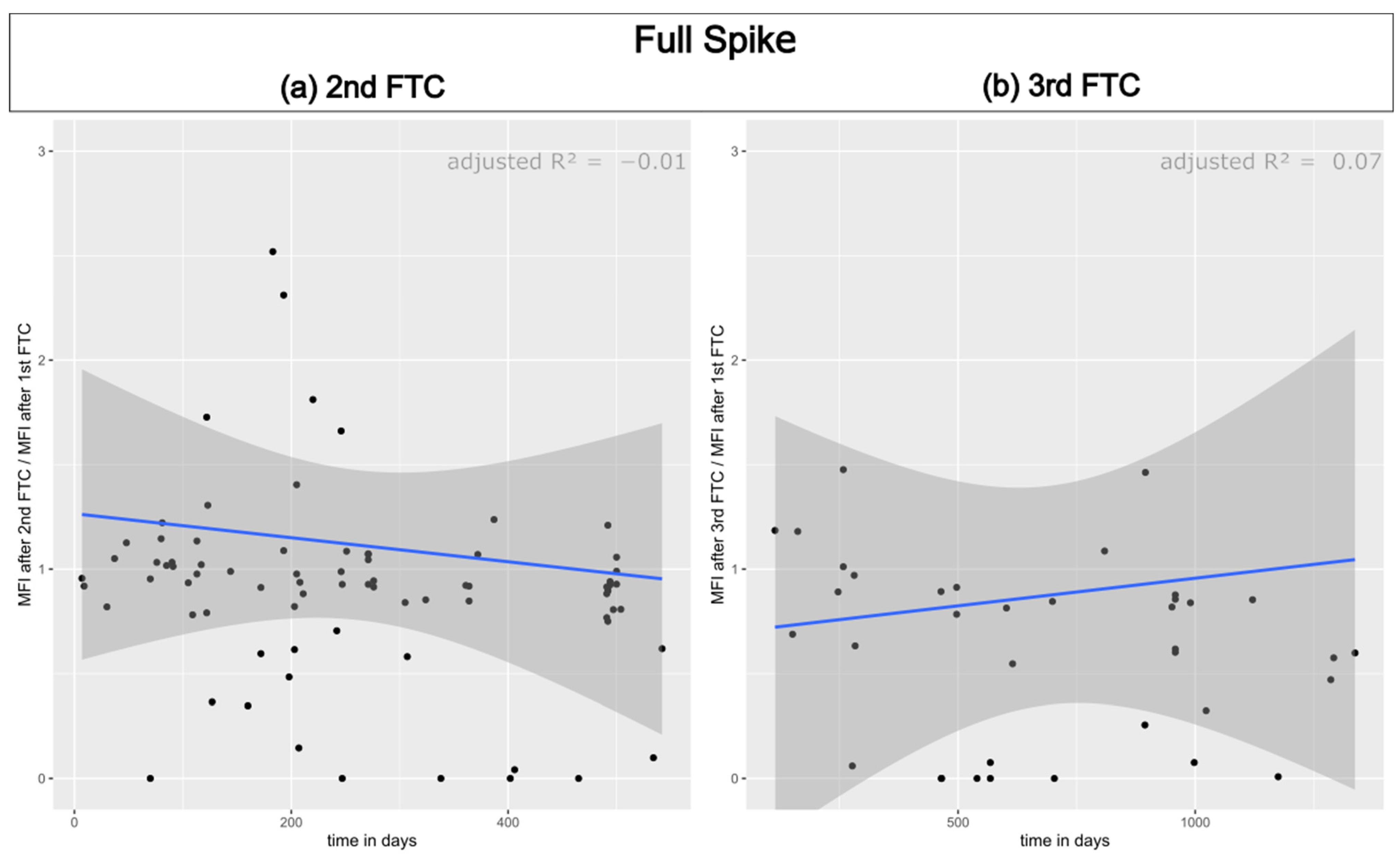
3.1. Reproducibility of Antibody Measurement Using the Luminex Test and Its Dependence on the Number of Freeze–Thaw Cycles
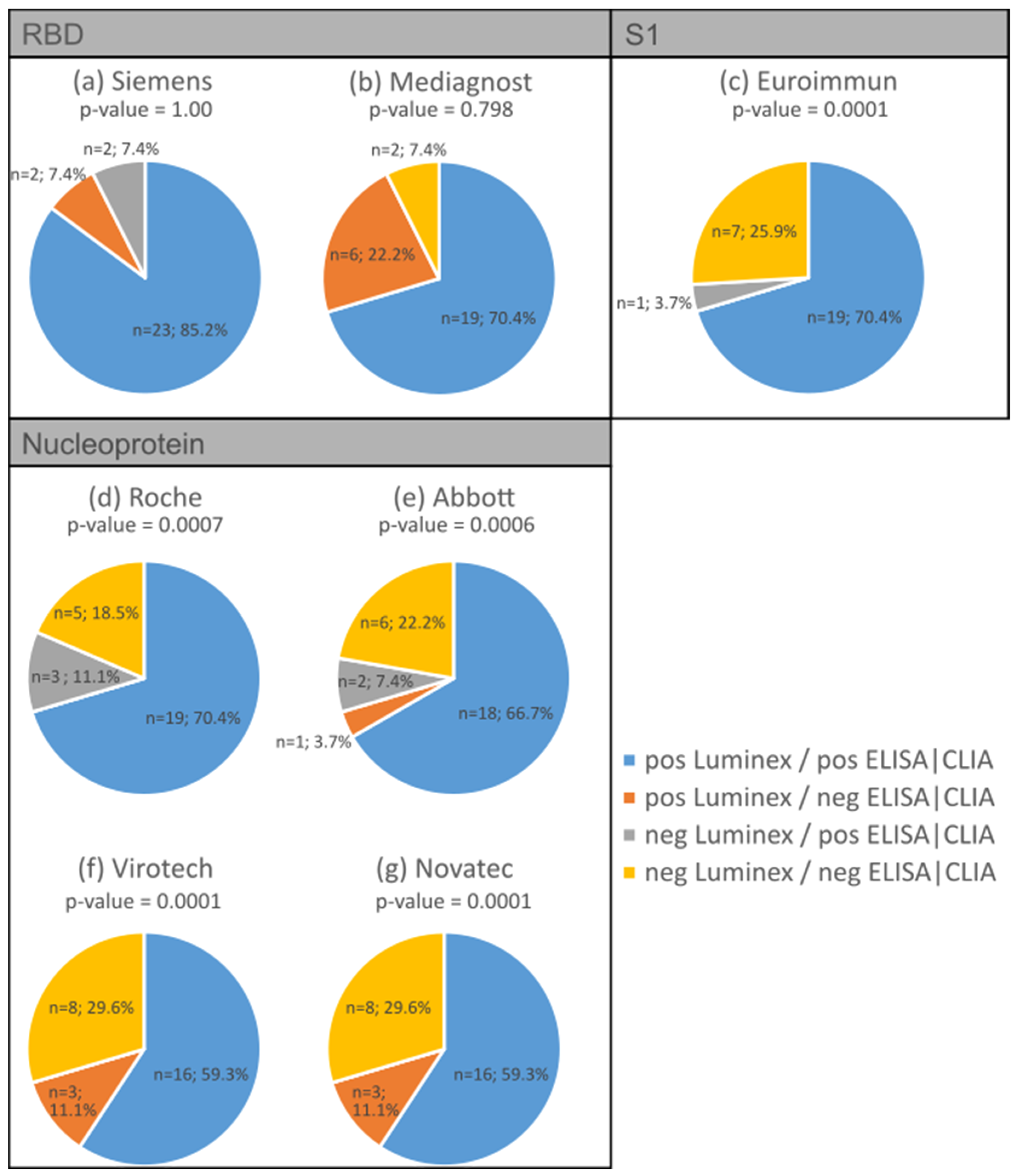
3.2. Validation of Antibody Measurement Using Luminex with Various Established ELISA/CLIA Tests
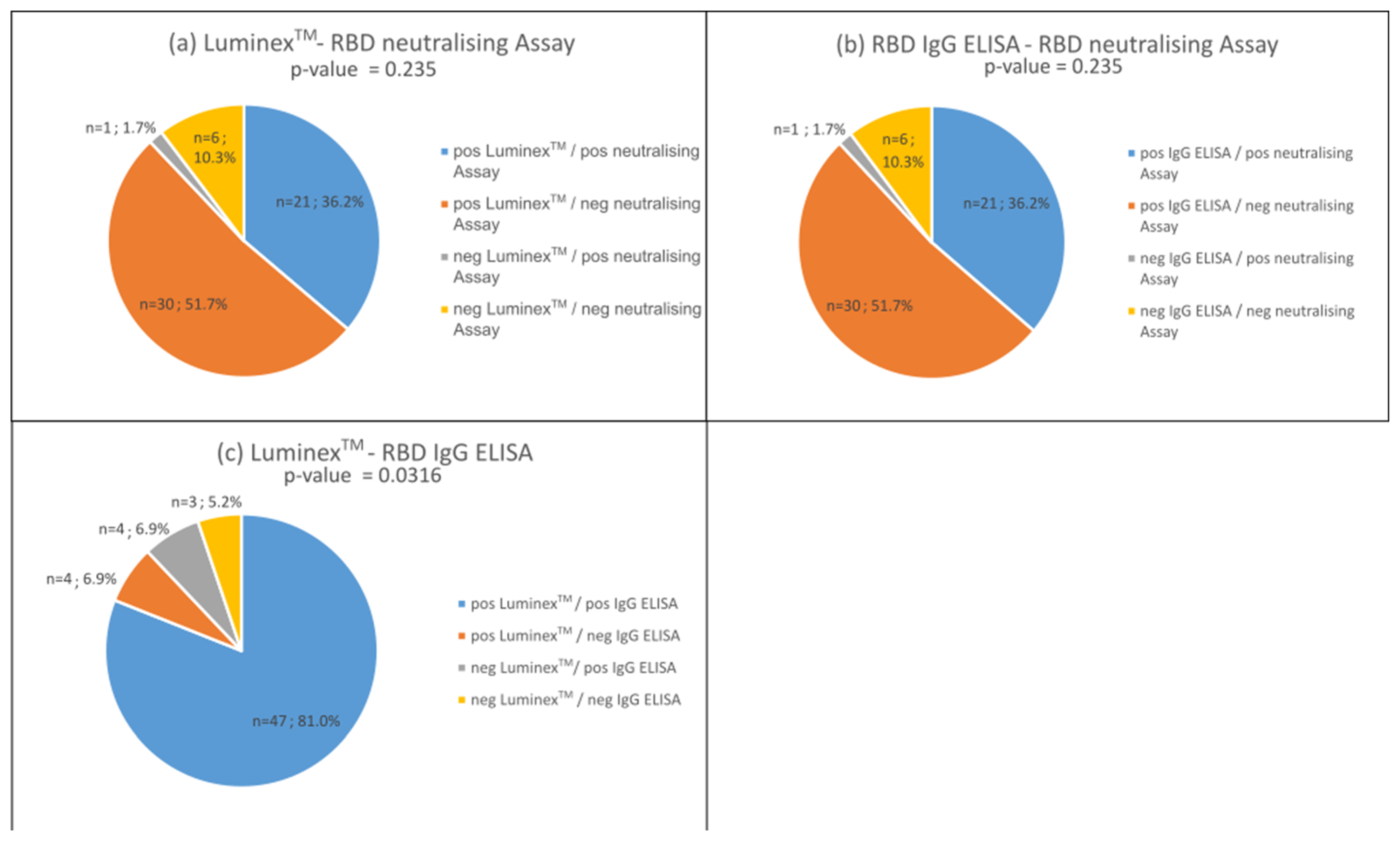
3.3. Proportion of Neutralising Antibodies in the Bead-Based Multiplex Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Impfung Gegen COVID-19. Available online: https://www.infektionsschutz.de/coronavirus/schutzimpfung/impfung-gegen-covid-19/ (accessed on 17 February 2024).
- Press Releases—Antibodies Post-SARS-CoV-2 Infection—New Insights into the Sensitivity and Detection Duration of Antibody Tests. Available online: https://www.pei.de/EN/newsroom/press-releases/year/2022/03-antibodies-post-sars-cov-2-infection-new-insights-sensitivity-detection-duration-antibody-tests.html (accessed on 17 February 2024).
- Devi, M.J.; Gaffar, S.; Hartati, Y.W. A review post-vaccination SARS-CoV-2 serological test: Method and antibody titer response. Anal. Biochem. 2022, 658, 114902. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, P.; Murawski, C.; Ehmen, C.; von Possel, R.; Pekarek, N.; Oestereich, L.; Duraffour, S.; Pahlmann, M.; Struck, N.; Eibach, D.; et al. Limited specificity of commercially available SARS-CoV-2 IgG ELISAs in serum samples of African origin. Trop. Med. Int. Health 2021, 26, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, P.; von Possel, R.; Hemmer, C.J.; Fritzsche, C.; Geerdes-Fenge, H.; Menge, B.; Messing, C.; Borchardt-Lohölter, V.; Deschermeier, C.; Steinhagen, K. Longitudinal detection of SARS-CoV-2-specific antibody responses with different serological methods. J. Med. Virol. 2021, 93, 5816–5824. [Google Scholar] [CrossRef] [PubMed]
- Kannenberg, J.; Schnurra, C.; Reiners, N.; Henschler, R.; Buhmann, R.; Kaiser, T.; Biemann, R.; Hönemann, M.; Ackermann, G.; Trawinski, H.; et al. Sensitivity of SARS-CoV-2 antibody tests with late convalescent sera. J. Clin. Virol. Plus 2021, 1, 100038. [Google Scholar] [CrossRef] [PubMed]
- Scheiblauer, H.; Nübling, C.M.; Wolf, T.; Khodamoradi, Y.; Bellinghausen, C.; Sonntagbauer, M.; Esser-Nobis, K.; Filomena, A.; Mahler, V.; Maier, T.J.; et al. Antibody response to SARS-CoV-2 for more than one year—Kinetics and persistence of detection are predominantly determined by avidity progression and test design. J. Clin. Virol. 2022, 146, 105052. [Google Scholar] [CrossRef] [PubMed]
- Von Possel, R.; Menge, B.; Deschermeier, C.; Fritzsche, C.; Hemmer, C.; Geerdes-Fenge, H.; Loebermann, M.; Schulz, A.; Lattwein, E.; Steinhagen, K.; et al. Performance Analysis of Serodiagnostic Tests to Characterize the Incline and Decline of the Individual Humoral Immune Response in COVID-19 Patients: Impact on Diagnostic Management. Viruses 2024, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Schnurra, C.; Reiners, N.; Biemann, R.; Kaiser, T.; Trawinski, H.; Jassoy, C. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J. Clin. Virol. 2020, 129, 104544. [Google Scholar] [CrossRef]
- Dou, X.; Wang, E.; Hu, J.; Zong, Z.; Jiang, R.; Wang, M.; Kan, L.; Zhang, X. Comparison of three automatic chemiluminescent immunoassays for monitoring dynamic profile of SARS-CoV-2 IgG and IgM. J. Clin. Lab. Anal. 2021, 35, e23681. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.C.; Risch, M.; Thiel, S.L.; Grossmann, K.; Nigg, S.; Wohlwend, N.; Lung, T.; Hillmann, D.; Ritzler, M.; Ferrara, F.; et al. Characteristics of Three Different Chemiluminescence Assays for Testing for SARS-CoV-2 Antibodies. Dis. Markers 2021, 2021, 8810196. [Google Scholar] [CrossRef]
- Bray, R.A.; Lee, J.-H.; Brescia, P.; Kumar, D.; Nong, T.; Shih, R.; Woodle, E.S.; Maltzman, J.S.; Gebel, H.M. Development and Validation of a Multiplex, Bead-based Assay to Detect Antibodies Directed Against SARS-CoV-2 Proteins. Transplantation 2021, 105, 79–89. [Google Scholar] [CrossRef]
- Heaney, C.D.; Pisanic, N.; Randad, P.R.; Kruczynski, K.; Howard, T.; Zhu, X.; Littlefield, K.; Patel, E.U.; Shrestha, R.; Laeyendecker, O.; et al. Comparative performance of multiplex salivary and commercially available serologic assays to detect SARS-CoV-2 IgG and neutralization titers. J. Clin. Virol. 2021, 145, 104997. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, J.; Wen, F.; Liu, K.; Chen, Y. Detection methods and dynamic characteristics of specific antibodies in patients with COVID-19: A review of the early literature. Heliyon 2024, 10, e24580. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.-C.; Tiu, C.; Hu, Z.; Chen, V.C.-W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Morales-Núñez, J.J.; Muñoz-Valle, J.F.; Torres-Hernández, P.C.; Hernández-Bello, J. Overview of Neutralizing Antibodies and Their Potential in COVID-19. Vaccines 2021, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Zhang, N.; Richardson, S.A.; Wu, J.V. Rapid lateral flow tests for the detection of SARS-CoV-2 neutralizing antibodies. Expert Rev. Mol. Diagn. 2021, 21, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jiang, X.; Yang, L.; Chen, L.; Zeng, X.; Liu, G.; Tang, Y.; Qian, C.; Wang, X.; Cheng, F.; et al. SARS-CoV-2-specific immune response in COVID-19 convalescent individuals. Signal Transduct. Target. Ther. 2021, 6, 256. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liang, J.; Hu, H.; Wu, M.; Ma, J.; Ma, Z.; Ji, J.; Chen, H.; Li, X.; Wang, Z.; et al. Development of an Effective Neutralizing Antibody Assay for SARS-CoV-2 Diagnosis. Int. J. Nanomed. 2023, 18, 3125–3139. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, M.; Krizsan, A.; Brakel, A.; Pohl, F.; Volke, D.; Hoffmann, R. Cross-Reactivity of IgG Antibodies and Virus Neutralization in mRNA-Vaccinated People Against Wild-Type SARS-CoV-2 and the Five Most Common SARS-CoV-2 Variants of Concern. Front. Immunol. 2022, 13, 915034. [Google Scholar] [CrossRef]
- Lehmann, C.; Loeffler-Wirth, H.; Balz, V.; Enczmann, J.; Landgraf, R.; Lakowa, N.; Gruenewald, T.; Fischer, J.C.; Doxiadis, I. Immunogenetic Predisposition to SARS-CoV-2 Infection. Biology 2022, 12, 37. [Google Scholar] [CrossRef]
- Rottmayer, K.; Loeffler-Wirth, H.; Gruenewald, T.; Doxiadis, I.; Lehmann, C. Individual Immune Response to SARS-CoV-2 Infection-The Role of Seasonal Coronaviruses and Human Leukocyte Antigen. Biology 2023, 12, 1293. [Google Scholar] [CrossRef]
- Kannenberg, J.; Trawinski, H.; Henschler, R.; Buhmann, R.; Hönemann, M.; Jassoy, C. Antibody Course and Memory B-Cell Response in the First Year After Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2022, 226, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, M.; Luo, J.; Brakel, A.; Krizsan, A.; Lakowa, N.; Grünewald, T.; Lehmann, C.; Wolf, J.; Borte, S.; Milkovska-Stamenova, S.; et al. Evaluation of S- and M-Proteins Expressed in Escherichia coli and HEK Cells for Serological Detection of Antibodies in Response to SARS-CoV-2 Infections and mRNA-Based Vaccinations. Pathogens 2022, 11, 1515. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Stevens, M.; Kallon, D.; Gupta, A.; White, E. Comparative evaluation of Luminex based assays for detection of SARS-CoV-2 antibodies in a transplantation laboratory. J. Immunol. Methods 2023, 517, 113472. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.-A. Humoral Responses and Serological Assays in SARS-CoV-2 Infections. Front. Immunol. 2020, 11, 610688. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, B.S.; Bogner, R.H.; Pikal, M.J. Protein stability during freezing: Separation of stresses and mechanisms of protein stabilization. Pharm. Dev. Technol. 2007, 12, 505–523. [Google Scholar] [CrossRef] [PubMed]
- Horn, J.; Jena, S.; Aksan, A.; Friess, W. Freeze/thaw of IGG solutions. Eur. J. Pharm. Biopharm. 2019, 134, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Maelegheer, K.; Devreese, K.M.J. The impact of repeated freeze-thaw cycles on antiphospholipid antibody titer. Res. Pract. Thromb. Haemost. 2018, 2, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Kanji, J.N.; Bailey, A.; Fenton, J.; Robbin Lindsay, L.; Dibernardo, A.; Toledo, N.P.; Waitt, B.; Lecocq, N.; Osiowy, C.; Giles, E.; et al. Stability of SARS-CoV-2 IgG in multiple laboratory conditions and blood sample types. J. Clin. Virol. 2021, 142, 104933. [Google Scholar] [CrossRef]
- Inada, M.; Togano, T.; Terada, M.; Shiratori, K.; Tsuzuki, S.; Takamatsu, Y.; Saito, S.; Hangaishi, A.; Morioka, S.; Kutsuna, S.; et al. Preserved SARS-CoV-2 neutralizing IgG activity of in-house manufactured COVID-19 convalescent plasma. Transfus. Apher. Sci. 2023, 62, 103638. [Google Scholar] [CrossRef]
- Santano, R.; Barrios, D.; Crispi, F.; Crovetto, F.; Vidal, M.; Chi, J.; Izquierdo, L.; Gratacós, E.; Moncunill, G.; Dobaño, C. Agreement between commercially available ELISA and in-house Luminex SARS-CoV-2 antibody immunoassays. Sci. Rep. 2021, 11, 18984. [Google Scholar] [CrossRef]
- Aguilar, R.; Li, X.; Crowell, C.S.; Burrell, T.; Vidal, M.; Rubio, R.; Jiménez, A.; Hernández-Luis, P.; Hofmann, D.; Mijočević, H.; et al. RBD-Based ELISA and Luminex Predict Anti-SARS-CoV-2 Surrogate-Neutralizing Activity in Two Longitudinal Cohorts of German and Spanish Health Care Workers. Microbiol. Spectr. 2023, 11, e0316522. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, I.; Dahma, H.; Wolff, F.; Dauby, N.; Delaunoy, S.; Wuyts, M.; Detemmerman, C.; Duterme, C.; Vandenberg, O.; Martin, C.; et al. Neutralizing antibody responses following natural SARS-CoV-2 infection: Dynamics and correlation with commercial serologic tests. J. Clin. Virol. 2021, 144, 104988. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.S.; Pérez-Alós, L.; Armenteros, J.J.A.; Hansen, C.B.; Fjallsbak, J.P.; Larsen, S.; Hansen, J.L.; Jarlhelt, I.; Kristiansen, M.F.; Við Streym, F.; et al. Factors influencing the immune response over 15 months after SARS-CoV-2 infection: A longitudinal population-wide study in the Faroe Islands. J. Intern. Med. 2023, 293, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Gillot, C.; Di Chiaro, L.; Eucher, C.; Elsen, M.; van Eeckhoudt, S.; David, C.; Morimont, L.; Dogné, J.-M.; Douxfils, J. Neutralizing Antibodies in COVID-19 Patients and Vaccine Recipients after Two Doses of BNT162b2. Viruses 2021, 13, 1364. [Google Scholar] [CrossRef] [PubMed]
- Tea, F.; Ospina Stella, A.; Aggarwal, A.; Ross Darley, D.; Pilli, D.; Vitale, D.; Merheb, V.; Lee, F.X.Z.; Cunningham, P.; Walker, G.J.; et al. SARS-CoV-2 neutralizing antibodies: Longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021, 18, e1003656. [Google Scholar] [CrossRef] [PubMed]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claër, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, C.J.; Bulman, C.A.; Yun, C.; Lynch, K.L.; Wu, A.H.B.; Whitman, J.D. Detection of IgM, IgG, IgA and neutralizing antibody responses to SARS-CoV-2 infection and mRNA vaccination. J. Med. Microbiol. 2023, 72, 001632. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, Y.; Omata, K.; Shimizu, Y.; Kinoshita-Iwamoto, N.; Terada, M.; Suzuki, T.; Morioka, S.; Uemura, Y.; Ohmagari, N.; Maeda, K.; et al. SARS-CoV-2-Neutralizing Humoral IgA Response Occurs Earlier but Is Modest and Diminishes Faster than IgG Response. Microbiol. Spectr. 2022, 10, e0271622. [Google Scholar] [CrossRef]
- Sullivan, H.C.; Gebel, H.M.; Bray, R.A. Understanding solid-phase HLA antibody assays and the value of MFI. Hum. Immunol. 2017, 78, 471–480. [Google Scholar] [CrossRef]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, C.; Thiaucourt, M.; Kittel, M.; Becker, C.; Ast, V.; Hetjens, M.; Neumaier, M.; Haselmann, V. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int. J. Infect. Dis. 2021, 107, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Naderi Ghale-Noie, Z.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef] [PubMed]
- Alamri, S.S.; Alsaieedi, A.; Khouqeer, Y.; Afeef, M.; Alharbi, S.; Algaissi, A.; Alghanmi, M.; Altorki, T.; Zawawi, A.; Alfaleh, M.A.; et al. The importance of combining serological testing with RT-PCR assays for efficient detection of COVID-19 and higher diagnostic accuracy. PeerJ 2023, 11, e15024. [Google Scholar] [CrossRef]
- Espejo, A.P.; Akgun, Y.; Al Mana, A.F.; Tjendra, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Review of Current Advances in Serologic Testing for COVID-19. Am. J. Clin. Pathol. 2020, 154, 293–304. [Google Scholar] [CrossRef]
| Antigen | MFI Cut-Off Batch 001 | MFI Cut-Off Batch 002 |
|---|---|---|
| SARS-CoV-2 Spike | 7500 | 6800 |
| SARS-CoV-2 Spike S1 | 4000 | 2700 |
| SARS-CoV-2 Spike RBD | 3500 | 5800 |
| SARS-CoV-2 Spike S2 | 1900 | 3200 |
| SARS-CoV-2 Nucleocapsid Protein | 3500 | 5900 |
| HCoV-229E Spike S1 | 3068 | 8012 |
| HCoV-HKU1 Spike S1 | 2614 | 4235 |
| HCoV-NL63 Spike S1 | 1043 | 4407 |
| HCoV-OC43 Spike S1 | 3127 | 3599 |
| MERS-CoV Spike S1 | 10 | 21 |
| SARS-CoV Spike S1 | 92 | 41 |
| Negative Control Bead (Bead-ID 1) | Positive Control Bead (Bead-ID 2) | SARS-CoV-2 Bead | Sample |
|---|---|---|---|
| − | + | + | positive |
| − | + | − | negative |
| − | − | +/− | invalid test (retested) |
| + | + | +/− | invalid test (retested) |
| + | − | +/− | invalid test (retested) |
| Spike | 2nd Freeze–Thaw Cycle | 3rd Freeze–Thaw Cycle |
|---|---|---|
| positive–positive | 64 | 22 |
| positive–negative | 4 | 8 |
| negative–positive | 2 | 1 |
| negative–negative | 12 | 9 |
| S1 | 2nd freeze–thaw cycle | 3rd freeze–thaw cycle |
| positive–positive | 62 | 21 |
| positive–negative | 5 | 9 |
| negative–positive | 1 | 1 |
| negative–negative | 14 | 9 |
| RBD | 2nd freeze–thaw cycle | 3rd freeze–thaw cycle |
| positive–positive | 59 | 20 |
| positive–negative | 5 | 7 |
| negative–positive | 3 | 1 |
| negative–negative | 13 | 12 |
| S2 | 2nd freeze–thaw cycle | 3rd freeze–thaw cycle |
| positive–positive | 55 | 16 |
| positive–negative | 11 | 12 |
| negative–positive | 3 | 2 |
| negative–negative | 13 | 10 |
| Nc | 2nd freeze–thaw cycle | 3rd freeze–thaw cycle |
| positive–positive | 17 | 6 |
| positive–negative | 6 | 7 |
| negative–positive | 1 | 0 |
| negative–negative | 58 | 27 |
| SARS-CoV-2 Domain | ELISA/CLIA Test | Spearman’s Correlation Coefficient |
|---|---|---|
| RBD | Siemens * | 0.87 |
| Mediagnost | 0.88 | |
| S1 | Euroimmun | 0.95 |
| Nucleoprotein | Roche * | 0.68 |
| Abbott * | 0.79 | |
| Virotech | 0.85 | |
| Novatec | 0.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rottmayer, K.; Schwarze, M.; Jassoy, C.; Hoffmann, R.; Loeffler-Wirth, H.; Lehmann, C. Potential of a Bead-Based Multiplex Assay for SARS-CoV-2 Antibody Detection. Biology 2024, 13, 273. https://doi.org/10.3390/biology13040273
Rottmayer K, Schwarze M, Jassoy C, Hoffmann R, Loeffler-Wirth H, Lehmann C. Potential of a Bead-Based Multiplex Assay for SARS-CoV-2 Antibody Detection. Biology. 2024; 13(4):273. https://doi.org/10.3390/biology13040273
Chicago/Turabian StyleRottmayer, Karla, Mandy Schwarze, Christian Jassoy, Ralf Hoffmann, Henry Loeffler-Wirth, and Claudia Lehmann. 2024. "Potential of a Bead-Based Multiplex Assay for SARS-CoV-2 Antibody Detection" Biology 13, no. 4: 273. https://doi.org/10.3390/biology13040273





