Multiomics Picture of Obesity in Young Adults
Abstract
:Simple Summary
Abstract
1. Introduction
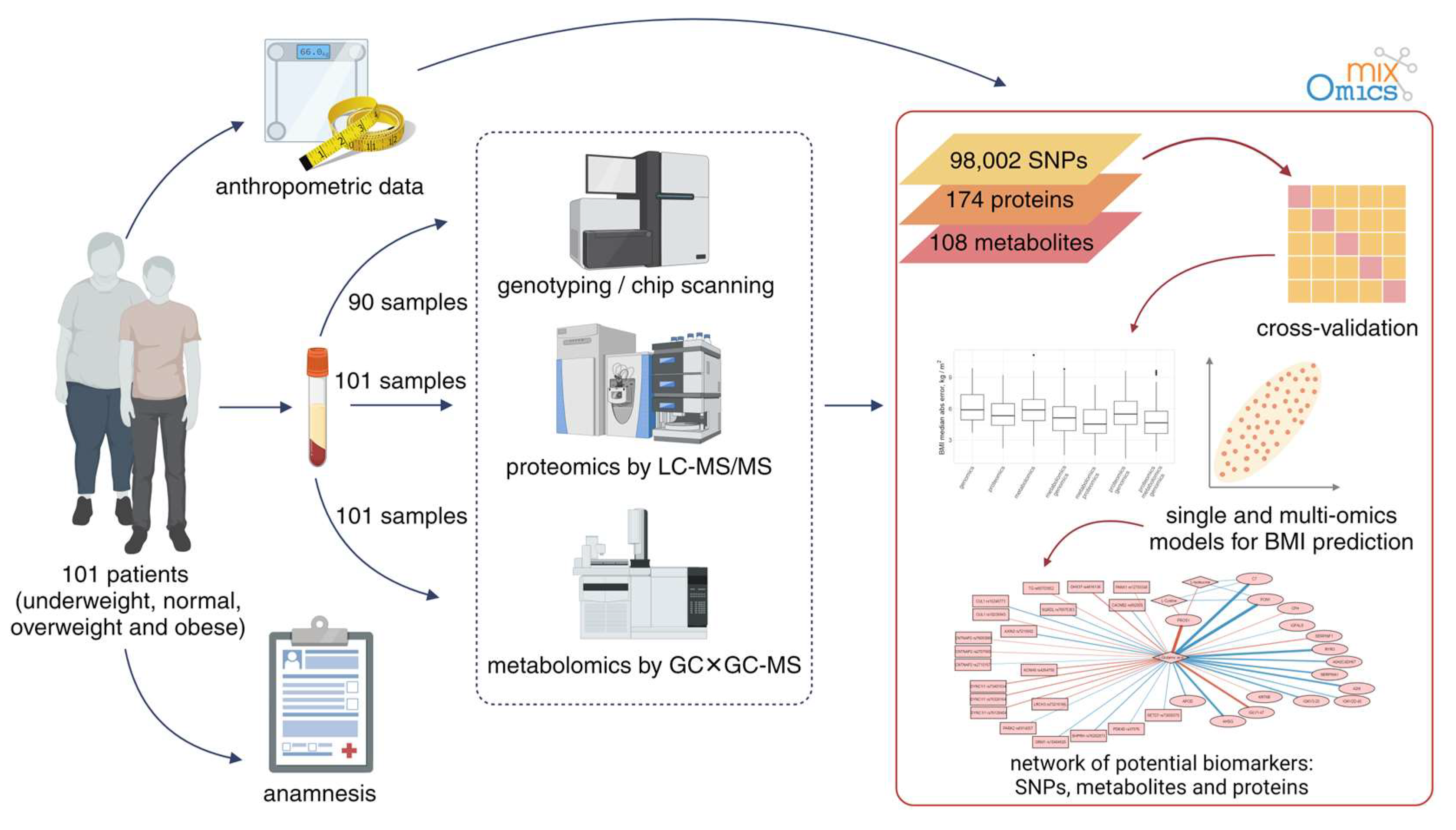
2. Materials and Methods
2.1. Subjects
2.2. Genomic Analysis
2.3. Proteomic Analysis
2.4. Metabolomic Analysis
2.4.1. Sample Preparation for Metabolomic Analysis
2.4.2. GC×GC–MS Analysis
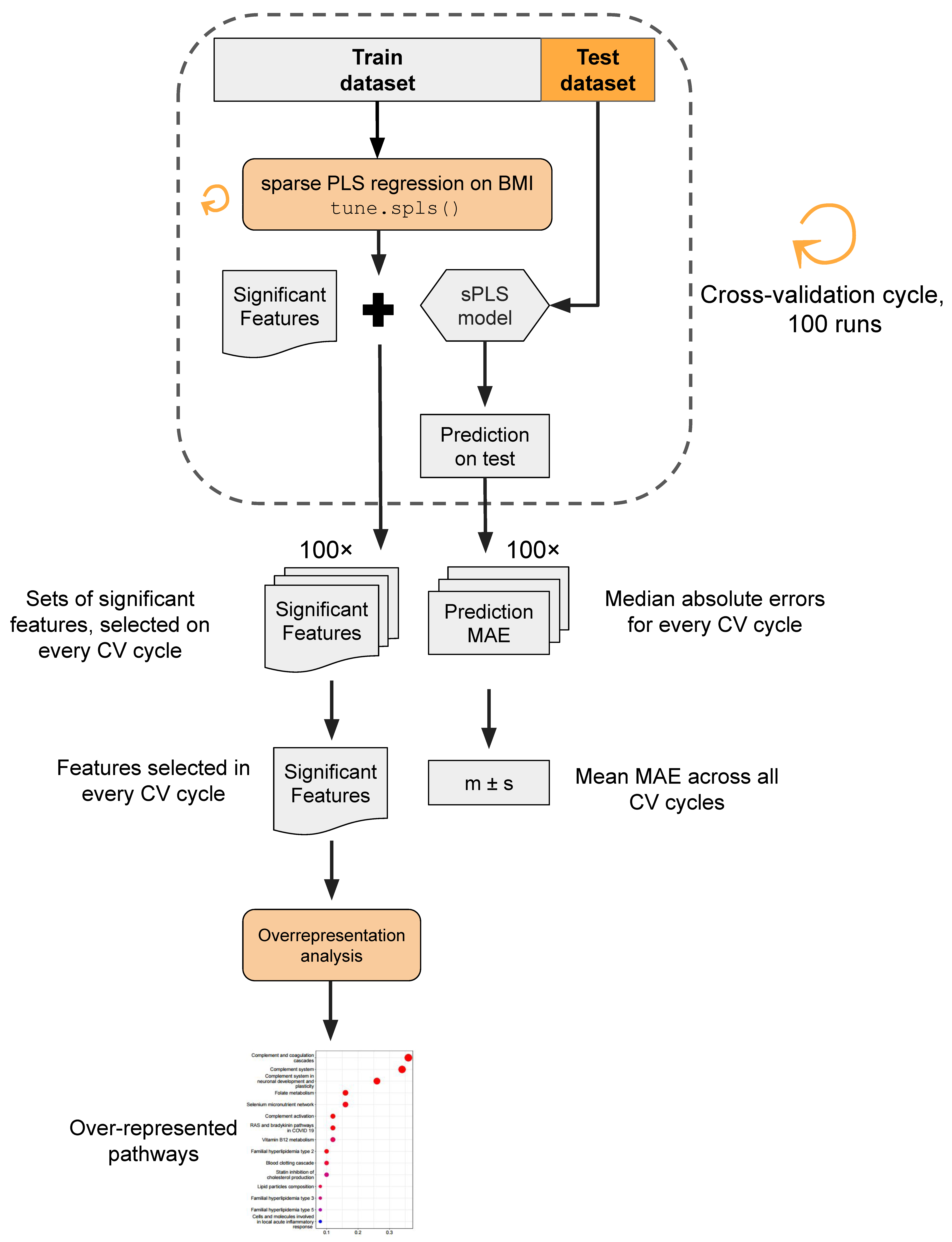
2.5. Data Processing and Statistical Analysis
3. Results
3.1. Genome Analysis
3.2. Proteomic Patterns
3.2.1. Key Proteins in Predictive Models
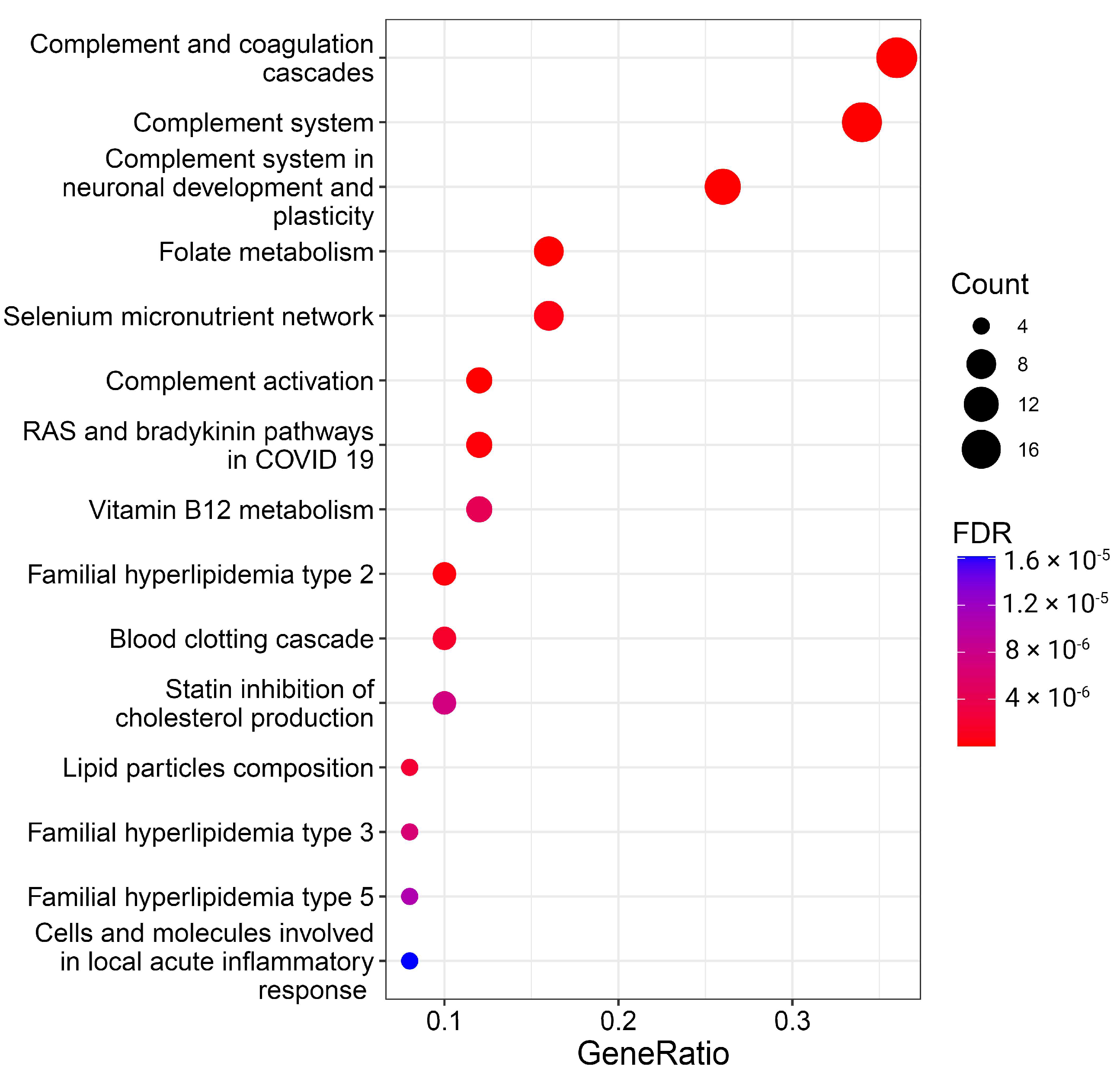
3.2.2. Overrepresentation of Certain Proteins
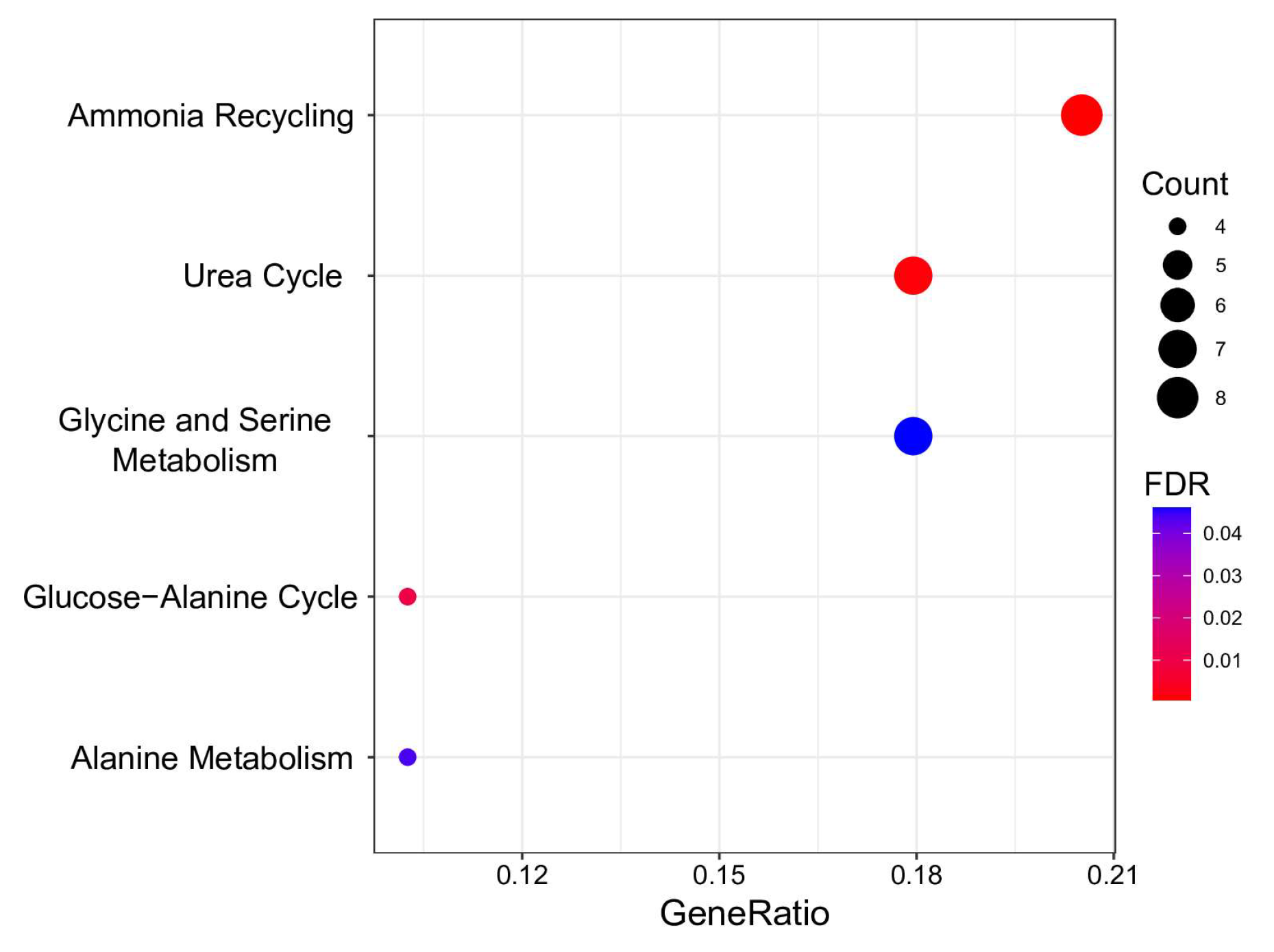
3.3. Metabolome Analysis
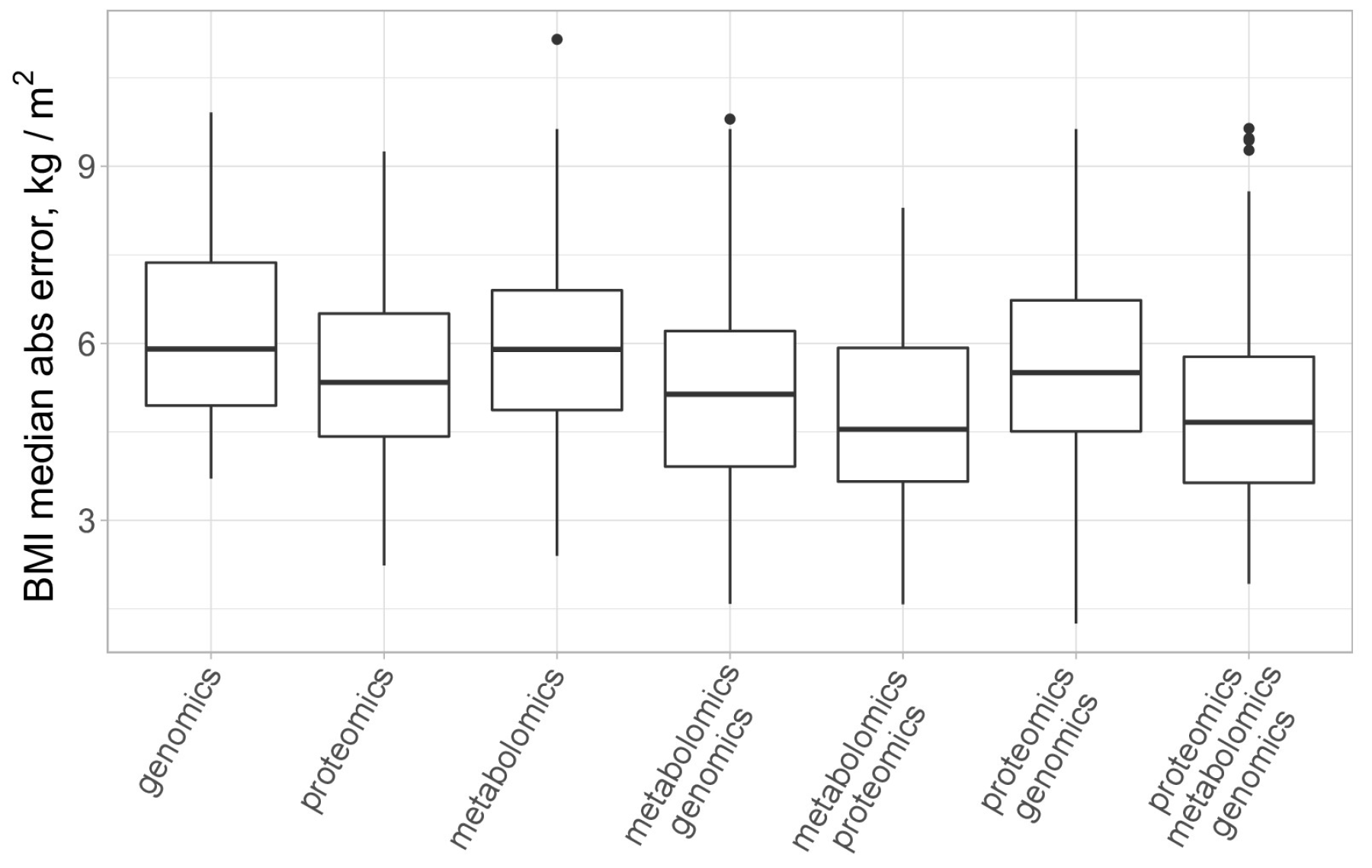
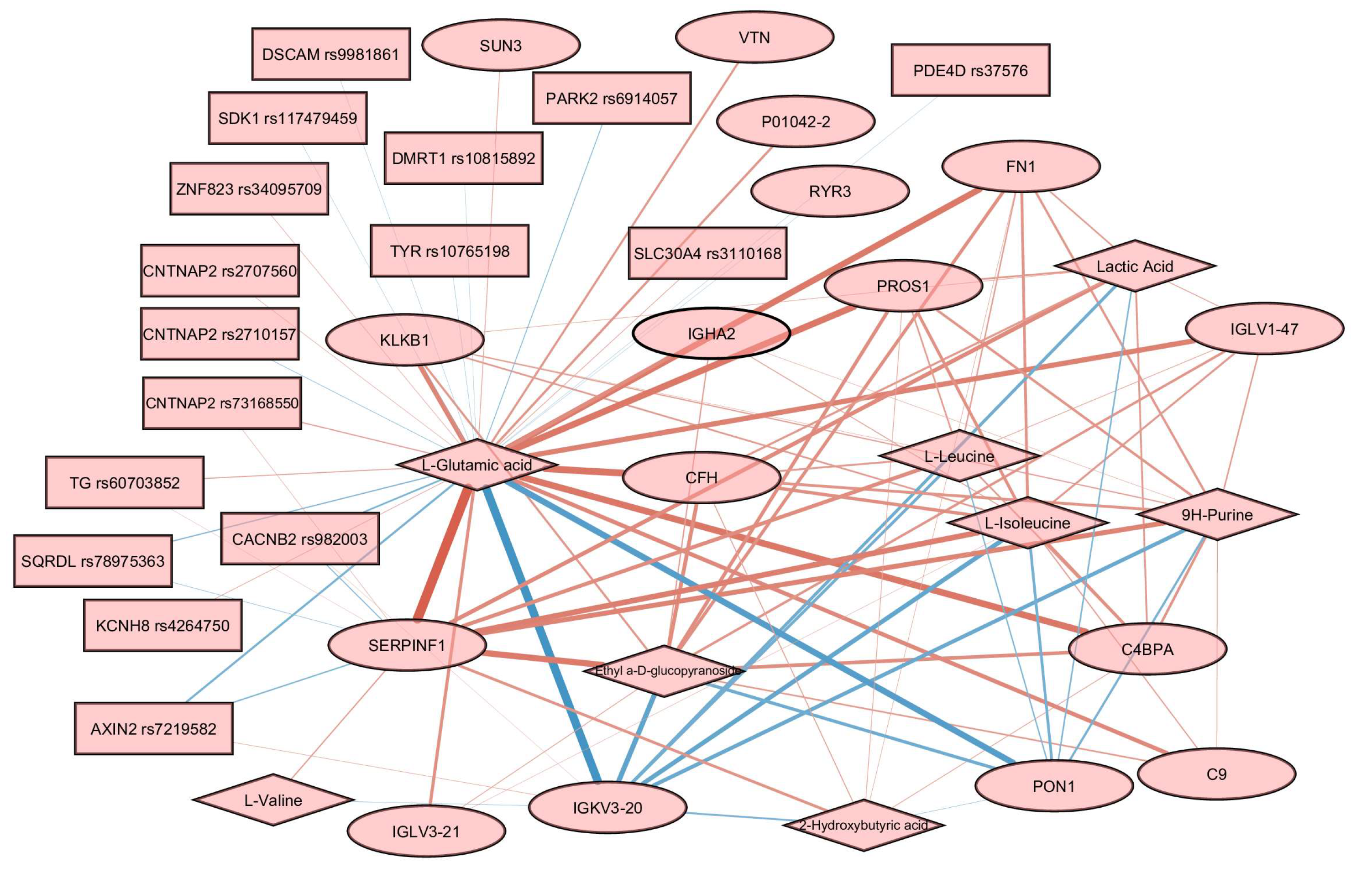
3.4. Multiomics Analysis for BMI Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, M.E. Body Mass Index and Heart Failure Mortality: More Is Less? JACC Heart Fail. 2018, 6, 243–245. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Egea Rodrigues, C.; Floegel, A.; Ahrens, W. Omics Biomarkers in Obesity: Novel Etiological Insights and Targets for Precision Prevention. Curr. Obes. Rep. 2020, 9, 219–230. [Google Scholar] [CrossRef]
- Thomas, D.M.; Weedermann, M.; Fuemmeler, B.F.; Martin, C.K.; Dhurandhar, N.V.; Bredlau, C.; Heymsfield, S.B.; Ravussin, E.; Bouchard, C. Dynamic Model Predicting Overweight, Obesity, and Extreme Obesity Prevalence Trends. Obesity 2014, 22, 590–597. [Google Scholar] [CrossRef]
- Cottam, D.R.; Mattar, S.G.; Barinas-Mitchell, E.; Eid, G.; Kuller, L.; Kelley, D.E.; Schauer, P.R. The Chronic Inflammatory Hypothesis for the Morbidity Associated with Morbid Obesity: Implications and Effects of Weight Loss. Obes. Surg. 2004, 14, 589–600. [Google Scholar] [CrossRef]
- Lee, M.-K.; Kim, Y.M.; Sohn, S.-Y.; Lee, J.-H.; Won, Y.J.; Kim, S.H. Evaluation of the Relationship of Subclinical Hypothyroidism with Metabolic Syndrome and Its Components in Adolescents: A Population-Based Study. Endocrine 2019, 65, 608–615. [Google Scholar] [CrossRef]
- Pivonello, R.; Isidori, A.M.; De Martino, M.C.; Newell-Price, J.; Biller, B.M.K.; Colao, A. Complications of Cushing’s Syndrome: State of the Art. Lancet Diabetes Endocrinol. 2016, 4, 611–629. [Google Scholar] [CrossRef]
- Gambineri, A.; Pelusi, C.; Vicennati, V.; Pagotto, U.; Pasquali, R. Obesity and the Polycystic Ovary Syndrome. Int. J. Obes. 2002, 26, 883–896. [Google Scholar] [CrossRef]
- Lucchese, M.; Maggi, M. Hypogonadism as a New Comorbidity in Male Patient’s Selection for Bariatric Surgery: Towards an Extended Concept of Metabolic Surgery? Obes. Surg. 2013, 23, 2018–2019. [Google Scholar] [CrossRef]
- Salvatori, R. Growth Hormone Deficiency in Patients with Obesity. Endocrine 2015, 49, 304–306. [Google Scholar] [CrossRef]
- Haslam, D.W.; James, W.P.T. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Hecker, J.; Freijer, K.; Hiligsmann, M.; Evers, S.M.A.A. Burden of Disease Study of Overweight and Obesity; the Societal Impact in Terms of Cost-of-Illness and Health-Related Quality of Life. BMC Public Health 2022, 22, 46. [Google Scholar] [CrossRef]
- Huynh, K. Vascular Disease: Cell Therapy Lowers P.A.D. Amputation Risk. Nat. Rev. Cardiol. 2017, 14, 130. [Google Scholar] [CrossRef]
- Akimoto, N.; Ugai, T.; Zhong, R.; Hamada, T.; Fujiyoshi, K.; Giannakis, M.; Wu, K.; Cao, Y.; Ng, K.; Ogino, S. Rising Incidence of Early-Onset Colorectal Cancer—A Call to Action. Nat. Rev. Clin. Oncol. 2021, 18, 230–243. [Google Scholar] [CrossRef]
- Watanabe, K.; Wilmanski, T.; Diener, C.; Earls, J.C.; Zimmer, A.; Lincoln, B.; Hadlock, J.J.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T.; et al. Multiomic Signatures of Body Mass Index Identify Heterogeneous Health Phenotypes and Responses to a Lifestyle Intervention. Nat. Med. 2023, 29, 996–1008. [Google Scholar] [CrossRef]
- Woldemariam, S.; Dorner, T.E.; Wiesinger, T.; Stein, K.V. Multi-Omics Approaches for Precision Obesity Management: Potentials and Limitations of Omics in Precision Prevention, Treatment and Risk Reduction of Obesity. Wien. Klin. Wochenschr. 2023, 135, 113–124. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic Studies of Body Mass Index Yield New Insights for Obesity Biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New Genetic Loci Link Adipose and Insulin Biology to Body Fat Distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M.; et al. Meta-Analysis of Genome-Wide Association Studies for Height and Body Mass Index in ∼700000 Individuals of European Ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Loos, R.J.F.; Yeo, G.S.H. The Genetics of Obesity: From Discovery to Biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef]
- Oller Moreno, S.; Cominetti, O.; Núñez Galindo, A.; Irincheeva, I.; Corthésy, J.; Astrup, A.; Saris, W.H.M.; Hager, J.; Kussmann, M.; Dayon, L. The Differential Plasma Proteome of Obese and Overweight Individuals Undergoing a Nutritional Weight Loss and Maintenance Intervention. Proteomics Clin. Appl. 2018, 12, 1600150. [Google Scholar] [CrossRef]
- Larsen, T.M.; Dalskov, S.; van Baak, M.; Jebb, S.; Kafatos, A.; Pfeiffer, A.; Martinez, J.A.; Handjieva-Darlenska, T.; Kunesová, M.; Holst, C.; et al. The Diet, Obesity and Genes (Diogenes) Dietary Study in Eight European Countries-a Comprehensive Design for Long-Term Intervention. Obes. Rev. 2010, 11, 76–91. [Google Scholar] [CrossRef]
- Haupt, A.; Thamer, C.; Machann, J.; Kirchhoff, K.; Stefan, N.; Tschritter, O.; Machicao, F.; Schick, F.; Häring, H.-U.; Fritsche, A. Impact of Variation in the FTO Gene on Whole Body Fat Distribution, Ectopic Fat, and Weight Loss. Obesity 2008, 16, 1969–1972. [Google Scholar] [CrossRef]
- Zhang, Q.; Meng, X.-H.; Qiu, C.; Shen, H.; Zhao, Q.; Zhao, L.-J.; Tian, Q.; Sun, C.-Q.; Deng, H.-W. Integrative Analysis of Multi-Omics Data to Detect the Underlying Molecular Mechanisms for Obesity in Vivo in Humans. Hum. Genom. 2022, 16, 15. [Google Scholar] [CrossRef]
- Kiseleva, O.I.; Arzumanian, V.A.; Poverennaya, E.V.; Pyatnitskiy, M.A.; Ilgisonis, E.V.; Zgoda, V.G.; Plotnikova, O.A.; Sharafetdinov, K.K.; Lisitsa, A.V.; Tutelyan, V.A.; et al. Does Proteomic Mirror Reflect Clinical Characteristics of Obesity? J. Pers. Med. 2021, 11, 64. [Google Scholar] [CrossRef]
- Dyussenbayev, A. Age Periods Of Human Life. Adv. Soc. Sci. Res. J. 2017, 4. [Google Scholar] [CrossRef]
- Barsnes, H.; Vaudel, M. SearchGUI: A Highly Adaptable Common Interface for Proteomics Search and de Novo Engines. J. Proteome Res. 2018, 17, 2552–2555. [Google Scholar] [CrossRef]
- Kiseleva, O.; Poverennaya, E.; Shargunov, A.; Lisitsa, A. Proteomic Cinderella: Customized Analysis of Bulky MS/MS Data in One Night. J. Bioinform. Comput. Biol. 2018, 16, 1740011. [Google Scholar] [CrossRef]
- Bubis, J.A.; Levitsky, L.I.; Ivanov, M.V.; Tarasova, I.A.; Gorshkov, M.V. Comparative Evaluation of Label-Free Quantification Methods for Shotgun Proteomics. Rapid Commun. Mass. Spectrom. 2017, 31, 606–612. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Kurbatov, I.; Kiseleva, O.; Arzumanian, V.; Dolgalev, G.; Poverennaya, E. Some Lessons Learned on the Impact of the Storage Conditions, Syringe Wash Solvent, and the Way of GC-MS Injection on the Reproducibility of Metabolomic Studies. Metabolites 2023, 13, 75. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLOS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next Generation Pathway Database. Nucleic Acids Res 2023, 52, D679–D689. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- WHO. Expert Consultation Appropriate Body-Mass Index for Asian Populations and Its Implications for Policy and Intervention Strategies. Lancet 2004, 363, 157–163. [Google Scholar] [CrossRef]
- Oniszczenko, W.; Stanisławiak, E. Association between Sex and Body Mass Index as Mediated by Temperament in a Nonclinical Adult Sample. Eat. Weight. Disord. 2019, 24, 291–298. [Google Scholar] [CrossRef]
- Tareen, S.H.K.; Kutmon, M.; Adriaens, M.E.; Mariman, E.C.M.; de Kok, T.M.; Arts, I.C.W.; Evelo, C.T. Exploring the Cellular Network of Metabolic Flexibility in the Adipose Tissue. Genes. Nutr. 2018, 13, 17. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Garratt, E.S.; Titcombe, P.; Melton, P.E.; Murray, R.J.S.; Barton, S.J.; Clarke-Harris, R.; Costello, P.M.; Holbrook, J.D.; Hopkins, J.C.; et al. Differential SLC6A4 Methylation: A Predictive Epigenetic Marker of Adiposity from Birth to Adulthood. Int. J. Obes. 2019, 43, 974–988. [Google Scholar] [CrossRef]
- Li, F.; Cui, X.; Jing, J.; Wang, S.; Shi, H.; Xue, B.; Shi, H. Brown Fat Dnmt3b Deficiency Ameliorates Obesity in Female Mice. Life 2021, 11, 1325. [Google Scholar] [CrossRef]
- Motahari-Rad, H.; Subiri, A.; Soler, R.; Ocaña, L.; Alcaide, J.; Rodríguez-Capitan, J.; Buil, V.; el Azzouzi, H.; Ortega-Gomez, A.; Bernal-Lopez, R.; et al. The Effect of Sex and Obesity on the Gene Expression of Lipid Flippases in Adipose Tissue. J. Clin. Med. 2022, 11, 3878. [Google Scholar] [CrossRef]
- Benot-Dominguez, R.; Cimini, A.; Barone, D.; Giordano, A.; Pentimalli, F. The Emerging Role of Cyclin-Dependent Kinase Inhibitors in Treating Diet-Induced Obesity: New Opportunities for Breast and Ovarian Cancers? Cancers 2022, 14, 2709. [Google Scholar] [CrossRef]
- Mlodzik-Czyzewska, M.A.; Malinowska, A.M.; Chmurzynska, A. Low Folate Intake and Serum Levels Are Associated with Higher Body Mass Index and Abdominal Fat Accumulation: A Case Control Study. Nutr. J. 2020, 19, 53. [Google Scholar] [CrossRef]
- Chan, C.-W.; Chan, P.-H.; Lin, B.-F. Folate Deficiency Increased Lipid Accumulation and Leptin Production of Adipocytes. Front. Nutr. 2022, 9, 852451. [Google Scholar] [CrossRef]
- Köse, S.; Sözlü, S.; Bölükbaşi, H.; Ünsal, N.; Gezmen-Karadağ, M. Obesity Is Associated with Folate Metabolism. Int. J. Vitam. Nutr. Res. 2020, 90, 353–364. [Google Scholar] [CrossRef]
- Fontenelle, L.C.; Cardoso de Araújo, D.S.; da Cunha Soares, T.; Clímaco Cruz, K.J.; Henriques, G.S.; do Nascimento Marreiro, D. Nutritional Status of Selenium in Overweight and Obesity: A Systematic Review and Meta-Analysis. Clin. Nutr. 2022, 41, 862–884. [Google Scholar] [CrossRef]
- Tinkov, A.A.; Ajsuvakova, O.P.; Filippini, T.; Zhou, J.-C.; Lei, X.G.; Gatiatulina, E.R.; Michalke, B.; Skalnaya, M.G.; Vinceti, M.; Aschner, M.; et al. Selenium and Selenoproteins in Adipose Tissue Physiology and Obesity. Biomolecules 2020, 10, 658. [Google Scholar] [CrossRef]
- Ortega, R.M.; Rodríguez-Rodríguez, E.; Aparicio, A.; Jiménez-Ortega, A.I.; Palmeros, C.; Perea, J.M.; Navia, B.; López-Sobaler, A.M. Young Children with Excess of Weight Show an Impaired Selenium Status. Int. J. Vitam. Nutr. Res. 2012, 82, 121–129. [Google Scholar] [CrossRef]
- Rex, D.A.B.; Deepak, K.; Vaid, N.; Dagamajalu, S.; Kandasamy, R.K.; Flo, T.H.; Keshava Prasad, T.S. A Modular Map of Bradykinin-Mediated Inflammatory Signaling Network. J. Cell Commun. Signal. 2022, 16, 301–310. [Google Scholar] [CrossRef]
- Barros, C.C.; Haro, A.; Russo, F.J.; Schadock, I.; Almeida, S.S.; Reis, F.C.; Moraes, M.R.; Haidar, A.; Hirata, A.E.; Mori, M.; et al. Bradykinin Inhibits Hepatic Gluconeogenesis in Obese Mice. Lab. Investig. 2012, 92, 1419–1427. [Google Scholar] [CrossRef]
- Boachie, J.; Adaikalakoteswari, A.; Samavat, J.; Saravanan, P. Low Vitamin B12 and Lipid Metabolism: Evidence from Pre-Clinical and Clinical Studies. Nutrients 2020, 12, 1925. [Google Scholar] [CrossRef]
- Baltaci, D.; Kutlucan, A.; Turker, Y.; Yilmaz, A.; Karacam, S.; Deler, H.; Ucgun, T.; Kara, I.H. Association of Vitamin B12 with Obesity, Overweight, Insulin Resistance and Metabolic Syndrome, and Body Fat Composition; Primary Care-Based Study. Med. Glas 2013, 10, 203–210. [Google Scholar]
- Takashina, C.; Tsujino, I.; Watanabe, T.; Sakaue, S.; Ikeda, D.; Yamada, A.; Sato, T.; Ohira, H.; Otsuka, Y.; Oyama-Manabe, N.; et al. Associations among the Plasma Amino Acid Profile, Obesity, and Glucose Metabolism in Japanese Adults with Normal Glucose Tolerance. Nutr. Metab. 2016, 13, 5. [Google Scholar] [CrossRef]
- Maltais-Payette, I.; Allam-Ndoul, B.; Pérusse, L.; Vohl, M.-C.; Tchernof, A. Circulating Glutamate Level as a Potential Biomarker for Abdominal Obesity and Metabolic Risk. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 1353–1360. [Google Scholar] [CrossRef]
- Wang, S.; Li, M.; Lin, H.; Wang, G.; Xu, Y.; Zhao, X.; Hu, C.; Zhang, Y.; Zheng, R.; Hu, R.; et al. Amino Acids, Microbiota-Related Metabolites, and the Risk of Incident Diabetes among Normoglycemic Chinese Adults: Findings from the 4C Study. Cell Rep. Med. 2022, 3, 100727. [Google Scholar] [CrossRef]
- JCM. High Plasma Cystine Levels Are Associated with Blood Pressure and Reversed by CPAP in Patients with Obstructive Sleep Apnea. Available online: https://www.mdpi.com/2077-0383/10/7/1387 (accessed on 19 November 2023).
- Elshorbagy, A.K.; Church, C.; Valdivia-Garcia, M.; Smith, A.D.; Refsum, H.; Cox, R. Dietary Cystine Level Affects Metabolic Rate and Glycaemic Control in Adult Mice. J. Nutr. Biochem. 2012, 23, 332–340. [Google Scholar] [CrossRef]
- Thomsen, K.L.; Eriksen, P.L.; Kerbert, A.J.; Chiara, F.D.; Jalan, R.; Vilstrup, H. Role of Ammonia in NAFLD: An Unusual Suspect. JHEP Rep. 2023, 5, 100780. [Google Scholar] [CrossRef]
- Chashmniam, S.; Hashemi Madani, N.; Nobakht Mothlagh Ghoochani, B.F.; Safari-Alighiarloo, N.; Khamseh, M.E. The Metabolome Profiling of Obese and Non-Obese Individuals: Metabolically Healthy Obese and Unhealthy Non-Obese Paradox. Iran. J. Basic. Med. Sci. 2020, 23, 186–194. [Google Scholar] [CrossRef]
- Alves, A.; Bassot, A.; Bulteau, A.-L.; Pirola, L.; Morio, B. Glycine Metabolism and Its Alterations in Obesity and Metabolic Diseases. Nutrients 2019, 11, 1356. [Google Scholar] [CrossRef]
- Palmnäs, M.S.A.; Kopciuk, K.A.; Shaykhutdinov, R.A.; Robson, P.J.; Mignault, D.; Rabasa-Lhoret, R.; Vogel, H.J.; Csizmadi, I. Serum Metabolomics of Activity Energy Expenditure and Its Relation to Metabolic Syndrome and Obesity. Sci. Rep. 2018, 8, 3308. [Google Scholar] [CrossRef]
- Huffman, K.M.; Shah, S.H.; Stevens, R.D.; Bain, J.R.; Muehlbauer, M.; Slentz, C.A.; Tanner, C.J.; Kuchibhatla, M.; Houmard, J.A.; Newgard, C.B.; et al. Relationships Between Circulating Metabolic Intermediates and Insulin Action in Overweight to Obese, Inactive Men and Women. Diabetes Care 2009, 32, 1678–1683. [Google Scholar] [CrossRef]
- Egami, R.; Kokaji, T.; Hatano, A.; Yugi, K.; Eto, M.; Morita, K.; Ohno, S.; Fujii, M.; Hironaka, K.; Uematsu, S.; et al. Trans-Omic Analysis Reveals Obesity-Associated Dysregulation of Inter-Organ Metabolic Cycles between the Liver and Skeletal Muscle. iScience 2021, 24, 102217. [Google Scholar] [CrossRef]
- Kob, R.; Bollheimer, L.C.; Bertsch, T.; Fellner, C.; Djukic, M.; Sieber, C.C.; Fischer, B.E. Sarcopenic Obesity: Molecular Clues to a Better Understanding of Its Pathogenesis? Biogerontology 2015, 16, 15–29. [Google Scholar] [CrossRef]
- Okun, J.G.; Rusu, P.M.; Chan, A.Y.; Wu, Y.; Yap, Y.W.; Sharkie, T.; Schumacher, J.; Schmidt, K.V.; Roberts-Thomson, K.M.; Russell, R.D.; et al. Liver Alanine Catabolism Promotes Skeletal Muscle Atrophy and Hyperglycaemia in Type 2 Diabetes. Nat. Metab. 2021, 3, 394–409. [Google Scholar] [CrossRef]
- Benson, M.D.; Eisman, A.S.; Tahir, U.A.; Katz, D.H.; Deng, S.; Ngo, D.; Robbins, J.M.; Hofmann, A.; Shi, X.; Zheng, S.; et al. Protein-Metabolite Association Studies Identify Novel Proteomic Determinants of Metabolite Levels in Human Plasma. Cell Metab. 2023, 35, 1646–1660.e3. [Google Scholar] [CrossRef]
- Kurbatov, I.; Dolgalev, G.; Arzumanian, V.; Kiseleva, O.; Poverennaya, E. The Knowns and Unknowns in Protein-Metabolite Interactions. Int. J. Mol. Sci. 2023, 24, 4155. [Google Scholar] [CrossRef]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis-a Framework for Unsupervised Integration of Multi-Omics Data Sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef]
- Mansuy-Aubert, V.; Zhou, Q.L.; Xie, X.; Gong, Z.; Huang, J.-Y.; Khan, A.R.; Aubert, G.; Candelaria, K.; Thomas, S.; Shin, D.-J.; et al. Imbalance between Neutrophil Elastase and Its Inhibitor A1-Antitrypsin in Obesity Alters Insulin Sensitivity, Inflammation, and Energy Expenditure. Cell Metab. 2013, 17, 534–548. [Google Scholar] [CrossRef]
- Böhm, A.; Ordelheide, A.-M.; Machann, J.; Heni, M.; Ketterer, C.; Machicao, F.; Schick, F.; Stefan, N.; Fritsche, A.; Häring, H.-U.; et al. Common Genetic Variation in the SERPINF1 Locus Determines Overall Adiposity, Obesity-Related Insulin Resistance, and Circulating Leptin Levels. PLoS ONE 2012, 7, e34035. [Google Scholar] [CrossRef]
- Geyer, P.E.; Wewer Albrechtsen, N.J.; Tyanova, S.; Grassl, N.; Iepsen, E.W.; Lundgren, J.; Madsbad, S.; Holst, J.J.; Torekov, S.S.; Mann, M. Proteomics Reveals the Effects of Sustained Weight Loss on the Human Plasma Proteome. Mol. Syst. Biol. 2016, 12, 901. [Google Scholar] [CrossRef]
- Shim, K.; Begum, R.; Yang, C.; Wang, H. Complement Activation in Obesity, Insulin Resistance, and Type 2 Diabetes Mellitus. World J. Diabetes 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Rensen, S.S.; Slaats, Y.; Driessen, A.; Peutz-Kootstra, C.J.; Nijhuis, J.; Steffensen, R.; Greve, J.W.; Buurman, W.A. Activation of the Complement System in Human Nonalcoholic Fatty Liver Disease. Hepatology 2009, 50, 1809–1817. [Google Scholar] [CrossRef]
- Subudhi, S.; Drescher, H.K.; Dichtel, L.E.; Bartsch, L.M.; Chung, R.T.; Hutter, M.M.; Gee, D.W.; Meireles, O.R.; Witkowski, E.R.; Gelrud, L.; et al. Distinct Hepatic Gene-Expression Patterns of NAFLD in Patients With Obesity. Hepatol. Commun. 2021, 6, 77–89. [Google Scholar] [CrossRef]
- Gabrielsson, B.G.; Johansson, J.M.; Lönn, M.; Jernås, M.; Olbers, T.; Peltonen, M.; Larsson, I.; Lönn, L.; Sjöström, L.; Carlsson, B.; et al. High Expression of Complement Components in Omental Adipose Tissue in Obese Men. Obes. Res. 2003, 11, 699–708. [Google Scholar] [CrossRef]
- King, B.C.; Blom, A.M. Complement in Metabolic Disease: Metaflammation and a Two-Edged Sword. Semin. Immunopathol. 2021, 43, 829–841. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Martínez-Barricarte, R.; Catalán, V.; Sabater, M.; Gómez-Ambrosi, J.; Ortega, F.J.; Ricart, W.; Blüher, M.; Frühbeck, G.; Rodríguez de Cordoba, S.; et al. Complement Factor H Is Expressed in Adipose Tissue in Association with Insulin Resistance. Diabetes 2010, 59, 200–209. [Google Scholar] [CrossRef]
- Li, J.; Shen, Y.; Tian, H.; Xie, S.; Ji, Y.; Li, Z.; Lu, J.; Lu, H.; Liu, B.; Liu, F. The Role of Complement Factor H in Gestational Diabetes Mellitus and Pregnancy. BMC Pregnancy Childbirth 2021, 21, 562. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, C.; Yang, P. Association of Single Nucleotide Polymorphisms in the 5’ Upstream Region of the C4BPA Gene with Essential Hypertension in a Northeastern Han Chinese Population. Mol. Med. Rep. 2017, 16, 1289–1297. [Google Scholar] [CrossRef]
- Ermert, D.; Blom, A.M. C4b-Binding Protein: The Good, the Bad and the Deadly. Novel Functions of an Old Friend. Immunol. Lett. 2016, 169, 82–92. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Tung, Y.-T.; Lin, Y.-K.; Liao, C.-C.; Chiu, C.-F.; Tung, T.-H.; Shabrina, A.; Huang, S.-Y. Effects of Caloric Restriction with Protein Supplementation on Plasma Protein Profiles in Middle-Aged Women with Metabolic Syndrome—A Preliminary Open Study. J. Clin. Med. 2019, 8, 195. [Google Scholar] [CrossRef]
- Natali, L.; Cavallini, A.; Cremonini, R.; Bassi, P.; Cionini, P.G. Amplification of Nuclear DNA Sequences during Induced Plant Cell Dedifferentiation. Cell Differ. 1986, 18, 157–161. [Google Scholar] [CrossRef]
- Ferrer, F.; Nazih, H.; Zaïr, Y.; Krempf, M.; Bard, J.M. Postprandial Changes in the Distribution of Apolipoprotein AIV between Apolipoprotein B- and Non Apolipoprotein B-Containing Lipoproteins in Obese Women. Metabolism 2003, 52, 1537–1541. [Google Scholar] [CrossRef]
- Culnan, D.M.; Cooney, R.N.; Stanley, B.; Lynch, C.J. Apolipoprotein A-IV, a Putative Satiety/Antiatherogenic Factor, Rises after Gastric Bypass. Obesity 2009, 17, 46–52. [Google Scholar] [CrossRef]
- Jiménez-Palomares, M.; Cózar-Castellano, I.; Ganfornina, M.D.; Sánchez, D.; Perdomo, G. Genetic Deficiency of Apolipoprotein D in the Mouse Is Associated with Nonfasting Hypertriglyceridemia and Hyperinsulinemia. Metabolism 2011, 60, 1767–1774. [Google Scholar] [CrossRef]
- Marcelin, G.; Gautier, E.L.; Clément, K. Adipose Tissue Fibrosis in Obesity: Etiology and Challenges. Annu. Rev. Physiol. 2022, 84, 135–155. [Google Scholar] [CrossRef]
- Sanchez, D.; López-Arias, B.; Torroja, L.; Canal, I.; Wang, X.; Bastiani, M.J.; Ganfornina, M.D. Loss of Glial Lazarillo, a Homolog of Apolipoprotein D, Reduces Lifespan and Stress Resistance in Drosophila. Curr. Biol. 2006, 16, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef] [PubMed]
- Adhe-Rojekar, A.; Mogarekar, M.R.; Rojekar, M.V. Paraoxonase Activity in Metabolic Syndrome in Children and Adolescents. Caspian J. Intern. Med. 2018, 9, 116–120. [Google Scholar] [CrossRef]
- Çayır, A.; Turan, M.İ.; Gurbuz, F.; Kurt, N.; Yildirim, A. The Effect of Lifestyle Change and Metformin Therapy on Serum Arylesterase and Paraoxonase Activity in Obese Children. J. Pediatr. Endocrinol. Metab. 2015, 28, 551–556. [Google Scholar] [CrossRef]
- Dahlbäck, B. Protein S and C4b-Binding Protein: Components Involved in the Regulation of the Protein C Anticoagulant System. Thromb. Haemost. 1991, 66, 49–61. [Google Scholar] [CrossRef]
- Rubio-Aliaga, I.; Marvin-Guy, L.F.; Wang, P.; Wagniere, S.; Mansourian, R.; Fuerholz, A.; Saris, W.H.M.; Astrup, A.; Mariman, E.C.M.; Kussmann, M. Mechanisms of Weight Maintenance under High- and Low-Protein, Low-Glycaemic Index Diets. Mol. Nutr. Food Res. 2011, 55, 1603–1612. [Google Scholar] [CrossRef]
- Otsuka, Y.; Ueda, M.; Nakazono, E.; Tsuda, T.; Jin, X.; Noguchi, K.; Sata, S.; Miyazaki, H.; Abe, S.; Imai, K.; et al. Relationship between Plasma Protein S Levels and Apolipoprotein C-II in Japanese Middle-Aged Obese Women and Young Nonobese Women. Blood Coagul. Fibrinolysis 2018, 29, 39–47. [Google Scholar] [CrossRef]
- Vandooren, J.; Itoh, Y. Alpha-2-Macroglobulin in Inflammation, Immunity and Infections. Front. Immunol. 2021, 12, 803244. [Google Scholar] [CrossRef]
- Netanel, C.; Goitein, D.; Rubin, M.; Kleinbaum, Y.; Katsherginsky, S.; Hermon, H.; Tsaraf, K.; Tachlytski, I.; Herman, A.; Safran, M.; et al. The Impact of Bariatric Surgery on Nonalcoholic Fatty Liver Disease as Measured Using Non-Invasive Tests. Am. J. Surg. 2021, 222, 214–219. [Google Scholar] [CrossRef]
- Deckmyn, O.; Poynard, T.; Bedossa, P.; Paradis, V.; Peta, V.; Pais, R.; Ratziu, V.; Thabut, D.; Brzustowski, A.; Gautier, J.-F.; et al. Clinical Interest of Serum Alpha-2 Macroglobulin, Apolipoprotein A1, and Haptoglobin in Patients with Non-Alcoholic Fatty Liver Disease, with and without Type 2 Diabetes, before or during COVID-19. Biomedicines 2022, 10, 699. [Google Scholar] [CrossRef]
- Dabrowska, A.M.; Tarach, J.S.; Wojtysiak-Duma, B.; Duma, D. Fetuin-A (AHSG) and Its Usefulness in Clinical Practice. Review of the Literature. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2015, 159, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Fritsche, A.; Weikert, C.; Boeing, H.; Joost, H.-G.; Häring, H.-U.; Schulze, M.B. Plasma Fetuin-A Levels and the Risk of Type 2 Diabetes. Diabetes 2008, 57, 2762–2767. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Hennige, A.M.; Staiger, H.; Machann, J.; Schick, F.; Kröber, S.M.; Machicao, F.; Fritsche, A.; Häring, H.-U. Alpha2-Heremans-Schmid Glycoprotein/Fetuin-A Is Associated with Insulin Resistance and Fat Accumulation in the Liver in Humans. Diabetes Care 2006, 29, 853–857. [Google Scholar] [CrossRef]
- Kaushik, S.V.; Plaisance, E.P.; Kim, T.; Huang, E.Y.; Mahurin, A.J.; Grandjean, P.W.; Mathews, S.T. Extended-Release Niacin Decreases Serum Fetuin-A Concentrations in Individuals with Metabolic Syndrome. Diabetes Metab. Res. Rev. 2009, 25, 427–434. [Google Scholar] [CrossRef]
- Brix, J.M.; Stingl, H.; Höllerl, F.; Schernthaner, G.H.; Kopp, H.-P.; Schernthaner, G. Elevated Fetuin-A Concentrations in Morbid Obesity Decrease after Dramatic Weight Loss. J. Clin. Endocrinol. Metab. 2010, 95, 4877–4881. [Google Scholar] [CrossRef]
| All | Underweight | Normal | Overweight | Obese | |
|---|---|---|---|---|---|
| Number of patients | 101 | 2 | 20 | 21 | 58 |
| Male to female ratio | 50/51 | 0/2 | 9/11 | 11/10 | 30/28 |
| Age (years) | 31.6 ± 6.7 | 29.0 ± 1.4 | 30.7 ± 5.6 | 32.9 ± 6.7 | 31.6 ± 7.2 |
| BMI (kg/m2) | 32.8 ± 9.2 | 17.8 ± 0.6 | 22.1 ± 1.5 | 27.5 ± 1.3 | 38.9 ± 7.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiseleva, O.I.; Pyatnitskiy, M.A.; Arzumanian, V.A.; Kurbatov, I.Y.; Ilinsky, V.V.; Ilgisonis, E.V.; Plotnikova, O.A.; Sharafetdinov, K.K.; Tutelyan, V.A.; Nikityuk, D.B.; et al. Multiomics Picture of Obesity in Young Adults. Biology 2024, 13, 272. https://doi.org/10.3390/biology13040272
Kiseleva OI, Pyatnitskiy MA, Arzumanian VA, Kurbatov IY, Ilinsky VV, Ilgisonis EV, Plotnikova OA, Sharafetdinov KK, Tutelyan VA, Nikityuk DB, et al. Multiomics Picture of Obesity in Young Adults. Biology. 2024; 13(4):272. https://doi.org/10.3390/biology13040272
Chicago/Turabian StyleKiseleva, Olga I., Mikhail A. Pyatnitskiy, Viktoriia A. Arzumanian, Ilya Y. Kurbatov, Valery V. Ilinsky, Ekaterina V. Ilgisonis, Oksana A. Plotnikova, Khaider K. Sharafetdinov, Victor A. Tutelyan, Dmitry B. Nikityuk, and et al. 2024. "Multiomics Picture of Obesity in Young Adults" Biology 13, no. 4: 272. https://doi.org/10.3390/biology13040272






