Biotechnology of Cold-Active Proteases
Abstract
:1. Introduction
2. Microbes Producing Cold-Active Proteases
| S. No. | Organisms | Properties of the proteases | Reference | ||
|---|---|---|---|---|---|
| Mol. weight (kDa) | TOpt (°C) | pHOpt. | |||
| 1 | Alcaligenes faecalis | - | 30 | 8.8 | [50] |
| 2 | Alkaliphilus transvaalensis | 30 | 40 | 12.6 | [51] |
| 3 | Alteromonas haloplanktis | 74–76 | 20 | 8–9 | [52] |
| 4 | Aspergillus ustus | 45 | 32 | 9 | [53] |
| 5 | Azospirillum sp. | 48.6 | 40 | 8.5 | [34] |
| 6 | Bacillus sp. | - | 30 | 9.6 | [54] |
| 7 | Bacillus spp. | - | 40 | 10.5–11 | [55] |
| 8 | Bacillus amyloliquefaciens S94 | 45 | - | 10 | [56] |
| 9 | Bacillus cereus | - | 20 | 9 | [57] |
| 10 | Bacillus licheniformis RKK-04 | 31 | 50 | 10 | [58] |
| 11 | Bacillus pumilus | - | 30 | 11.5 | [59] |
| 12 | Beauveria bassiana | - | 37 | 10 | [60] |
| 13 | Candida humicola | - | 37 | 10 | [61] |
| 14 | Clostridium sp. | 46 | 37 | 7 | [36] |
| 15 | Colwellia sp. | 60 | 35 | 8–9 | [62] |
| 16 | Colwellia psychrerythraea strain 34H | 71 | 19 | 6–8.5 | [63] |
| 17 | Curtobacterium luteum | 115 | 20 | 7 | [39] |
| 18 | Engyodontium album | - | 25 | 11 | [64] |
| 19 | Escherichia freundii | 55 | 25 | 10 | [65] |
| 20 | Exiguobacterium sp.SKPB5 | 36 | 40 | 8 | [40] |
| 21 | Flavobacterium YS-80 | 49 | 30 | 8–11 | [66] |
| 22 | Flavobacterium balustinum P104 | 70 | 40 | 7–9 | [67] |
| 23 | Leucosporidium antarcticum 171 | 34.4 | 30 | 8 | [68] |
| 24 | Pedobacter cryoconitis, | 27 | 40 | 8 | [41] |
| 25 | Penicillium chrysogenum FS010 | 41 | 35 | 9 | [42] |
| 26 | Planomicrobium sp. 547 | - | 35 | 9 | [69] |
| 27 | Pseudoalteromonas sp. D12-004 | 34 | 35 | 7–8 | [70] |
| 28 | Pseudoalteromonas sp. NJ276 | 28 | 30 | 8 | [37] |
| 29 | Pseudoalteromonas sp. P96-47 | - | 20 | 8 | [71] |
| 30 | Pseudoalteromonas sp. SM9913 | 65.84 | 25 | 9 | [72] |
| 31 | Pseudomonas sp Ele-2 | 45 | 40 | - | [73] |
| 32 | Pseudomonas sp. | - | 20 | [74] | |
| 33 | Pseudomonas strain DY-A | - | 40 | 10 | [43] |
| 34 | Pseudomonas aerugenosa MTCC 7926 | - | 40 | 9 | [75] |
| 35 | Pseudomonas lundensis | 48 | 30 | 10.5 | [76] |
| 36 | Pseudomonas fluorescens | - | 35 | 5 | [77] |
| 37 | Pseudomonas fluorescens 114. | 47 | 35-40 | 8 | [78] |
| 38 | Pycnoporus cinnabarinus ss3 | - | 30 | 4 | [79] |
| 39 | Roseobacter sp. [MMD040] | - | 37-40 | 8–9 | [80] |
| 40 | Serratia marcescens AP3801 | 58 | 40 | 6.5–8.0 | [81] |
| 41 | Serratia marcescens TS1 | 56 | 40 | 8 | [82] |
| 42 | Serratia proteamaculans 94 | 50 | 4-30 | 8 | [83] |
| 43 | Shewanella strain Ac10 | 44 | 5-15 | 9 | [84] |
| 44 | Stenotrophomonas sp. | 55 | 15 | 10 | [85] |
| 45 | Stenotrophomonas maltophilia MTCC 7528 | 75 | 20 | 10 | [49] |
| 46 | Streptomyces sp. | - | 30 | 10 | [86] |
| 47 | Streptomyces alboniger | - | 37 | 9–11 | [87] |
| 48 | Teredinobacter turnirae | - | 25 | 7 | [88] |
| 49 | Trichoderma atroviride | 24 | 25 | 6.2 | [89] |
| 50 | Vibrio sp. | 35 | 40 | 8.5–9.0 | [90] |
| 51 | Vibrio sp. PA-44 | 47 | 25 | 8.6 | [46] |
3. Classification of Proteases
4. Optimization of Fermentation Conditions for Production of Cold-Active Proteases
| Endoprotease | EC No. | Mol. Mas Range (kDa) | pHOpt. | TOpt. (°C) | Metal Ion Required | Active Site a Residues | Major Inhibitor(s) |
|---|---|---|---|---|---|---|---|
| Aspartic or Carboxyl proteases | 3.4.23 | 30–45 | 3–5 | 40–55 | Ca2+ | Aspartate or cysteine | Pepstatin |
| Cysteine or thiol proteases | 3.4.22 | 34–35 | 2–3 | 40–55 | - | Aspartate or cysteine | Indoacetamide, p-CMB |
| Metallo- proteases | 3.4.24 | 19–37 | 5–7 | 65–85 | Zn2+, Ca2+ | Phenylalanine or leucine | Chelating agents such as EDTA, EGTA |
| Serine proteases | 3.4.21 | 18–35 | 6–11 | 50–70 | Ca2+ | Serine, histidine and aspartate | PMSF, DIFP, EDTA, soybean trypsin inhibitor, phosphate buffers, indole, phenol, triamino acetic acid |
5. Purification of Cold-Active Proteases
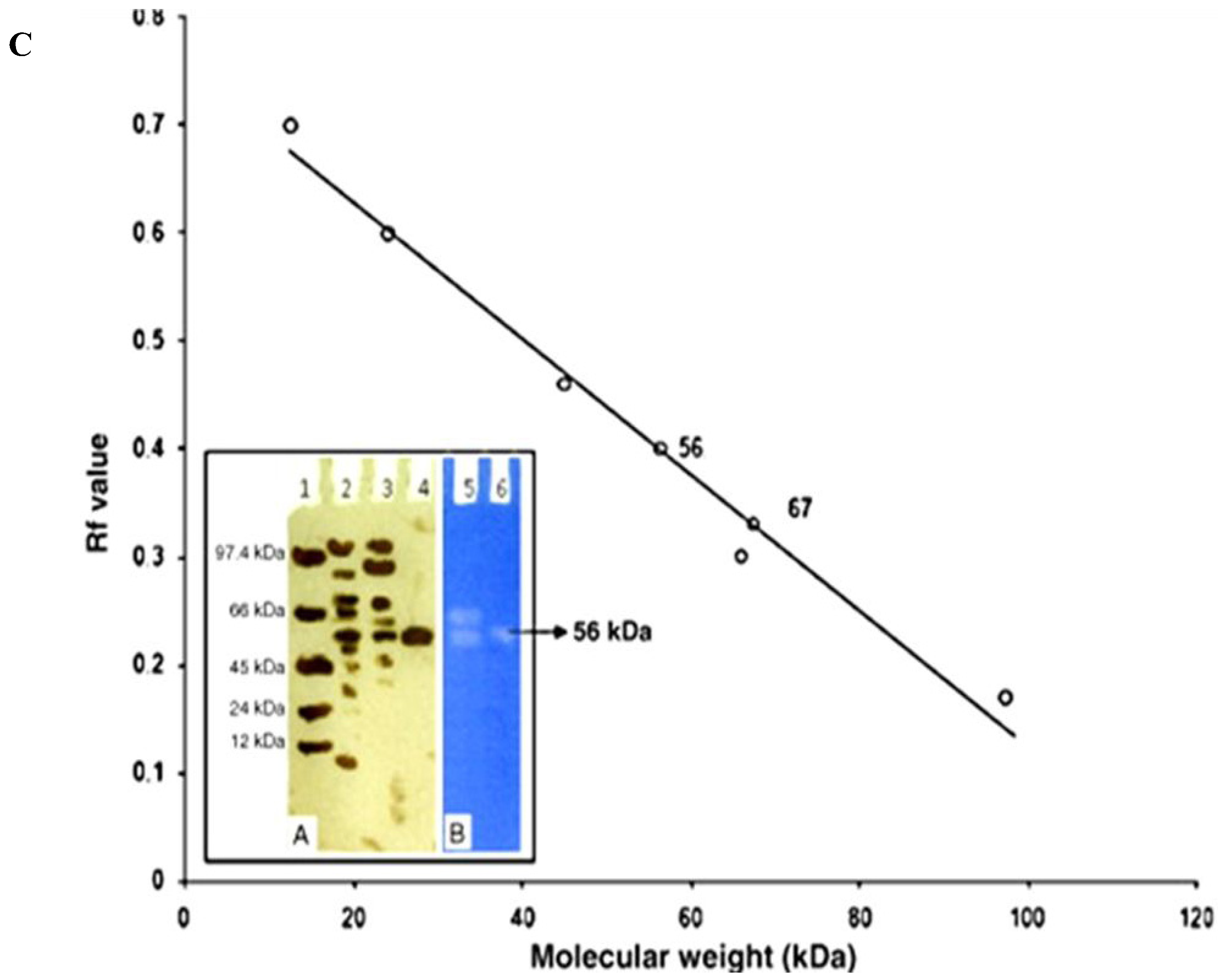
| Protease Source | Protease Type | Concentration Method | Column Matrices | Fold Purification | References |
|---|---|---|---|---|---|
| Alkaliphilus transvaalensis | Serine protease | Amicon Ultra-15 | DEAE Toyopearl 650M resin, CM-Toyopearl 650M | 96 | [58] |
| Clostridium species | Serine-type metalloenzyme | Ammonium sulfate precipitation | Sephadex G-100 | 12.7 | [36] |
| Colwellia psychrerythraea strain 34H. | Aminopeptidase | - | Sepharose Q, Hydroxyapatite, Resource Q | 460 | [63] |
| Curtobacterium luteum MTCC 7529 | Metalloprotease | Ammonium sulphate precipitation | DEAE- Cellulose | 34.1 | [39] |
| Escherichia freundii, | Neutral serine protease | Ammonium sulfate precipitate | CM-cellulose, DEAE-Sephadex A-50, Sephadex G-100 | - | [65] |
| Leucosporidium antarcticum 171 | Serine proteinase | Acetone precipitation | Sephadex G-75, Diethylaminoethyl-Sephacel, Sephacryl S-100 | 1,568 | [68] |
| Oerskovia xanthineolytica TK-1 | Serine protease | Ultrafiltration | Phenyl-Sepharose CL-4B, DEAE-Sephacel | 39.6 | [97] |
| Pedobacter cryoconitis | Metalloprotease | - | SP Sepharose, Syn-Chropak CM300 | - | [39] |
| Penicillium chrysogenum FS010 | Serine protease | Ammoniumsulfate precipitation | DEAE Sepharose, Sephadex G-100 | 103.2 | [42] |
| Planomicrobium species | Serine protease | Ammonium sulfate precipitation, Lyophilization | DEAE-52 | - | [69] |
| Pseudoalteromonas sp. NJ276 | Serine protease | Ammonium sulfate precipitation | DEAE-Sephadex A50, Sephadex G-75 | 22.5 | [37] |
| Pseudoalteromonas sp. SM9913 | Serine protease. | Ammonium sulfate precipitation, PEG 2000. | Sephadex G100 | - | [53] |
| Pseudomonas aeruginosa IFO 3455 | Metalloprotease | - | QAE-agarose | - | [74] |
| P. fluorescence 114 | Neutral metalloprotease | Ammoniumsulfate precipitation | DEAE Toyopearl 650 M, Superdex 200 HR 10/30 | - | [78] |
| Pseudomonas strain DY-A | Serine protease | Ammonium sulfate precipitation, | DEAE Sepharose CL-6B, Sephadex G-100 | 84.2 | [43] |
| Serratia marcescens AP3801 | Metalloprotease | Ammonium sulfate precipitation | Sephacryl S-100, Q Sepharose | 0.48 | [62] |
| S. marcescens TS1. | Metalloprotease | Ammonium sulphateation, acetone precipitation | DEAE-cellulose | - | [82] |
| S. proteamaculans | Trypsin-like protease | Ultrafiltration | Q-Sepharose, BPTI-Sepharose | - | [98] |
| S. proteamaculans | Serine trypsin-like and Zn-dependent protease. | - | BPTI-Sepharose | - | [100] |
| S. proteamaculans 94 | Cysteine protease | - | Arg-Silochrom Z-Gly- DL-Pro-Gly-Silochrom, Superise 12 HR 10/30 column | 3433 | [83] |
| Shewanella strain Ac10 | Alkaline serine protease | - | Bacitracin-Sepharose column | [84] | |
| Stenotrophomonas maltophilia | Serine proteases | Ultrafiltration | S-Sepharose | - | [101] |
| Stenotrophomonas sp. | Alkaline protease | Ammonium sulfate precipitation | DEAE-Sepharose | 18.45 | [85] |
| Marine psychrophilic strainPA-43 | Serine peptidase | - | Q Sepharose, Sephacryl S-300, PBE 94 | 25.0 | [102] |
| Vibrio sp. PA-44 | subtilisin-like proteinase | Ammonium sulfate precipitation | N-carbobenzoxy-d-phenylalanyl-triethylenetetramine-Sepharose, phenyl-sepharose | - | [46] |
6. Properties of Cold-Active Proteases
6.1. Temperature
6.2. pH
6.3. Metal Ions
6.4. Effect of Inhibitors and Other Reagents
6.5. Catalytic Efficiencies
6.6. Substrate Spectrum
7. Cloning and Expression of Cold-Active Proteases
8. Crystal Structure of Cold-Active Proteases

9. Cold Environment Metagenomics: Tapping Biodiversity
10. Enhancing Thermo-Stability of Cold-Active Proteases
11. Applications of Cold-Active Proteases

12. Conclusions and Future Perspectives
Acknowledgements
References
- D’Amico, S.; Collins, T.; Marx, J.; Feller, G.; Gerday, C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Gomes, J.; Steiner, W. The biocatalytic potential of extremophiles and extremozymes. Food Technol. Biotechnol. 2004, 42, 223–235. [Google Scholar]
- Rodrigues, D.F.; Tiedje, J.M. Coping with our cold planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F.; Marx, J.C.; Gerday, C. Psychrophiles: from Biodiversity to Biotechnology; Springer: Berlin, Heidelberg, 2008; pp. 211–224. [Google Scholar]
- Gounot, A. Bacterial life at low temperature: physiological aspects and biotechnological implications. J. Appl. Bacteriol. 1991, 71, 386–397. [Google Scholar] [CrossRef]
- Huston, A.L.; Krieger-Brockett, B.B.; Deming, J.W. Remarkably low temperature optima for extracellular enzyme activity from Arctic bacteria and sea ice. Environ. Microbiol. 2000, 2, 383–388. [Google Scholar] [CrossRef]
- Feller, G.; Bussy, O.L.; Gerday, C. Expression of psychrophilic genes in mesophilic hosts: assessment of the folding state of a recombinant α-amylase. Appl. Environ. Microbiol. 1998, 64, 1163–1165. [Google Scholar]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Crystallization and preliminary X-ray diffraction studies of α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Prot. Sci. 1996, 5, 2128–2129. [Google Scholar] [CrossRef]
- Jeon, J.H.; Kim, J.T.; Kim, Y.J.; Kim, H.K.; Lee, H.S.; Kang, S.G.; Kim, S.J.; Lee, J.H. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009, 81, 865–874. [Google Scholar] [CrossRef]
- Suzuki, T.; Nakayama, T.; Kurihara, T.; Nishino, T.; Esaki, N. Cold-active lipolytic activity of psychrotrophic Acinetobacter sp. strain no. 6. J. Biosci. Bioeng. 2001, 92, 144–148. [Google Scholar]
- Feller, G.; Amico, D.S.; Benotmane, A.M.; Joly, F.; Van Beeumen, J.; Gerday, C. Purification, characterization of nucleotide sequence of the thermolabile α–amylase from Antarctic psychrotroph Alteromonas haloplanktis A23. J. Biol. Chem. 1992, 267, 5217–5221. [Google Scholar]
- Villeret, V.; Chessa, J.P.; Gerday, C.; Van Beeumen, J. Preliminary crystal structure determination of the alkaline protease from Antarctic psychrophile Pseudomonas aeruginosa. Prot. Sci. 1997, 6, 2462–2464. [Google Scholar]
- Gerike, U.; Danson, M.J.; Hough, D.W. Sequencing and expression of the gene encoding a cold-active citrate synthase from an antarctic bacterium strain DS2–3R. Eur. J. Biochem. 1997, 248, 49–57. [Google Scholar]
- Feller, G.; Gerday, C. Psychrophilic enzymes: molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997, 53, 830–841. [Google Scholar] [CrossRef]
- Kim, S.Y.; Hwang, K.Y.; Kim, S.H.; Sung, H.C.; Han, Y.S.; Cho, Y. Structural basis of cold adaptation. Sequence, biochemical properties and crystal structure of malate dehydrogenase from a psychrophilic Aquaspirillum articum. J. Biol. Chem. 1999, 274, 11761–11767. [Google Scholar]
- Alvarez, M.; Johan, P.H.; Zeelen, J.P.; Veronique Mainfroid, V.; Joseph, A.; Martial, J.A. Triose phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. J. Biol. Chem. 1998, 273, 2199–2206. [Google Scholar]
- Georlette, D.; Blaise, V.; Collins, T.; D'Amico, S. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 2004, 28, 25–42. [Google Scholar] [CrossRef]
- Collins, T.; Meuwis, M.A.; Stals, I.; Claeyssens, M.; Feller, G.; Gerday, C. A novel family 8 xylanase, functional and physicochemical characterization. J. Biol. Chem. 2002, 277, 35133–35139. [Google Scholar]
- Russell, R.J.; Gericke, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure (Lond) 1998, 6, 351–361. [Google Scholar]
- Birgisson, H.; Delgado, O.; Arroyo, L.G.; Hatti-Kaul, R.; Mattiasson, B. Cold-adapted yeasts as producers of cold-active polygalacturonases. Extremophiles 2003, 7, 185–193. [Google Scholar]
- Akila, G.; Chandra, T.S. A novel cold-tolerant Clostridium strain PXYL1 isolated from a psychrophilic cattle manure digester that secretes thermolabile xylanase and cellulose. FEMS Microbiol. Lett. 2003, 219, 63–67. [Google Scholar] [CrossRef]
- Mavromatis, K.; Lorito, M.; Woo, S.L.; Bouriotis, V. Mode of action and antifungal properties of two cold-adapted chitinases. Extremophiles 2003, 7, 385–390. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ihara, H.; Kozakic, S.; Kawasaki, H. A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochim. Biophys. Acta 2003, 1624, 70–75. [Google Scholar]
- Nakagawa, T.; Nagaoka, T.; Taniguchi, S.; Miyaji, T.; Tomizuka, N. Isolation and characterization of psychrophilic yeasts producing cold-adapted pectinolytic enzymes. Lett. Appl. Microbiol. 2004, 38, 383–387. [Google Scholar] [CrossRef]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar]
- Godfrey, T.; West, S. Introduction to industrial enzymology. In Industrial Enzymology, 2nd; Godfrey, W., Ed.; Macmillan Press: London, UK, 1996; pp. 1–8. [Google Scholar]
- Gaur, S.; Agrahari, S.; Wadhwa, N. Purification of Protease from Pseudomonas thermaerum GW1 Isolated from poultry waste site. Open Microbiol. J. 2010, 4, 67–74. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–161. [Google Scholar]
- Deming, J.W. Psychrophiles and polar regions. Curr. Opin. Biotechnol. 2002, 5, 301–309. [Google Scholar]
- Margesin, R.; Feller, G.; Gerday, C.; Russell, N. Cold-adapted microorganisms: adaptation strategies and biotechnological potential. In The Encyclopedia of Environmental Microbiology; Bitton, Ed.; Wiley: New York, 2002; pp. 871–885. [Google Scholar]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georiette, D.; Hoyoux, A.; Lonhience, T.; Meuwis, M. Cold-adapted enzymes, from fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Morita, R.J. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar]
- Feller, G. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. 2003, 60, 648–662. [Google Scholar] [CrossRef]
- Oh, K.H.; Seong, C.S.; Lee, S.W.; Kwon, O.S.; Park, Y.S. Isolation of a psychrotrophic Azospirillum sp. and characterization of its extracellular protease. FEMS Microbiol. Lett. 1999, 174, 173–178. [Google Scholar] [CrossRef]
- Baghel, V.S.; Tripathi, R.D.; Ramteke, R.W.; Gopal, K.; Dwivedi, S.; Jain, R.K.; Rai, U.N.; Singh, S.N. Psychrotrophic proteolytic bacteria from cold environments of Gangotri glacier, Westren Himalaya India. Enzyme Microbial. Technol. 2005, 36, 654–659. [Google Scholar] [CrossRef]
- Alam, S.I.; Dube, S.; Reddy, G.S.N.; Bhattacharya, B.K.; Shivaji, S.; Singh, L. Purification and characterization of extracellular protease produced by Clostridium sp. from Schirmacher oasis, Antarctica. Enzyme Microbial. Technol. 2005, 36, 824–831. [Google Scholar] [CrossRef]
- Wang, Q.; Hou, Y.; Xu, Z.; Miao, J.; Li, G. Optimization of cold-active protease production by the psychrophilic bacterium Colwellia sp NJ341 with response surface methodology. Biores. Technol. 2008, 99, 1926–1931. [Google Scholar] [CrossRef]
- Olivera, N.L.; Sequeiros, C.; Nievas, M.L. Diversity and enzyme properties of protease-producing bacteria isolated from sub- Antarctic sediments of Isla de Los Estados, Argentina. Extremophiles 2007, 11, 517–526. [Google Scholar] [CrossRef]
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum. (MTCC 7529), enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392. [Google Scholar] [CrossRef]
- Kasana, R.C.; Yadav, S.K. Isolation of a psychrotrophic Exiguobacterium sp SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr. Microbiol. 2007, 54, 224–229. [Google Scholar] [CrossRef]
- Margesin, R.; Dieplinger, H.; Hofmann, J.; Sarg, B.; Lindner, H. A cold-active extracellular metalloprotease from Pedobacter cryoconitis-production and properties. Res. Microbiol. 2005, 156, 499–505. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Tian, Y.; Hou, Y.H.; Wang, T.H. Purification and characterization of the cold-active alkaline protease from marine cold-adaptive Penicillium chrysogenum FS010. Mol. Biol. Rep. 2009, 36, 2169–2174. [Google Scholar] [CrossRef]
- Zeng, R.; Zhang, R.; Zhao, J.; Lin, N. Cold-active serine alkaline protease from the psychrophilic bacterium Pseudomonas strain DY-A: enzyme purification and characterization. Extremophiles 2003, 7, 335–337. [Google Scholar] [CrossRef]
- Denner, E.B.; Mark, B.; Busse, H.J.; Turkiewicz, M.; Lubitz, W. Psychrobacter proteolyticus sp. Nov., a psychrotrophic, halotolerant bacterium isolated from the Antarctic krill Euphausia superba Dana, excreting a cold-adapted metalloprotease. Syst. Appl. Microbiol. 2001, 24, 44–53. [Google Scholar] [CrossRef]
- Larsen, A.L.; Moe, E.; Helland, R.; Gjellesvik, D.R.; Willassen, N.P. Characterization of a recombinantly expressed proteinase K-like enzyme from a psychrotrophic Serratia sp. FEBS J. 2006, 273, 47–60. [Google Scholar] [CrossRef]
- Kristjansson, M.M.; Magnusson, O.T.; Gudmundsson, H.M.; Alfredsson, G.A.; Matsuzawa, H. Properties of a subtilisin-like proteinase from a psychrotrophic Vibrio species comparison with proteinase K and aqualysin I. Eur. J. Biochem. 1999, 260, 752–760. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. Characterization of a metalloprotease from psychrophilic Xanthomonas maltophilia. FEMS Microbiol. Lett. 1991, 79, 257–262. [Google Scholar] [CrossRef]
- Yu, Y.; Li, H.R.; Zeng, Y.X.; Chen, B. Bacterial diversity and bioprospecting for cold-active hydrolytic enzymes from culturable bacteria associated with sediment from Nella Fjord, Eastern Antarctica. Mar. Drugs 2011, 9, 184–195. [Google Scholar] [CrossRef]
- Kuddus, M.; Ramteke, P.W. Production optimization of an extracellular cold-active alkaline protease from Stenotrophomonas maltophilia MTCC 7528 and its application in detergent industry. Afr. J. Microbiol. Res. 2011, 7, 809–816. [Google Scholar]
- Thangam, E.B.; Rajkumar, G.S. Studies on the production of extracellular protease by Alcaligenes faecalis. World J. Microb. Biot. 2000, 16, 663–666. [Google Scholar] [CrossRef]
- Kobayashi, T.; Lu, J.; Li, Z.; Hung, V.S.; Kurata, A.; Hatada, Y.; Takai, K.; Ito, S.; Horikoshi, K. Extremely high alkaline protease from a deep-subsurface bacterium, Alkaliphilus transvaalensis. Appl. Microbiol. Biotechnol. 2007, 75, 71–80. [Google Scholar] [CrossRef]
- Suzuki, S.; Odagami, T. Low-temperature-active thiol protease from marine bacterium Alteromonas haloplanktis. J. Biotechnol. 1997, l5, 230–233. [Google Scholar]
- Damare, C.; Raghukuma, C.; Muraleedharan, U.D.; Raghukumar, S. Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enzyme Microb. Tech. 2006, 39, 172–181. [Google Scholar] [CrossRef]
- Kaur, S.; Vohra, R.M.; Kapoor, M.; Beg, Q.K.; Hoondal, G.S. Enhanced production and characterization of a highly thermostable alkaline protease from Bacillus sp. P-2. World J. Microb. Biot. 2001, 17, 125–129. [Google Scholar] [CrossRef]
- Okuda, M.; Sumitomo, N.; Takimura, Y.; Ogawa, A.; Saeki, K.; Kawai, S.; Kobayashi, T.; Ito, S. A new subtilisin family: nucleotide and deduced amino acid sequences of new high-molecular-mass alkaline proteases from Bacillus spp. Extremophiles 2008, 4, 229–235. [Google Scholar]
- Son, E.S.; Kim, J.I. Multicatalytic alkaline serine protease from the psychrotrophic Bacillus amyloliquefaciens S94. J. Microbiol. 2003, 41, 58–62. [Google Scholar]
- Joshi, G.K.; Kumar, S.; Sharma, V. Production of moderately halotolerant, SDS stable alkaline protease from Bacillus cereus MTCC 6840 isolated from lake Nainital, Uttaranchal state, India. Braz. J. Microbiol. 2007, 38, 773–779. [Google Scholar] [CrossRef]
- Toyokawa, Y.; Takahara, H.; Reungsang, A.; Fukuta, M.; Hachimine, Y.; Tachibana, S.; Yasuda, M. Purification and characterization of a halotolerant serine proteinase from thermotolerant Bacillus licheniformis RKK-04 isolated from Thai fish sauce. Appl. Microbiol. Biotechnol. 2010, 86, 1867–1875. [Google Scholar] [CrossRef]
- Kumar, C.G. Purification and characterization of a thermostable alkaline protease from alkalophilic Bacillus pumilus. Lett. Appl. Microbiol. 2002, 34, 13–17. [Google Scholar] [CrossRef]
- Rao, Y.K.; Lu, S.C.; Liu, B.L.; Tzeng, Y.M. Enhanced production of an extracellular protease from Beauveria bassiana by optimization of cultivation processes. Biochem. Eng. J. 2006, 28, 57–66. [Google Scholar]
- Ray, M.K.; Devi, K.U.; Kumar, G.S.; Shivaji, S. Extracellular protease from the Antarctic yeast Candida humicola. Appl. Environ. Microbiol. 1992, 58, 1918–1923. [Google Scholar]
- Wang, Q.; Miao, J.L.; Hou, Y.H.; Ding, Y.; Wang, G.D.; Li, G.Y. Purification and characterization of an extracellular cold–active serine protease from the psychrophilic bacterium Colwellia sp. NJ341. Biotech. Lett. 2005, 27, 1195–1198. [Google Scholar] [CrossRef]
- Huston, A.L.; Methe, B.; Deming, J.W. Purification, characterization, sequencing of an extracellular cold–active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 2004, 70, 2321–2328. [Google Scholar]
- Chellappan, S.; Jasmin, C.; Basheer, S.M.; Elyas, K.K.; Bhat, S.G.; Chandrasekaran, M. Production, purification and partial characterization of a novel protease from marine Engyodontium album BTMFS10 under solid state fermentation. Process Biochem. 2006, 41, 956–961. [Google Scholar] [CrossRef]
- Nakajima, M.; Mizusawa, K.; Yoshida, F. Purification and properties of an extracellular proteinase of psychrophilic Escherichia freundii. Eur. J. Biochem. 1974, 44, 87–96. [Google Scholar] [CrossRef]
- Zhang, S.C.; Sun, M.; Li, T.; Wang, Q.H.; Hao, J.H. Structure analysis of a new psychrophilic marine protease. PLoS One 2011. [Google Scholar] [CrossRef]
- Morita, Y.; Hasan, Q.; Sakaguchi, T.; Murakami, Y.; Yokoyama, K.; Tamiya, E. Properties of a cold–active protease from psychrotrophic Flavobacterium balustinum P104. Appl. Microbiol. Biotechnol. 1998, 50, 669–675. [Google Scholar] [CrossRef]
- Turkiewicz, M.; Pazgier, M.; Kalinowska, H.; Bielecki, S. A cold adapted extracellular serine protease of the yeast. Leucosporidium antarcticum. Extremophiles 2003, 7, 435–442. [Google Scholar] [CrossRef]
- Sheng, Y.X.; Lin, C.X.; Zhong, X.U.X.; Ying, Z.R. Cold-adaptive alkaline protease from the psychrophilic Planomicrobium sp. 547: Enzyme characterization and gene cloning. Adv. Polar Sci. 2011, 22, 49–54. [Google Scholar]
- Xiong, H.; Song, S.; Xu, Y.; Tsoi, M.Y.; Dobretsov, S.; Qian, P.Y. Characterization of proteolytic bacteria from the Aleutian deep-sea and their proteases. J. Ind. Microbiol. Biot. 2007, 34, 63–71. [Google Scholar]
- Vazquez, S.C.; Hernández, E.; Mac Cormack, W.P. Extracellular proteases from the Antarctic marine Pseudoalteromonas sp. P96–47 strain. Rev. Argent. Microbiol. 2008, 40, 63–71. [Google Scholar]
- Chen, X.L.; Xie, B.B.; Lu, J.T.; He, H.L.; Zhang, Y. A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913: Catalytic and structural properties of deseasin MCP-01. Microbiology 2007, 153, 2116–2125. [Google Scholar] [CrossRef]
- Vazquez, S.C.; Coria, S.H.; Mac Cormack, W.P. Extracellular proteases from eight psychrotolerant Antarctic strains. Microbiol. Res. 2004, 159, 157–166. [Google Scholar] [CrossRef]
- Chessa, J.P.; Petrescu, I.; Bentahir, M.; Beeumen, J.V.; Gerday, C. Purification, physico-chemical characterization and sequence of a heat labile alkaline metalloprotease isolated from a psychrophilic Pseudomonas species. Biochim. Biophys. Acta (BBA) - Protein Structure and Molecular Enzymology 2000, 1–2, 265–274. [Google Scholar]
- Patil, U.; Chaudhari, A. Optimal production of alkaline protease from solvent- tolerant alkaliphilic Pseudomonas aeruginosa MTCC 7926. Indian J. Biotechnol. 2011, 10, 329–339. [Google Scholar]
- Yang, C.; Yang, F.; Hao, J.; Zhang, K.; Yuan, N.; Sun, M. Identification of a proteolytic bacterium HW08 and characterization of its extracllular cold-Active alkaline metalloprotease ps5. Biosci. Biotechnol. Biochem. 2010, 74, 1220–1225. [Google Scholar] [CrossRef]
- Koka, R.; Weimer, B.C. Isolation and characterization of a protease from Pseudomonas fluorescens RO98. J. Appl. Microbiol. 2000, 89, 280–288. [Google Scholar] [CrossRef]
- Hamamato, T.; Kaneda, M.; Horikoshi, K.; Kudo, T. Characterization of a Protease from a psychrotroph, Pseudomonas fluorescens 114. Appl. Environ. Microbiol. 1994, 60, 3878–3880. [Google Scholar]
- Meza, J.C.; Auria, R.; Lomascolo, A.; Sigoillot, J.C.; Casalot, L. Role of ethanol on growth, laccase production and protease activity in Pycnoporus cinnabarinus ss3. Enzyme Microb. Tech. 2007, 41, 162–168. [Google Scholar] [CrossRef]
- Shanmughapriya, S.; Krishnaveni, J.; Selvin, J.; Gandhimathi, R.; Arunkumar, M.; Thangavelu, T.; Kiran, G.S.; Natarajaseenivasan, K. Optimization of extracellular thermotolerant alkaline protease produced by marine Roseobacter sp. (MMD040). Bioprocess Biosyst. Eng. 2008, 31, 427–433. [Google Scholar]
- Tariq, A.L.; Reyaz, A.L.; Prabakaran, J.J. Purification and characterization of 56 kDa cold-active protease from Serratia marcescens. Afr. J. Microbiol. Res. 2011, 5, 5841–5847. [Google Scholar]
- Morita, Y.; Kondoha, K.; Hasanb, Q.; Sakaguchia, T.; Murakamia, Y.; Yokoyamaa, K.; Tamiyaa, E. Purification and characterization of a cold-Active protease from psychrotrophic Serratia marcescens AP3801. J. Am. Oil Chem. Soc. 1997, 11, 1377–1383. [Google Scholar]
- Mozhina, N.V.; Burmistrova, O.A.; Pupov, D.V.; Rudenskaya, G.N.; Dunaevsky, Y.E.; Demiduk, I.V.; Kostrov, S.V. Isolation and properties of Serratia proteamaculans 94 cysteine protease. Russ. J. Bioorg. Chem. 2008, 34, 274–279. [Google Scholar] [CrossRef]
- Kulakova, L.; Galkin, A.; Kurihara, T.; Yoshimura, T.; Esaki, N. Coldactive serine alkaline protease from the psychrotrophic bacterium Shewanella strain ac10, gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 1999, 65, 611–617. [Google Scholar]
- Saba, I.; Qazi, P.H.; Rather, S.A.; Dar, R.A.; Qadri, Q.A.; Ahmad, N.; Johri, S.; Taneja, S.S.S. Purification and characterization of a cold-active alkaline protease from Stenotrophomonas sp., isolated from Kashmir, India. World J. Microbiol. Biotechnol. 2012, 28, 1071–1079. [Google Scholar] [CrossRef]
- Tokiwa, Y.; Kitagawa, M.; Fan, H.; Raku, T.; Hiraguri, Y.; Shibatani, S.; Kurane, R. Synthesis of vinyl arabinose ester catalyzed by protease from Streptomyces sp. Biotechnol. Tech. 1999, 13, 173–176. [Google Scholar]
- Lopes, A.; Coelho, R.R.R.; Meirelles, M.N.L.; Branquinha, M.H.; Vermelho, A.B. Extracellular serine proteinase isolated from Streptomyces alboniger: Partial characterization and effect of aprotinin on cellular structure. Mem. Inst. Oswaldo Cruz. 1999, 94, 763–770. [Google Scholar]
- Elibol, M.; Moreira, A.R. Optimizing some factors affecting alkaline protease production by a marine bacterium Teredinobacter turnirae under solid state fermentation. Process Biochem. 2005, 40, 1951–1956. [Google Scholar] [CrossRef]
- Kredics, L.; Terecskei, K.; Antal, Z.; Szekeres, A.; Hatvani, L.; Manczinger, L.; Vagvolgyi, C. Purification and preliminary characterization of a cold–adapted extracellular proteinase from Trichoderma atroviride. Acta Biol. Hung. 2008, 59, 259–268. [Google Scholar] [CrossRef]
- Hamamato, T.; Kaneda, M.; Kudo, T.; Horikoshi, K. Characterization of a protease from a psychrophilic Vibrio sp Strain 5709. J. Mar. Biotechnol. 1995, 2, 219–222. [Google Scholar]
- Ward, O.P. Proteolytic Enzymes. In : Comprehensive Biotechnology; Moo-Young, Ed.; Pergamon Press: Oxford, UK, 1985; Volume 3, pp. 789–818. [Google Scholar]
- Morihara, K. Comparative specificity of microbial proteinases. Adv. Enzymol. 1974, 41, 179–243. [Google Scholar]
- Hartley, B.S. Proteolytic enzymes. Annu. Rev. Biochem. 1960, 29, 45–72. [Google Scholar] [CrossRef]
- Joo, H.S.; Kumar, C.G.; Park, G.C.; Paik, S.R.; Chang, C.S. Oxidant and SDS-stable alkaline protease from Bacillus clausii I–52, production and some properties. J. Appl. Microbiol. 2003, 95, 267–272. [Google Scholar] [CrossRef]
- Vazquez, S.C.; MacCormack, W.P.; Rios Merino, L.N.; Fraile, E.R. Factors influencing protease production by two Antarctic strains of Stenotrophomonas maltophilia. Rev. Argent. Microbiol. 2000, 32, 53–62. [Google Scholar]
- Dube, S.; Singh, L.; Alam, S.I. Proteolytic anaerobic bacteria from lake sediment of Antarctica. Enzyme Microb. Tech. 2001, 20, 114–118. [Google Scholar] [CrossRef]
- Saeki, K.; Iwata, J.; Watanabe, Y.; Tamai, Y. Purification and characterization of an alkaline protease from Oerskovia xanthineolytica TK-1. J. Ferment. Bioeng. 1994, 77, 554–556. [Google Scholar] [CrossRef]
- Khairullin, R.F.; Mikhailova, A.G.; Sebyakina, T.Y.; Lubenets, N.L.; Ziganshin, R.H.; Demidyuk, I.V.; Gromova, T.Y.; Kostrov, S.V.; Rumsh, L.D. Oligopeptidase B from Serratia proteamaculans. I. Determination of primary structure, isolation, and purification of wild-type and recombinant enzyme variants. Biochem. (Moscow) 2009, 74, 1164–1172. [Google Scholar] [CrossRef]
- Zambare, V.; Nilegaonkar, S.; Kanekar, P. A novel extracellular protease from Pseudomonas aeruginosa MCM B-327: Enzyme production and its partial characterization. New Biotechnol. 2011, 28, 173–181. [Google Scholar]
- Mikhailova, A.G.; Likhareva, V.V.; Khairullin, R.F.; Lubenets, N.L.; Rumsh, L.D.; Demidyuk, I.V.; Kostrov, S.V. Psychrophilic trypsin-type protease from Serratia proteamaculans. Biochem. (Moscow) 2006, 71, 563–570. [Google Scholar] [CrossRef]
- Vazquez, S.; Ruberto, L.; Cormack, W.M. Properties of extracellular proteases from three psychrotolerant Stenotrophomonas maltophilia isolated from Antarctic soil. Polar Biol. 2005, 28, 319–325. [Google Scholar] [CrossRef]
- Irwin, J.A.; Alfredesson, G.A.; Lanzetti, A.J.; Haflidi, M.; Gudmundsson, H.M.; Engel, P.C. Purification and characterization of a serine peptidase from the marine psychrophile strain PA-43. FEMS Microbiol. Lett. 2001, 201, 285–290. [Google Scholar] [CrossRef]
- Feller, G.; Narinx, E.; Arpingy, J.L.; Aittaleb, M.; Baise, E.; Genicot, S.; Gerday, C. Enzymes from psychrophilic organisms. FEMS Microbiol. Rev. 1996, 18, 189–202. [Google Scholar] [CrossRef]
- Feller, G.; Payan, F.; Theys, F.; Qian, M.; Haser, R.; Gerday, C. Stability and structural analysis of α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 1994, 222, 441–447. [Google Scholar] [CrossRef]
- Yan, B.Q.; Chen, X.L.; Hou, X.Y.; He, H.L.; Zhou, B.C.; Zhang, Y.Z. Molecular analysis of the gene encoding a cold-adapted halophilic subtilase from deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913: cloning, expression, characterization and function analysis of the C-terminal PPC domains. Extremophiles 2009, 13, 725–733. [Google Scholar] [CrossRef]
- Patel, T.R.; Jackman, D.M.; Bartlett, F.M. Heat-Stable protease from Pseudomonas fluorescens T16, purification by affinity column chromatography and characterization. Appl. Environ. Microbiol. 1983, 46, 333–337. [Google Scholar]
- Kim, J.; Lee, S.M.; Jung, H.J. Characterization of calcium-activated bifunctional peptidase of the psychrotrophic Bacillus cereus. J. Microbiol. 2005, 43, 237–243. [Google Scholar]
- Ni, X.; Yue, L.; Chi, Z.; Li, J.; Wang, X.; Madzak, C. Alkaline protease gene cloning from the marine yeast Aureobasidium pullulans HN2–3 and the protease surface display on Yarrowia lipolytica for bioactive peptide production. J. Mar. Biotechnol. 2009, 11, 81–89. [Google Scholar] [CrossRef]
- Wintrode, P.L.; Miyazaki, K.; Arnold, F.H. Cold adaptation of a mesophilic subtilisin-like protease by laboratory evolution. J. Biochem. 2000, 275, 31635–31640. [Google Scholar]
- Taguchi, S.; Ozaki, A.; Momose, H. Engineering of a Cold-adapted protease by sequential random mutagenesis and a screening system. Appl. Environ. Microbiol. 1998, 64, 492–495. [Google Scholar]
- Parrilli, E.; Vizio, D.D.; Cirulli, C.; Tutino, M.L. Development of an improved Pseudoalteromonas haloplanktis TAC125 strain for recombinant protein secretion at low temperature. Microb. Cell Fact. 2008. [Google Scholar] [CrossRef]
- Huston, A.L. Psychrophiles: From Biodiversity to Biotechnology; Springer: Heidelberg, Germany, 2008; pp. 347–363. [Google Scholar]
- Almog, O.; González, A.; Godin, N.; de Leeuw, M.; Mekel, M.J.; Klein, D.; Braun, S. The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins 2009, 74, 489–496. [Google Scholar] [CrossRef]
- Trimbur, D.E.; Gutshall, K.R.; Prema, P.; Brenchley, J.E. Characterization of a psychrotrophic Arthrobacter gene and its cold-active beta galactosidase. Appl. Environ. Microbiol. 1994, 60, 4544–4552. [Google Scholar]
- Feller, G.; Thiry, M.; Gerday, C. Nucleotide sequence of the lipase gene lip2 from the Antarctic psychrotroph Moraxella TA 144 and site-specific mutagenesis of the conserved serine and histidine residues. DNA Cell Biol. 1991, 10, 381–388. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Feller, G.; Gerday, C. Cloning, sequence and structural features of a lipase from the Antarctic facultative psychrophilic Psychrobacter immobilis B10. Biochim. Biophys. Acta 1993, 1171, 331–333. [Google Scholar]
- Dong, D.; Ihara, T.; Motoshima, H.; Watanabe, K. Crystallization and preliminary X-ray crystallographic studies of a psychrophilic subtilisin-like protease Apa1 from Antarctic Pseudoalteromonas sp. strain AS-11. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 308–311. [Google Scholar] [CrossRef]
- Chevrier, B.; Schalk, C.; D'Orchymont, H.; Rondeau, J.M.; Moras, D.; Tarnus, C. Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family. Structure 1994, 2, 283–291. [Google Scholar] [CrossRef]
- Papaleo, E.; Pasi, M.; Tiberti, M.; De Gioia, L. Molecular dynamics of mesophilic-like mutants of a cold-adapted enzyme: Insights into distal effects induced by the mutations. PLoS ONE 2011. [Google Scholar] [CrossRef]
- Spiwok, V.; Lipovova, P.; Skalova, T.; Duskova, J.; Dohnalek, J.; Haaek, J.; Russell, N.J.; Kralova, B. Cold-active enzymes studied by comparative molecular dynamics simulation. J. Mol. Model. 2007, 13, 485–497. [Google Scholar] [CrossRef]
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000, 297, 1015–1026. [Google Scholar] [CrossRef]
- Kasana, R.C. Proteases from psychrotrophs: An overview. Crit. Rev. Microbiol. 2010, 36, 134–145. [Google Scholar] [CrossRef]
- Couto, G.H.; Glogauer, A.; Faoro, H.; Chubatsu, L.S.; Souza, E.M.; Margesin, F.O. Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet. Mol. Res. 2010, 9, 514–523. [Google Scholar] [CrossRef]
- Roh, C.; Villatte, F. Isolation of a low-temperature adapted lipolytic enzyme from uncultivated micro-organism. J. Appl. Microbiol. 2008, 105, 116–123. [Google Scholar] [CrossRef]
- Voget, S.; Steele, H.L.; Streit, W.R. Characterization of a metagenome-derived halotolerant cellulase. J. Biotechnol. 2006, 126, 26–36. [Google Scholar] [CrossRef]
- Sharma, S.; Khan, F.G.; Qazi, G.N. Molecular cloning and characterization of amylase from soil metagenomic library derived from Northwestern Himalayas. Appl. Microbiol. Biotechnol. 2010, 86, 1821–1828. [Google Scholar] [CrossRef]
- Lee, C.C.; Kibblewhite-Accinelli, R.E.; Wagschal, K.; Robertson, G.H.; Wong, D.W.S. Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 2006, 10, 295–300. [Google Scholar] [CrossRef]
- Berlemont, R.; Pipers, R.; Delsaute, M.; Angiono, F.; Feller, G.; Galleni, M.; Power, P. Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev. Argent. Microbiol. 2011, 43, 94–103. [Google Scholar]
- Tondo, E.C.; Lakus, F.R.; Oliveira, F.A.; Brandelli, A. Identification of heat stable protease of Klebsiella oxytoca isolated from raw milk. Lett. Appl. Microbiol. 2004, 38, 146–150. [Google Scholar] [CrossRef]
- Secades, P.; Alvarez, B.; Guijarro, J.A. Purification and characterization of a psychrophilic calcium induced, growth-phase dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2001, 67, 2436–2444. [Google Scholar] [CrossRef]
- Matta, H.; Punj, V. Isolation and partial characterization of a thermostable extracellular protease of Bacillus polymyxa B–17. Int. J. Food Microbiol. 1998, 42, 139–145. [Google Scholar] [CrossRef]
- Zhang, Y.; Porcelli, M.; Cacciapuoti, G.; Ealick, S.E. The crystal structure of 5’-deoxy-5’-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds. J. Mol. Biol. 2006, 357, 252–262. [Google Scholar] [CrossRef]
- Storch, E.M.; Daggett, V.; Atkins, W.M. Engineering out motion: A surface disulfide bond alters the mobility of tryptophan 22 in cytochrome b5 as probed by time-resolved fluorescence and 1H NMR experiments. Biochem. 1999, 38, 5054–5064. [Google Scholar] [CrossRef]
- Matthews, B.W.; Nicholson, H.; Becktel, W.J. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. P. Natl. Acad. Sci. USA 1987, 84, 6663–6667. [Google Scholar] [CrossRef]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. T. Roy. Soc. B. 2002, 357, 917–925. [Google Scholar] [CrossRef]
- Boehr, D.D.; Dyson, H.J.; Wright, P.E. An NMR perspective on enzyme dynamics. Chem. Rev. 2006, 106, 3055–3079. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef]
- Tehei, M.; Zaccai, G. Adaptation to high temperatures through macromolecular dynamics by neutron scattering. FEBS J. 2007, 274, 4034–4043. [Google Scholar] [CrossRef]
- Adcock, S.A.; McCammon, J.A. Molecular Dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 2006, 106, 1589–1615. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Bakowies, D.; Baron, R.; Chandrasekhar, I.; Christen, M.; Daura, X.; Gee, P.; Geerke, D.P.; Glattli, A.; Hunenberger, P.H. Biomolecular modeling: Goals, problems, perspective. Angewandte Chemie International Edition in English 2006, 45, 4064–4092. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Dolenc, J.; Mark, A.E. Molecular simulation as an aid to experimentalists. Curr. Opin. Struc. Biol. 2008, 18, 149–153. [Google Scholar] [CrossRef]
- Pantoliano, M.W.; Whitlow, M.; Wood, J.F.; Dodd, S.W.; Hardman, K.D.; Rollence, M.L.; Bryan, P.N. Large increases in general stability for the subtilisin BPN’ through incremental changes in the free energy of unfolding. Biochem. 1989, 28, 7205–7213. [Google Scholar] [CrossRef]
- Strausberg, S.L.; Alexander, P.A.; Gallagher, D.T.; Gilliland, G.L.; Barnett, B.L.; Bryan, P.N. Directed evolution of a subtilisin with calcium-independent stability. Biotechnology 1995, 13, 669–673. [Google Scholar]
- Shao, Z.; Zhao, H.; Giver, L.; Arnold, F.H. Random-priming in vitro recombination: An effective tool for directed evolution. Nucleic Acids Res. 1998, 26, 681–683. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, L.; Yang, S.; Zhu, L.; Wu, Y.; Li, Z. A mutant subtilisin E with enhanced thermostability. World J. Microb. Biot. 2000, 16, 249–251. [Google Scholar] [CrossRef]
- Siguroardottir, A.G.; Arnorsdottir, J.; Thorbjarnardottir, S.H.; Eggertsson, G.; Suhre, K.; Kristjansson, M.M. Characteristics of mutants designed to incorporate a new ion pair into the structure of a cold adapted subtilisin-like serine proteinase. Biochim. Biophys. Acta. 512–518.
- Narinx, E.; Baise, E.; Gerday, C. Subtilisin from psychrophilic antarctic bacteria: characterization and site-directed mutagenesis of residues possibly involved in the adaptation to cold. Prot. Eng. 1997, 10, 1271–1279. [Google Scholar] [CrossRef]
- Zhao, H.; Arnold, F.H. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Engi. 1999, 12, 47–53. [Google Scholar]
- He, H.L.; Chen, X.L.; Zhang, X.Y.; Sun, C.Y.; Zou, B.C.; Zhang, Y.Z. Novel Use for the Osmolyte Trimethylamine N-oxide, Retaining the psychrophilic characters of cold–adapted protease Deseasin MCP-01 and simultaneously improving its thermostability. Mar. Biotechnol. 2009, 11, 710–716. [Google Scholar] [CrossRef]
- Vazquez, S.C.; MacCormack, W.P. Effect of isolation temperature on the characteristics of extracellular proteases produced by Antarctic bacteria. Polar Res. 2002, 21, 63–71. [Google Scholar] [CrossRef]
- Nielson, M.H.; Jepsen, S.J.; Outrup, H. Enzymes for lower temperature washing. J. Am. Oil Chem. Soc. 1981, 58, 644–649. [Google Scholar] [CrossRef]
- Pawar, R.; Zambare, V.; Barve, Z.; Paratkar, G. Application of protease isolated from Bacillus sp158 in enzymatic cleansing of contact lenses. Biotechnology 2009, 8, 276–280. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Joshi, S.; Satyanarayana, T. Biotechnology of Cold-Active Proteases. Biology 2013, 2, 755-783. https://doi.org/10.3390/biology2020755
Joshi S, Satyanarayana T. Biotechnology of Cold-Active Proteases. Biology. 2013; 2(2):755-783. https://doi.org/10.3390/biology2020755
Chicago/Turabian StyleJoshi, Swati, and Tulasi Satyanarayana. 2013. "Biotechnology of Cold-Active Proteases" Biology 2, no. 2: 755-783. https://doi.org/10.3390/biology2020755
APA StyleJoshi, S., & Satyanarayana, T. (2013). Biotechnology of Cold-Active Proteases. Biology, 2(2), 755-783. https://doi.org/10.3390/biology2020755




