Recent Advances in the Analysis of Macromolecular Interactions Using the Matrix-Free Method of Sedimentation in the Analytical Ultracentrifuge
Abstract
:1. Introduction
2. Analytical Ultracentrifugation
3. Sedimentation Velocity
- (i)
- A correction to the sedimentation coefficient distribution for diffusion broadening (this assumes all particles in the distribution have the same frictional ratio f/fo (ratio of the friction coefficient of a macromolecule to the friction coefficient of a sphere of the same mass and anhydrous volume)—the resulting corrected distribution is known as a c(s) vs. s distribution.
- (ii)
- Conversion of g(s) vs. s or c(s) vs. s for a discrete distribution (i.e., of a small number of resolvable components) to a molecular weight distribution (again, assuming particles of the same shape/frictional ratio), known as g(M) vs. M or c(M) vs. M.
- (iii)
- For continuous distributions, a recently published algorithm known as the Extended Fujita algorithm [8] was incorporated into SEDFIT.
4. Sedimentation Equilibrium
5. Protein Self-Association
5.1. Tetanus Toxoid Protein


5.2. Yeast Exoribonuclease Rrp6 Associated Cofactor Rrp47
5.3. Mini-Agrin
6. Carbohydrate Self-Association
6.1. Xyloglucans
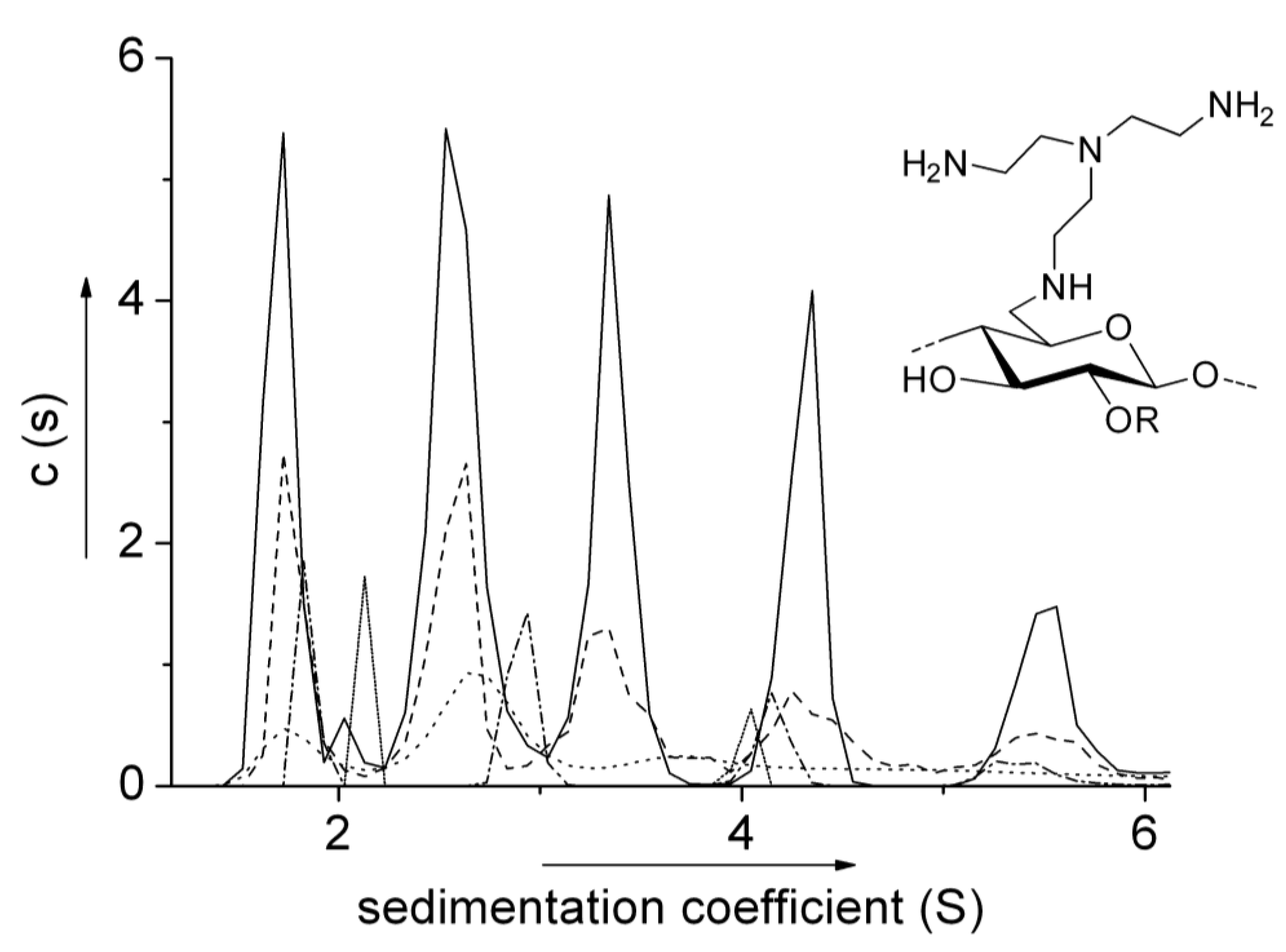
6.2. Aminocelluloses
7. Carbohydrate-Protein Interactions

8. Nucleic Acid-Carbohydrate Interactions

9. Carbohydrate-Carbohydrate Interactions
9.1. Chitosan-Xanthan Interactions
9.2. PGX®
10. Concluding Remarks
Acknowledgements
Conflicts of Interest
References
- Harding, S.E.; Rowe, A.J. Insight into protein-protein interactions from analytical ultracentrifugation. Biochem. Soc. Trans. 2010, 38, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.E. Mucoadhesive interactions. Biochem. Soc. Trans. 2003, 31, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.E. Trends in mucoadhesive analysis. Trends Food Sci. Tech. 2006, 17, 255–262. [Google Scholar] [CrossRef]
- Harding, S.E.; Rowe, A.J.; Horton, J.C. Analytical Ultracentrifugation in Biochemistry and Polymer Science; Royal Society of Chemistry: Cambridge, UK, 1992. [Google Scholar]
- Scott, D.J.; Harding, S.E.; Rowe, A.J. Analytical Ultracentrifugation. Techniques and Methods; Royal Society of Chemistry: Cambridge, UK, 2005. [Google Scholar]
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000, 78, 1606–1619. [Google Scholar] [CrossRef] [PubMed]
- Dam, J.; Schuck, P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Meth. Enzymol. 2004, 384, 185–212. [Google Scholar] [PubMed]
- Harding, S.E.; Schuck, P.; Abdelhammed, A.S.; Adams, G.; Kök, M.S.; Morris, G.A. Extended Fujita approach to the molecular weight distribution of polysaccharides and other polymer systems. Methods 2011, 54, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Gillis, R.B.; Adams, G.G.; Wolf, B.; Berry, M.; Besong, T.M.D.; Corfield, A.; Kök, S.M.; Sidebottom, R.; Lafond, D.; Rowe, A.J.; et al. Molecular weight distribution analysis by ultracentrifugation: Adaptation of a new approach for mucins. Carbohyd. Polym. 2003, 93, 178–183. [Google Scholar] [CrossRef]
- Creeth, J.M.; Harding, S.E. Some observations on a new type of point average molecular weight. J. Biochem. Biophys. Methods 1982, 7, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Schuck, P.J.; Gillis, R.B.; Besong, D.; Almutairi, F.; Adams, G.G.; Rowe, A.J.; Harding, S.E. SEDFIT-MSTAR: Molecular weight and molecular weight distribution analysis of polymers by sedimentation equilibrium in the ultracentrifuge. Analyst 2014, 139, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Gillis, R.B.; Adams, G.G.; Heinze, T.; Nikolajski, M.; Harding, S.E.; Rowe, A.J. MultiSig: A new high-precision approach to the analysis of complex biomolecular systems. Europ. Biophys. J. 2013, 42, 777–786. [Google Scholar] [CrossRef]
- Heinze, T.; Nikolajski, M.; Daus, S.; Besong, T.M.D.; Michaelis, N.; Berlin, P.; Morris, G.A.; Rowe, A.J.; Harding, S.E. Protein-like oligomerisation of carbohydrates. Ang. Chem. Int. Ed. 2011, 50, 8602–8604. [Google Scholar] [CrossRef]
- Nikolajski, M.; Adams, G.G.; Gillis, R.B.; Besong, D.T.; Rowe, A.J.; Heinze, T.; Harding, S.E. Protein-like fully reversible tetramerisation and super-association of an aminocellulose. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Rallison, J.M.; Harding, S.E. Excluded volumes for pairs of triaxial ellipsoids at dominant Brownian motion. J. Coll. Int. Sci. 1985, 103, 284–289. [Google Scholar] [CrossRef]
- Harding, S.E.; Horton, J.C.; Jones, S.; Thornton, J.M.; Winzor, D.J. COVOL: An interactive program for evaluating second virial coefficients from the triaxial shape or dimensions of rigid macromolecules. Biophys. J. 1999, 76, 2432–2438. [Google Scholar] [CrossRef] [PubMed]
- Astronomo, R.D.; Burton, D.R. Carbohydrate vaccines: Developing sweet solutions to sticky situations? Nat. Rev. Drug Discov. 2010, 9, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, A.S.; Morris, G.A.; Adams, G.G.; Rowe, A.J.; Laloux, O.; Cerny, L.; Bonnier, B.; Duvivier, P.; Conrath, K.; Lenfant, C.; et al. An asymmetric and slightly dimerized structure for the tetanus toxoid protein used in glycoconjugate vaccines. Carbohyd. Polym. 2012, 90, 1831–1835. [Google Scholar] [CrossRef]
- Feigenbutz, M.; Jones, R.; Besong, T.D.; Harding, S.E.; Mitchell, P. Assembly of the yeast exoribunuclease Rrp6 with its associated cofactor Rrp47 occurs in the nucleus and is critical for the controlled expression of Rrp47. J. Biol. Chem. 2013, 288, 15959–15970. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.R.; Besong, T.M.D.; Patel, N.; Meier, M.; Harding, S.E.; Winzor, D.J.; Stetefeld, J. Evidence for self-association of a miniaturized version of agrin from hydrodynamic and small-angle X-ray scattering measurements. J. Phys. Chem. B 2011, 115, 11286–11293. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.R.; Harding, S.E.; Ebringerova, A.; Deszczynski, M.; Hromadkova, Z.; Togola, A.; Paulsen, B.S.; Morris, G.A.; Rowe, A.J. Weak self-association in a carbohydrate system. Biophys. J. 2007, 93, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.; Berlin, P. New water-soluble and film-forming aminocellulose tosylates as enzyme support matrices with Cu2+-chelating properties. Cellulose 2005, 12, 67–84. [Google Scholar] [CrossRef]
- Kök, M.S.; Gillis, R.; Ang, S.; Lafond, D.; Tatham, A.S.; Adams, G.; Harding, S.E. Can dietary fibre help provide safer food products for sufferers of gluten intolerance? A well-established biophysical probe may help towards providing an answer. BMC Biophys. 2012, 5. [Google Scholar] [CrossRef]
- Gillis, R. Protein Polysaccharide Complexes: Permanent/Nonpermanent Interactions between Polysaccharides and Polypeptides. PhD Thesis, University of Nottingham, Nottingham, UK, 2015. [Google Scholar]
- Almutairi, F.M.; Erten, T.; Adams, G.G.; Hayes, M.; McGloughlin, P.; Kök, M.S.; Mackie, A.R.; Rowe, A.J.; Harding, S.E. Hydrodynamic characterisation of chitosan and its interaction with two polyanions: DNA and xanthan. Carbohyd. Polym. 2015. [Google Scholar] [CrossRef]
- Dumitriu, S.; Magny, P.; Montane, D.; Vidal, P.; Chornet, E. Polyionic hydrogels obtained by complexation between xanthan and chitosan: Their properties as supports for enzyme immobilization. J. Bioact. Compat. Polym. 1994, 9, 184–209. [Google Scholar] [CrossRef]
- Martínez-Ruvalcaba, A.; Chornet, E.; Rodrigue, D. Viscoelastic properties of dispersed chitosan/xanthan hydrogels. Carbohyd. Polym. 2007, 67, 586–595. [Google Scholar] [CrossRef]
- Shchipunov, Y.; Sarin, S.; Kim, I.; Ha, C.-S. Hydrogels formed through regulated self-organization of gradually charging chitosan in solution of xanthan. Green Chem. 2010, 12, 1187–1195. [Google Scholar] [CrossRef]
- Erten, T.; Adams, G.G.; Foster, T.J.; Harding, S.E. Comparative heterogeneity, molecular weights and viscosities of xanthans of different pyruvate and acetate content. Food Hydrocoll. 2014, 42, 335–341. [Google Scholar] [CrossRef]
- Abdelhameed, A.S.; Ang, S.; Morris, G.A.; Smith, I.; Lawson, C.; Gahler, R.; Wood, S.; Harding, S.E. An analytical ultracentrifuge study on ternary mixtures of konjac glucomannan supplemented with sodium alginate and xanthan gum. Carbohyd. Polym. 2010, 81, 145–148. [Google Scholar] [CrossRef]
- Harding, S.E.; Smith, I.H.; Lawson, C.J.; Gahler, R.J.; Wood, S. Studies on macromolecular interactions in ternary mixtures of konjac glucomannan, xanthan gum and sodium alginate. Carbohyd. Polym. 2011, 83, 329–338. [Google Scholar]
- Smith, I.H.; Lawson, C.J.; Harding, S.E.; Gahler, R.J.; Lyon, M.R.; Wood, S. Viscosity development during aqueous dispersion and dissolution: A comparison of PGX® with other dietary supplements and individual polysaccharides. Food Hydrocoll. 2014, 38, 152–162. [Google Scholar] [CrossRef]
- Rivas, G.; Minton, A. Beyond the second virial coefficient: Sedimentation equilibrium in highly non-ideal solutions. Methods 2011, 54, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Luchinat, C.; Parigi, G.; Ravera, E.; Reif, B.; Turano, P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc. Nat. Acad. Sci. USA 2011, 108, 10396–10399. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Luchinat, C.; Parigi, G.; Ravera, E. SedNMR: On the edge between solution and solid-state NMR. Acc. Chem. Res. 2013, 46, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harding, S.E.; Gillis, R.B.; Almutairi, F.; Erten, T.; Kök, M.Ş.; Adams, G.G. Recent Advances in the Analysis of Macromolecular Interactions Using the Matrix-Free Method of Sedimentation in the Analytical Ultracentrifuge. Biology 2015, 4, 237-250. https://doi.org/10.3390/biology4010237
Harding SE, Gillis RB, Almutairi F, Erten T, Kök MŞ, Adams GG. Recent Advances in the Analysis of Macromolecular Interactions Using the Matrix-Free Method of Sedimentation in the Analytical Ultracentrifuge. Biology. 2015; 4(1):237-250. https://doi.org/10.3390/biology4010237
Chicago/Turabian StyleHarding, Stephen E., Richard B. Gillis, Fahad Almutairi, Tayyibe Erten, M. Şamil Kök, and Gary G. Adams. 2015. "Recent Advances in the Analysis of Macromolecular Interactions Using the Matrix-Free Method of Sedimentation in the Analytical Ultracentrifuge" Biology 4, no. 1: 237-250. https://doi.org/10.3390/biology4010237
APA StyleHarding, S. E., Gillis, R. B., Almutairi, F., Erten, T., Kök, M. Ş., & Adams, G. G. (2015). Recent Advances in the Analysis of Macromolecular Interactions Using the Matrix-Free Method of Sedimentation in the Analytical Ultracentrifuge. Biology, 4(1), 237-250. https://doi.org/10.3390/biology4010237








