Abstract
Warren Weaver, writing about the function that science should have in mankind’s developing future, ideas and ideals, proposed to classify scientific problems into ‘problems of simplicity’, ‘problems of disorganised complexity’, and ‘problems of organised complexity’—the huge complementary class to which all biological, human, and social problems belong. Problems of simplicity have few components and variables and have been extensively addressed in the last 400 years. Problems of disorganised complexity have a huge number of individually erratic components and variables, but possess collective regularities that can be analysed by resourcing to stochastic methods. Yet, ‘problems of organised complexity’ do not yield easily to classical or statistical treatment. Interrelations among phenomenon elements change during its evolution alongside commonly used state variables. This invalidates independence and additivity assumptions that support reductionism and affect behaviour and outcome. Moreover, organisation, the focal point in this complementary class, is still an elusive concept despite gigantic efforts undertaken since a century ago to tame it. This paper addresses the description, representation and study of phenomena in the ‘problems of organised complexity’ class, arguing that they should be treated as a collection of interacting organisations. Furthermore, grounded on relational mathematical constructs, a formal theoretical framework that provides operational definitions, schemes for representing organisations and their changes, as well as interactions of organisations is introduced. Organisations formally extend the general systems concept and suggest a novel perspective for addressing organised complexity phenomena as a collection of interacting organisations.
1. Introduction
Natural phenomena stem from a collection of things that interact and, while interacting, change the aspects we observe in them. No change, no phenomenon. No interaction, no change. This observation can be used to make explicit who interacts and what is exchanged in the interactions that give rise to a phenomenon. This line of reasoning allows for classifying different phenomena by means of different concepts in an integrated manner [1,2,3]. Warren Weaver in 1948 [1] classified natural phenomena yielding to scientific investigation into three groups grounded on characteristics of ‘things’ and their interactions, but also on methods used to investigate them: ‘problems of simplicity’, ‘problems of disorganised complexity’ and ‘problems of organised complexity’.
Phenomena in the first class have a small number of determinable things and variables. The motion of two or three balls on a billiard table, oscillations of two interacting pendulums, the motion of planets around the sun are typical examples of those. If the number of things and observable aspects increase, these problems become intractable not because of theoretical hindrances but due to computational or operational difficulties. For instance, when considering a large number of pendulums, billiard balls, or many planets simultaneously. Note that, even in these latter cases, understanding have been achieved by considering just two things at a time, in all sensible combinations.
Phenomena in the second class possess a huge number of erratic things and variables that interact in the same manner or whose interactions cannot be distinguished through changes in the observed aspects. Like in the first class, there is no restriction on who interacts with whom and the interaction possibilities remain unchanged throughout the course of the phenomena. These problems present collective regularities that can be investigated employing stochastic and statistical methods. Typical examples include: the motion of atoms in a volume of matter or gas, the motion of stars in the Universe, heredity, thermal behaviour, and traffic. In them, there are observable aspects, like temperature, pressure, vorticity, or birth-rates that only make sense for collectivities. While concerning motions and exchanges of energy and momenta, problems in these first two classes and methods for solving them have been the subject of physics.
In the ‘organised complexity’ class, not only the attributes of things change but also the number of interacting things and the nature and possibilities of their interactions change; what affects the dimension of the state-space and the description of the interactions [4,5,6,7]. Following Weaver, the singular characteristic of phenomena in the last class lies in the fact that they show the essential feature of organisation in both: components and interactions. To date, there are but a few definitions of organisation; none widely accepted nor used to explain life phenomena, the quintessence of ‘organised complexity problems’. Organisation is something easily recognised but difficult to grasp. Perusing available examples in Weaver’s ‘organised complexity’ class, some characteristics show up:
- The number of variables is moderate but it is not possible to hold all but two or three variables with fixed values;
- There are fundamental aspects that are non-quantitative or elude identification and measurement;
- Aspects observed are entangled, invalidating assumptions about superposition and independence;
- A collection of things interrelated in a stable and individuated manner may act as an aggregate thing, a whole, interacting with other things at the same or at different aggregation levels;
- Interrelations and possible channels of interaction among elements change, affecting the phenomenon propensities or behaviour; and
- Units of interaction ‘adapt’, ‘learn’, and ‘fabricate’ [8,9] other units of interaction adding new factors, aspects and variables, to the phenomenon description.
The ‘problems of organised complexity’ class encompass virtually all biological, health and social problems, extending to any phenomena that involve living-things as components [10]. Franklin Harold [11] distinguishes living from non-living through their capacity of maintaining, reproducing and multiplying “states of matter characterised by an extreme degree of organisation” (see also [12]). The characterisation of this class, though, does not primarily depend on the number of things or variable aspects involved. In his own words and emphasis [1]:
They are problems which involve dealing simultaneously with a seizable number of factors which are interrelated into an organic whole.
This happens in a manner similar to composite systems and their collective dynamics, which leads to self-organisation, but involves also forbidden interactions, channels of interaction that change, and factors that are entangled and interdependent. These entangled factors encapsulate into units, adapt themselves and integrate several dynamic scales [6,8,13,14,15,16,17], leading to the picture described by Harold [11,14].
Quite a number of the problems enrolled as examples of phenomena in this class have been more or less successfully addressed nowadays. To this date, their mathematical description and treatment, that originated through associations with complex systems [18,19], employ methods akin to those used in the investigation of problems in Weaver’s ‘disorganised complexity’ class, centring on formalisms of thermodynamics, state transitions and critical phenomena [20]. Note that, as a state of matter, living things are as organised as a crystal, as fluid as a liquid, but in no case aleatory as a gas or plasma. Properly re-phrasing several approaches employed to study living and ‘organised complexity’ phenomena, things in them may be rightly named organised matter and living phenomena considered as the interplay and dynamics of (material) organisations.
Despite their utter beauty and usefulness in explaining the appearance of organisation, the compatibility of organised matter dynamics with physical laws of out-of-equilibrium systems, the tendency of physical phenomena to self-organise, and what happens at the border of the (extended) “criticality zone” [20], these methods are of limited utility to describe entailments and what happens within ‘organised complexity’ phenomena—that is, to explain how organised-matter maintains, entails and evolves organisation.
In scientific enquiries, we primevally choose to describe interacting things in a phenomenon as members of some archetypical thing-class, most of which are associated with formalisms that support representing (modelling) the phenomenon and reasoning about it. Depending on which aspects are observed, on what is exchanged in interactions, and on what is being asked, typical thing-classes may be particles, fields, substances, bodies, fluids, molecules, organisms, individuals, populations, firms, organisations (e.g., social entities and human associations), ecosystems, or a mixture of them. Thing-classes highlight characteristics considered important in studying a phenomenon and for its explanation, based on correlates of it and previous experience. Thing-classes act as spectacles we use to observe, model and understand a phenomenon. Concomitantly, they constrain which aspects make sense, what can be observed, which questions can be posed, as well as what can be referred to and studied [3,21]. Nevertheless, it is well acknowledged that general systems theory and methodology [22,23] provide a way of formally representing and handling phenomena independently of the thing-classes chosen or the application domain.
Choosing a thing-class implies making hypotheses about the behaviour of things and their interactions and about which aspects are relevant to understand a phenomenon. Treating molecules as particles, we hypothesise that their geometric attributes like form, volume etc, plus chemical-affinity are not relevant to understand what is being observed. These choices constrain and freeze the way we look into Nature. Thus, one should use several perspectives wisely for the same phenomenon, adjusting them to our evolving questions and observation procedures in order to enlarge our perception. For instance, billiard balls over a table can be a collection of particles or of spherical bodies. The phenomenon is the same, the stand varies. Each perspective highlights and enlightens different facets of what is observed in their moving. Each stand moulds the set of questions that can be posed and answered under the point-of-view imposed by its choice. Different things in a phenomenon may be represented by elements of different thing-classes. A collection of interacting things can be considered to be of more than one thing-class bringing new enlightenment to a phenomenon, as in the case of the interplay between thermodynamics and statistical mechanics ([24] section 4.4). That these visions are related in certain situations, however restrictive, is a real wonder.
In this paper, I contend that to develop a more extensive, integrated, and encompassing attack to ‘problems of organised complexity’, earning a less ad hoc knowledge about the underlying phenomena, we need to enlarge our present collection of thing-classes with thing-classes that conform to Weaver’s characterisation above and go beyond it; by interpreting phenomena as a collection of interacting organisations that alter the organisation of the things themselves and the connections between things (channels of allowed interactions) in a phenomenon while interacting. To accomplish this, organisation is approached from a novel and complementary stand that does not involve dynamics. Its definition and analysis employ relational mathematical tools grounded on ‘sizeable numbers’ and highlighting the role of interrelationships in the constitution of organisations, with no presuppositions about or reference to behaviour, context, and ‘function’.
The purpose of this writing is to propose a generalisation for the concept of systems that could better aid taming the inherent complexity of ‘organised complexity’ problems, introducing a concept of organisation as a candidate for such thing-class. From the formal point of view, organisations are obtained from general systems apposing hierarchy and encapsulation to them. This provides a mathematical definition and model for the “systems of systems” idea, allowing to treat “systems of systems” as units of interaction [25]. Consequently, it also suggests an enhanced general systems perspective, the organisation perspective. This perspective hypothesises that things in a phenomenon are organisations, rather than particles, substances, individuals etc, that interact predominantly exchanging signals and a specific kind of (see Section 3.2 and Section 5) that pictures . In this way, living things and phenomena may be seen as a fifth state of matter (organised-matter), that maintains, reproduces, multiplies, and enchains organisations [3], as proposed by F. Harold [11,14] and R. Rosen [8] (see also [3])
The investigation wherefrom this ideas emanate was initially inspired and driven by difficulties encountered while modelling ecological and biological systems with variable structure [4,5,6,7,13] whose extreme values, possible factors, or domain cannot be established in advance. Thus, examples and illustrations in this text are mostly centred on living subjects, despite being true that the idea of systems is subjacent to phenomena in all three Weaver classes, the ‘organised complexity’ class has an embracing character, and the concepts presented here have a wider application. Even with this restriction, the related literature is extremely vast. To keep bibliographic referring manageable, citations to work that focus on behaviour and analyse organisation and information from a dynamical stand were kept to a minimum. The historical account presented in Section 2 aims just to contextualise the present proposal and put bounds on what will be discussed rather than to picture past achievements. References supporting arguments are employed parsimoniously. Frequently, not all pertinent references were included in a citation. Finally, relations between dynamics and interaction graphs, a simple form of organisation as here addressed, were analysed in a previous writing of ours [26] and the references within this work should be accessed through it.
This work is structured as follows: (1) purpose, described in this introduction; (2) a short and far from exhaustive overview of occurrence of terms organisation and information in the literature of life systems and sciences with little reference to behaviour, in the next section; (3) the organisation concept and the accompanying framework, in the third section; (4) ontological considerations, justifications and exemplifications, in the fourth section; (5) seeing phenomena as interacting organisations, the organisation perspective, in the fifth section; (6) closing remarks, in the last.
2. Organisation and Information in Life Systems and Sciences
One of the most conspicuous characteristics of life phenomena is alternatively named architectural structure or organisation [11,12,27,28,29]. Both terms refer to the same idea—the relative position, connection, or interaction channel of things and thing components with respect to one another, that become hierarchically arranged as a consequence of encapsulation into wholes or units [30]. Organisation is a central characteristic of biological entities and biological phenomena [1,11,14,27,28,31,32]. Organisation appears everywhere. It may be a collective aspect, as in simple oscillatory chemical reactions, consensus bio-chemical setups of cells [33], and “self-organising” phenomena; or a structural, individual-centred aspect present in macro-molecules, cell compartments, and cellular functional modules [14,34,35].
Organisation is generally associated with material instances of biological entities: modules, organelles, vacuoles, tissues etc [32]. Nevertheless, biological processes manifest organisation as well, e.g., the cytoskeleton [36], network activation-deactivation assemblies [37], chromatin, chaperones complexes [38], dynamic self-assembly processes [14], etc. Organisations expand beyond organelles and cell inner structures into tissues and organs of multicellular organisms and further on into populations, societies and cultures. Despite this ubiquitousness, instantiation of analogous organisations at different scales present seemingly uncorrelated forms [2,39]. Organisation is also frequently associated with complexity in biological entities [1,27,40,41,42]. Despite its importance and the bridge it launches with the study of more general complex systems [43], this subject shall not be pursued here beyond the contents of Section 4.2. Organising, the process of arranging and evolving organisations, shall not be addressed either.
Organisation configurations in living systems are credited to affect interactions, outcomes, and properties such as stability, reactiveness, capture, infection, (organelle) multiplication, stasis and mitosis [11,14,44], as well as their own existence [29], even by those that approach the living from a molecular scale, without reference to scale integration. Cell components, like the cytoskeleton and its variants, encompass dynamical organisations that are continuously reassembled. That is, their elements and their stable relative positions and dependencies result from a well orchestrated collection of movements and rearrangements that continuously relocate components and substitute missing parts or create new organelles and structures, like vacuoles, lysosomes, filopodia, micro-tubules, or centroids [14,36,45,46].
Organisation, in its variegated though ill-defined to-be concepts, has been considered a distinctive characteristic of life entities and phenomena at least since the beginnings of last century [2,11,12,21,29,44,47]. Notwithstanding the impreciseness of the concept [48,49] organisation is central while considering quaternary structure of proteins, protein conformations and their effects on protein interactions [50], protein aggregates, motifs, and cellular functional units. It is helpful when considering spatial effects in biochemical networks and in many biological enquiries and explanations ([51], last sections), along with efforts to build a theoretical framework for biology [8,52,53,54,55,56,57] and chemistry [58].
An enormous amount of work has been produced in the last 100 years to refine the idea and identification of biological organisation, to understand its onset and to justify its possibility on physico-chemical grounds [2,21,22,37,39,40,44,59]. Explanatory efforts search to dissect organisation from several standpoints like being a consequence of self-organising dynamics [59,60], resulting from regulatory processes [52] or bursting out from information [40,61,62,63]. Only a handful of these efforts address organisation directly, trying to tame the concept by considering questions akin to “what is organisation?”, by searching models for organisation, or by constructing models and arguments based directly on its properties [8,27,28,29,30,52,62,63,64,65,66,67,68,69,70,71,72]. Albeit, none of them provides a clear concept or definition of organisation [16,48,73]. In the present text, I take for granted that organisation is a fact of Nature, present a working definition and a mathematical model for organisation, and discuss some consequences of this approach. This is a variant of Niels Bohr suggestion of taking life, like quanta, as a given fact of Nature [74], unexplainable in terms of other natural facts.
Another idea tightly intertwined with life phenomena since its earliest developments is information. Information, in Shannon-Brillouin sense [51], has dominated biological discourse, being considered a central characteristic of living systems and associated phenomena [52]. Nevertheless, it is not as conspicuous, directly observable, and recognizable as organisation. Despite this, information has been considered the key observable attribute of the living albeit always recognised only during the analysis of a phenomenon and its interpretation.
Ontologically, information in biological phenomena has been associated with transmission of hereditary characters [75,76], regulatory (feedback) assemblages [52], immunology [77], and ecology [13,78]. Theoretically, it has been associated with biological structure and function as well as their adaptation to various stimuli [8,52,65], among other phenomena.
When associated to structure and function, it reflects the ability of organisms to perform tasks in response to environmental stimuli, which is allegedly characterised by the information contend of its organisations [61]. Recently, information is being credited as the main vehicle of biological interactions [79,80] and can be observed operating at the cellular scale [81].
Information is employed while searching for explanations concerning the appearance and resilience of biological organisation [40], for defining it formally [65], for defining life [61,82,83], and the consequent efforts to build biological theories (see [41] for a more detailed and critical survey). Moreover, information intervenes directly or indirectly in all essays to explain the living through computational metaphors [41,61,84,85,86].
In its commonly used sense, the employ of information in biological explanations is highly debatable [87,88] and information is erroneously taken as the key biological observable [51]. This is due to the persistence of using the term information with different, non-commutable meanings [30] and the misleading association between organisation, information, complexity, and computation that this overloading in meaning induces [51]. The definition of presented in Section 3.2 is non-numeric, grounded on the organisation concept introduced in Section 3.1, and inspired by the inner workings of the cell.
3. Theoretical Framework: The Organisation Perspective
Abiding to the view about natural phenomena delineated earlier, where phenomena stem from a collection of interacting things that alter what is observed, this section introduces things (organisations) and discusses what is exchanged in their interactions (). The framework to be presented in this section supports the representation of phenomena as a collection of interacting organisations that interact exchanging and changing their own organisation.
Aligning with the spirit of this special issue, this section shortens technical details in favour of examples and clarifications. The presentation of this framework will employ less formal and more illustrative arguments, in the hope of throwing light on basal concepts, letting the reader more at easy with a new form of seeing natural phenomena. Profiting from discrete and relational mathematical constructs, formal elements will be often introduced with the aid of figures, less formally but no less rigourously. Figures are symbols that may be more easily accessed by a multi-disciplinary audience.
3.1. Biological Organisation: A Minimalist Snapshot
As argued in Section 2, organisation is an important and widely used concept that has nevertheless no consensual definition. Organisation is recognised through patterns that appear among Weaver’s sizable number of factors and are believed to form what he called organic wholes. Representations of this sort of phenomena are often called composite systems even when they behave linearly ([89] chapter 9) and do not present the aggregate behaviour and patterns that may be taken as “organic wholes”.
At the beginnings of (general) system theory, organisation was mostly attached to the systems structure [10,23,30], conspicuous in its reaction term, slowly drifting to their dynamical behaviour with the maturation of concepts like attractors, basins of attraction, slow and center manifolds, homeostasis, homeomorphic indexes, perturbations, fluctuations etc [49,59,70,90]. That is, organisation came to be associated with self-organisation and emergent patterns in the dynamical behaviour of many-particle or many-component composite systems and, by extension, with complexity ([19] preface, [49] chapters 6 and 7). Collaterally and supported by the maturation of these concepts, a wealth of methods to analyse and illuminate these systems form various perspectives have been developed [19].
This approach, however, cannot easily handle hierarchical systems of variable structure so common in biology, ecology and other domains [4,5,6,7,66,91], that adapt to stimuli by changing the number of their state-variables, the quality and nature of their interactions, and where properties within a level cannot be observed or explained from information rooted in its lower-levels. These characteristics are the essence of Weaver’s ‘problems of organised complexity’, underlying and justifying the approach presented in the sequel.
The organisation concept advanced below (Section 3.1.1 and Section 3.1.2) is not a consequence of dynamics nor is it associated with the idea of function (biological or ecological) in any manner. It supports reasoning about organisations on their own, independently of any correlated phenomenon, accommodating organisation transformation and comparison (Section 3.1.4 and Section 4.2). Nevertheless, organisations are tightly associated with dynamics, having the usual (dynamic) systems as special cases (see Section 3.1.3 below).
3.1.1. Organisations
Consider a large enough heap of bricks indistinguishable with respect to all relevant attributes, characteristics and factors. The bricks are thus identical although not the same. This heap constitutes a set of bricks. Choose 20 of these bricks at random. These 20 chosen bricks are still a set of bricks. They form a subset of the heap, if you don’t take them apart from the heap. If you do, there will be two heaps (sets) and their connection may have become imperceptible.
Ensuite, pick all bricks in this heap and build a house. The house has 6 pieces: a living room, a dining room, two bedrooms, a bathroom and a kitchen. In the middle, there is a hall giving access to the bedrooms and the bathroom. The kitchen is accessed from the dining room that is accessed from the living room. Walls, made with the bricks, divide and delimit each piece. Some walls have doors, some other have windows, a few may have both. After the house is built, there are no more bricks but walls and rooms. Is a house a set of bricks? Is a brick in a wall identical to a brick in another wall or to another brick somewhere else in the same wall?
Consider any of the walls of the house. Mark 20 bricks in it at random. Are these bricks as indistinguishable as before? They may be side by side and taking them out will make a hole in the wall; luckily the hole may become a window or a door, depending on its height, shape, and localisation. They may also be scattered throughout the wall; what may be a refreshing strategy. Or they may concentrate in a junction of two walls, weakening its structure and ruining the house. The bricks are in consequence not indistinguishable anymore. They are interchangeable (any two bricks may be swapped) but not indistinguishable; the effect of taking any (or a collection) of them out of the walls is not anymore the same. They became parts of a wall, which in turn became a part of the pieces of the house, and these latter parts of the house.
Organisation is what distinguishes a house from a set of bricks. It may be informally defined in the following way. An atom here refers to an epistemological or cognitive atom—something we cannot, or do not want to, inspect or subdivide.
Definition 1.
An is one of the following:
- an atom;
- a set of organisations;
- a group of organisations put somehow in relation to one another;
- nothing else.
This definition purposefully lacks a better clarification of the expression somehow in relation with one another, since this may be instantiated in several ways. Notwithstanding, it includes as organisations sets of atoms, organisations, or both. In the case of the above example, for instance, bricks are atoms. Even if one is taken apart in the building process, we do not want to describe or analyse of what and how they are made of, independently of the relevance of their attributes. Nevertheless, we may consider them associated in many ways. A brick may be associated to another if they are in contact, sharing a face or a portion of a face, if they belong to the same wall, or yet to a specified region of a wall.
This idea of organisation may be applied even to the heap itself. A brick may be associated to another brick if they are close enough together and groups of bricks may be considered separately. The ones at the top of the heap or at its base, for instance. In this case, however, shaking the heap a little could provoke a complete upheaval in this organisation. In terms of organisation, the heap is not as stable as a house, although the heap is pretty stable as a set.
Note that this concept of organisation can distinguish facts which are difficult to be traced by current descriptions.
The two houses depicted in Figure 1 differ only by the placement of a door in two distinct walls which are in contact. The number of bricks that occupy the volumes of the doors is the same, assuming that they have the same form and size. Moreover, their relative positions with respect to the walls of the house are very close to each other. Differences like these are difficult to be distinguished by either dynamical or statistical approaches. The energy and (statistical) information required to build either of the houses is the same. Moreover, the same set of bricks may have gone to one or another wall to erase the would-be door. The trajectories undergone by each brick from the heap to the walls can be traced by dynamical systems. Small perturbations in their dynamics led them from the same initial condition, the bricks heap, to different final conditions, the two houses.
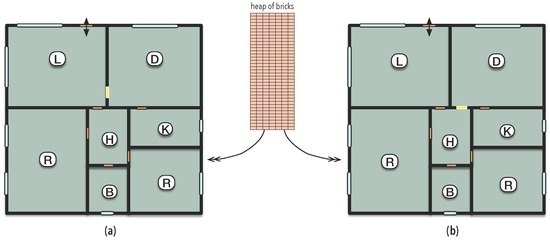
Figure 1.
Two distinct houses obtained from exactly the same bricks. Due to a minimal position change, the yellow door connects the living and dining rooms in (a) and the hall and the dining room in (b).
Although similar, the organisation of both houses, given by the connectivity between rooms, is different; making one better suited for certain purposes than the other. For instance, the house in Figure 1a could be more easily used as a restaurant than the other one due to the accessibility of the kitchen from the dining room and of the dining room from the entrance via the living room, what clearly appears in the diagrams of Figure 2. Furthermore, the living-room may also be used to attend and direct costumers of the restaurant.
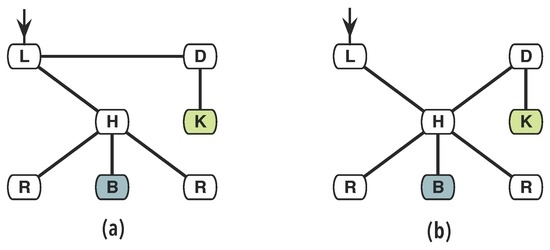
Figure 2.
The ground access organisation of the two houses in Figure 1, respectively graphs (a,b). The new position of the door substantially changed their basic organisation.
3.1.2. A Mathematical Model for Organisations
Molecules, the ground stuff of living entities and processes [39], possess the characteristics of Weaver’s ‘organised complexity’ class. Simple molecules may be understood through a small number of parameters and variables but not proteins nor any of the large biochemical molecules intervening in life phenomena. Protein molecules act as wholes when catalysing reactions, regulating biochemical processes, or chaperoning protein-folding although their parts are exchanged in protein construction and degradation processes. Spatial and chemical interrelations are crucial non-quantitative aspects of molecules. Protein dynamics (Vinson, 2009; Blanchoin et al., 2014) describe conformal changes in molecules that affect how a molecule interacts with other molecules or respond to stimuli from its environment.
Molecules are one of the simplest instances of organisation in Nature and fully conform to Definition 1, where chemical atoms are organisational atoms while chemical bonds instantiate the property ‘in relation to one another’, as hinted in Figure 3. Mathematically, molecules are depicted as graphs since long [92,93]; where nodes stand for chemical atoms and arcs for chemical bonds. Notwithstanding, we often describe molecules not by their atoms but in terms of other molecules, atom-groups or ions, to highlight chemical and structural properties, the way they interact chemically, or how they fit in a context. We refer to the hydroxyl group in alcohols, the carboxyl group in amino and other carboxylic acids, amino-acids in proteins and so on, treating them as units. This mode of describing molecules (Figure 3) is the essence of graphs (-graphs), the model for organisations described below.
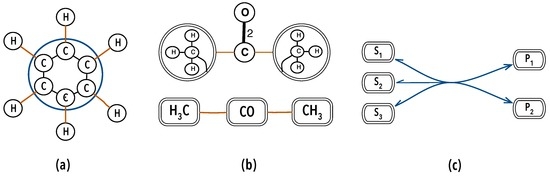
Figure 3.
Molecular hyper-graphs: (a) the benzene ring (nodes, arcs, hyper-edge); (b) acetone (hyper-nodes, arcs); (c) a (bio)chemical reaction (hyper-nodes, 1 hyper-edge).
To make these terms more precise, we need to restrain what may be considered as organisational atoms in modelling phenomena. Atoms in -graphs are required to be elements of a finite admissible set [94], , established in advance. Admissible sets allow for treating certain sets as atomic organisations more conveniently [95]. Sets in are not meant to be ‘inspected’ and differ from sets constructed by clause 2 of Definition 1. For instance, a brick as a set of clay particles is an element of and as single element (atomic) organisation it is a member of the set ‘heap of bricks’, a trivial organisation under the definition that is being introduced here.
The set need not to be the same in different representations, unless the organisations being represented interact or are related. may also be typed; that is, its elements may be of different types, as when considering chemical substances like carbon, hydrogen, oxygen, nitrogen, etc. There is no restriction on the number of elements in , or of one of its types. For instance, may contain all (chemical) atoms in the Universe, letters of an alphabet, parts of a car, modules [96] and organelles of a cell etc. One may consider also an consisting of both letters and chemical atoms if this helps understanding the phenomena in hand.
As hinted in Figure 3 and Figure 4b, associations will be represented by hyper-graphs [97,98]. Arcs of graphs can connect at most two nodes. Edges of hyper-graphs may connect any number of nodes. The choice for hyper-graphs is justified for two reasons at least. First, it provides a simple and uniform description of delocalised bonds, or electron bonds embracing more than two atoms. For instance, the benzene ring is representable as a hyper-graph with 12 nodes , 6 carbons and 6 hydrogens and 13 edges: six binding hydrogens to carbons , six binding subsequent carbons , where , in the ring and one connecting all six carbon atoms , as shown in Figure 3a.
Figure 4.
(a) The same hyper-graph drawn: as a Venn diagram (); with type I Hyper-edges (); with type II Hyper-edges (); as a bipartite graph (); (b) the benzene ring (Figure 3a) shown as a bipartite graph. Its strict sense hyper-edge is shown in blue.
Second, chemical reactions are typical hyper-edge candidates, since they in general transform several substrates into several products. A minimal reaction takes 2 substrates into one product or vice-versa (Figure 3c) due to mass conservation, unless conformal changes occur during chemical reactions. The enchainment of biochemical reactions results in networks that are indeed hyper-graphs [99].
Mathematically, a hyper-graph h is a pair of sets where N is the set of nodes and A the edge-set. The edge-set is a collection of subsets of N, that is, where denotes the power set of the set S, satisfying:
Hypergraphs generalize graphs, in the sense that graphs are hypergraphs which edges have just 2 incident nodes, that is: [97]. To establish the framework this and all other definitions will be as generic and encompassing as possible. Restrictions, if any, shall be imposed in their instantiations at each phenomenon being modelled.
Hyper-graphs can be depicted with Venn diagrams, with hyper-edges, or as bi-partite graphs , as indicated in Figure 4. Bi-partite graphs have two groups of nodes and arcs connect nodes from one group to nodes of the other. When expressing hyper-graphs, one group of nodes is the hyper-graph node-set and the other the names of the hyper-edges.
To handle wholes and parts and the recursion of Definition 1, we will need some meta-elements besides and (hyper)graphs: an enumerable set of meta-variables and a collection of special undistinguished elements . Meta-variables represent voids: places where organisations may grow, associate with other organisations, or detail their representation. For instance, docking sites in proteins, ion-binding sites, or polymerisation sites at the tip of filopodia [36] should be voids. Elements are unreachable constructive elements. They are hidden-names or ‘hooks’ of organisations that may become part of organisations with voids. Naming an organisation transforms it into a whole. Wholes being unreachable allows for swapping indistinguishable sub-organisations while preserving the overall organisation. Like when replacing bricks in a wall or when repairing DNA, proteins and organelles.
Hence, nodes will now be taken from the extended universal set . They may be organisational atoms, meta-variables (voids) or the element (hook). Hyper-graphs with nodes from will be called extended hyper-graphs. Any hyper-graph may become extended by adding at least a meta-variable or a hook to it, as in Figure 5. The collection of all hyper-graphs will be denoted by , while the collection of extended hyper-graphs by . That is,
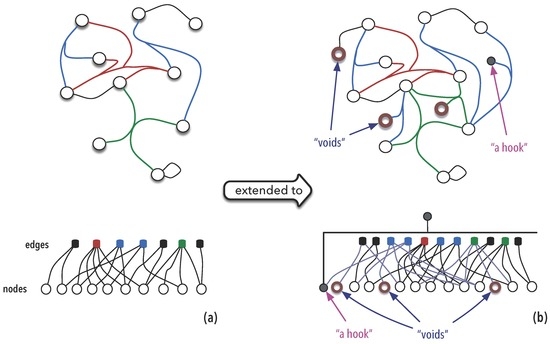
Figure 5.
Extending hyper-graphs: the hyper-graph shown in (a) both with hyper-edges and as a bipartite graph is extended (b) by adding a ‘hook’ and some meta-variables (voids).
Clearly, .
Whole-part graphs are constructed by binding extended hyper-graphs with hooks or hidden-names to voids. This is achieved by “assigning” the generic element of a hyper-graph to a meta-variable (void) in another hyper-graph . This construction starts from the following operator prototype (see Figure 6):
where denotes the class of all extended hyper-graphs that have meta-variables as nodes, denotes the class of all extended hyper-graphs that have the special meta-element as a node, and ni is the mirror writing of , a predicate identifying an element as a node of h.
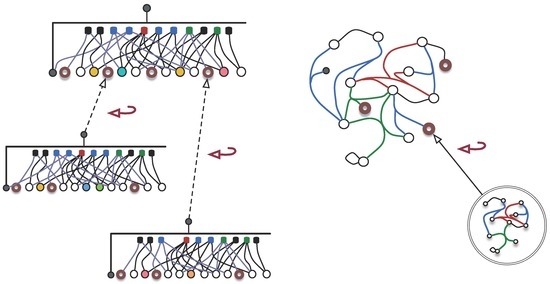
Figure 6.
Binding extended hyper-graphs: extended hyper-graphs become “nodes” of extended hyper-graphs.
An extended hyper-graph has just one element as node (one hidden-name) but may have many meta-variables as nodes. Moreover, if several hyper-graphs are to be bound to , there is in principle no special reason to privilege any binding order. Thus, the binding operator should be taken as a collateral binding of hyper-graphs to :
where m is the number of meta-variables in , is the number of extended hyper-graphs to be bound to . Without loss of generality, it is supposed that binds to .
Using the representation of hyper-graphs as bipartite graphs [98,100], binding hyper-graphs may be depicted pictorially, as in Figure 6.
The binding operator has two possible interpretations suggested by common usage: as an ‘encapsulator’, enclosing organisations as elements of another organisation, or as ‘hierarchy constructors’ (Figure 7).
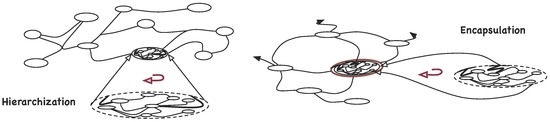
Figure 7.
Binding Operator: as a hierarchy constructor and as an encapsulator.
To complete the construction of -graphs observe that, while there are still unbound meta-variables as nodes of any hyper-graph in the assemblage resulting from binding extended hyper-graphs to one another, it is possible to continue binding other hyper-graphs to them. These assemblages are the -graphs and are formalised by the following recursive definition:
Definition 2.
An object γ is a -graph, that is, if and only if:
- 1.
- ,
- 2.
- ,
- 3.
- nothing else.
The fact that and a fixed point theorem for structures [101] warrant that Γ is non-void and well defined [95]. Note that Definition 2 makes no reference to sets nor atoms. The following lemma shows that Definition 2 indeed consider both, taking into account that atoms in Γ should be elements of . For the arguments it will be convenient to denote as the collection of all subsets of S with exactly k elements, that is, .
Lemma 1.
Γ has the following properties:
- 1.
- 2.
Proof.
(Ideas used in proofing case (1) intersperse the reasoning in (2).)
- Note that since then . Moreover, can be identified with and with through the following injectionand the immersionshows that .
- If , then . Let be a set of k meta-variables and let be identified to by the procedure in step 1. Thenis an element of Γ.
Moreover, and are identified by arguments analogous to those of step 1. ☐
The possibility of binding indefinitely new -graphs to meta-variables and the extensibility of any means that -graphs may grow forever and that adding details is unbounded in principle. Figure 8 displays typical elements (points) of Γ.
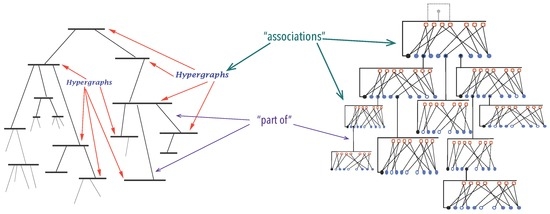
Figure 8.
Typical -graph: localisation of associations and the part of relations in elements of Γ.
3.1.3. Synexions
The elements of can be a variety of things: letters, chemical atoms, molecules, forms, objects, concepts, ideas etc. Whole-part graphs from Definition 2 may represent organisations whose atoms may be concrete, abstract or imaginary: words and paragraphs, molecules and macro-molecules, cultural entities, sketches, cognitive maps, pictures, mythological beings, ideas, etc. Otherwise, synexions are meant to represent organisations having existence reflected in terms of physical aspects. Straightforwardly, if a whole-part graph represents a molecule, a corresponding synexion would represent it with all volumes, angles, energy and vibrations possessed or defined by its parts and atoms. This intuition will guide the initial presentation of the concept. Nevertheless, it will be seen at the end of this section that usual dynamical systems are instances of synexions.
It is worth noting that the same object may be represented by way of different organisations, different elements of Γ (see Section 4.2), depending on what should be distinguished and on the questions addressed. For instance, the common organisation of a text is grounded on sequences of letters, words, phrases, sentences, paragraphs, sections etc. This organisation is fine for reading a text. However, if we are interested in mining key ideas in the text and associations among them, a better organisation could be as a cognitive map or a hypertext. The cell is another example. Its organisation may be based on its compartments and topological space-time relations among them or as a superposition of several bio-chemical networks grounded exclusively on chemical affinity. Molecules provide outstanding examples about organising an object in different ways, by grouping its atoms differently (Figure 3b).
The same subject of study may be seen as different organisations concurrently. Emotions may be concomitantly abstract and concrete—abstract while described in words or concrete if we consider the bio-chemical alterations and oscillations entailed with them. The organisations that may be associated with these two aspects of emotions are profoundly different.
Molecules, and all concrete organisations, occupy volumes in space and recognition of their organisation is moulded by our perception of spatial arrangements, dynamical stabilities-instabilities, and functional dependencies. Concrete organisations also vibrate and oscillate around a stable state, which means that they have extension in time as well as in space, being cylinders in space-time. Moreover, fluctuations in physico-chemical attributes may prevent, hamper, or facilitate the existence of certain organisations in favour of others (see Section 4.1.2). Therefore, we need means to represent organisations “embodied” in physical spaces. This will be achieved by associating volumes with -graphs in a way that preserves whole-part relations, as suggested in Figure 9.
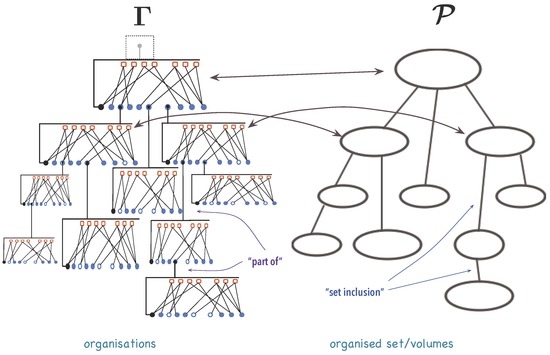
Figure 9.
Synexions or Organised Sets (the relation is defined in Section 3.1.4).
The results of these associations are the (This term synthesises the key features of organised volumes. It is formed from the Greek verb , which means to hold or maintain as a whole, and the particle , that means occurrence, instance. An alternative terminology could be syntheions, from , that means composed by the union of parts, and , see [41,102].) or organised volumes. Synexions are associations between a and volumes or subsets of a “physical” space where set inclusion preserve whole-part relations. That is, if is a sub-organisation of then the volume associated with is contained in the volume associated with .
Mathematically, let be a collection of physical attributes including space and time that is endowed with the classical space-time structure [103]. That is, a space where molecules or any physical structure will not suffer conformal deformations when rotated or displaced. Synexions, denoted by , are recursive associations between volumes and sub-organisations of such that
where .
Note that is a one-to-many relation. To each , there are many families of subsets of that may be associated with it satisfying the constraint enforced by Equation (9). Any family of sets associated with γ may be uniformly expanded (fattened) or contracted (shrank) and still satisfy constraint (9), for instance, by changing temperature (vibration of parts), or by coherently displacing and deforming them in space-time, while preserving γ. The synexion space, , is the class of all associations between and subsets of , for any . That is,
Synexions are not sets in the usual sense. Subsets of must also conform to conditions established by (9) and be formed from subsets of , where β is a part of γ. Thus, we may have as organised volumes, even though as usual subsets of .
This property of synexions allows them to represent movements and deformations of organised things more effectively because it is possible to impose a kinematic behaviour to points in that is constrained by Equation (9) to conform to the whole-part relation inherent to γ. This kinematic behaviour selectively changes and moves the volumes associated with parts of γ while preserving its organisational identity. That is, if are two parts of γ, . Moreover, if then and, conversely, if then . Otherwise, . Cell-motion [45,104] is a good example of this feature, since organelles and cell-parts deform and move with the cell and within the cell without intercepting themselves, nor destroying the inner organisation of the cell. Synexions support the disentanglement of physical from organisational changes, which has far reaching consequences. They provide a bridge between organisation and (usual) physico-chemical dynamics. They enforce dynamical coherence: characteristic times and distances of parts are smaller than those of wholes. A cell cannot undergo mitosis before all its parts are duplicated, including the nucleus and the outer membrane [14].
Remark 1 (Elements of ).
This rather informal and rigourless note presents some of the simplest elements of . The association between dynamical systems and interaction graphs is discussed in detail and with the due rigour in [26]. As discussed in this work, any dynamical system
can be associated with a graph , the set of all graphs with n nodes. Since for any atom-set containing the names of variables in the dynamical system given by (11), . Furthermore, the collection of orbits of a dynamical system of the class (11) is a subset of whenever the dynamical system represents a natural phenomenon. The specific nature of is intimately dependent on but is often a variety or a set of chronicles [8], and is tightly related to properties of [26].
From another stand, the mapping that associates a dynamical system with its interaction graph is not injective and there are many such that . Then
is an element of , since and , satisfying the first case of Equation (9). Clearly, also belongs to , for any dynamical system given by (11). That is, interaction graphs are (simple) organisations and the orbits of dynamical systems associated with it are -volumes in the sense considered above.
If depends on a parameter τ, is an “orbit” in and exemplifies transformations of synexions. However, parameter dependency in dynamical systems do not in general affect its dimension n. When it does, it does in general by reducing the value of n while trapping orbits into sub-varieties of . Generic transformations are not subject to this constrain and systems of variable structure [4,5,6,7,66,91] can be straightforwardly represented with the concourse of transformations in .
3.1.4. Further Basic Constructions
Spaces Γ and have interesting properties. Synexions are material organisations but not the only “concrete” organisations. Special organisations like sets, lists, trees, S-expressions and other data-structures can be identified as sub-classes of Γ [105]. Transformations, operations and relations can be defined in Γ and . Albeit a proper discussion about mathematical operations, predicates and properties of Γ and being outside the scope of this writing, developing a few of them here will better illustrate the -graph and synexion constructs. Those below are restricted to Γ and support arguments in some of the following sections. Analogous transformations can be defined in . Two of them are illustrated in Figure 10.
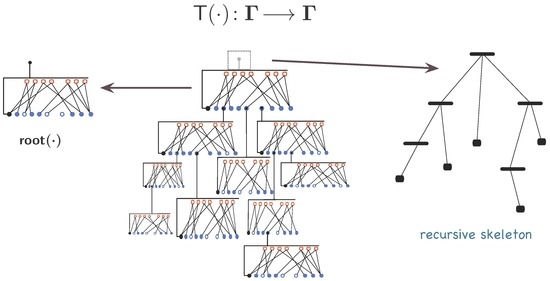
Figure 10.
Transformations in Γ: the and the recursive skeleton of γ.
Definition 2 implicitly defines a mapping in Γ, (see Figure 10). The operator is properly defined as:
To define equality, observe first that if , or , in the set-theoretical sense, that is, and as sets.
Definition 3 ((Equality)).
Two -graphs are equal () :
- 1.
- and , as elements of ;
- 2.
- and
- ,
- ,
Note that, following Definition 2, means that and . Furthermore, implies that , , and as elements of .
An important thing to remark about Definition 2 is that it induces a whole-part relationship in Γ, which is highlighted by the predicate , where the token should be taken as a single stropped (mathematical) symbol denoting this predicate. This naming method, common in the theory of programming languages, shall be used for creating symbols suggestive of their semantics for uncommon mathematical entities defined in Γ and . The relation is recursively defined as:
Definition 4.
The values of for are given by:
- 1.
- If , then , as elements of .
- 2.
- If then if either condition holds:
- or
- .
- 3.
- Else, .
Hierarchy is one of the most conspicuous characteristics of organisation. It was intentionally left out while developing Definition 1 and its mathematical model (Definition 2). Nevertheless, as suggested in Figure 8, Figure 9 and Figure 10, hierarchy is an intrinsic characteristic of any -graph induced by its recursive construction and attached to the whole-part relationship inherent in Γ -elements. The operator , where is the class of recursive trees in Γ [105], points it out. To define it, let be the set of all meta-variables (meta-nodes) used to construct γ (Definition 2). It is important to note that meta-variables in and in any , such that , are different even thought the notation may be duplicated to make the reading more intuitive.
Definition 5.
The recursive skeleton function of a -graph () is given by:
- 1.
- If , , where and ;
- 2.
- if , then
- 3.
- nothing else.
That is, is a replica of γ where all nodes from and related associations have been erased.
The last Γ-transformation to be introduced here is the connection network. It is in a certain sense a counterpart for , since it erases the meta-elements from γ leaving only nodes from and the associations relative to them. This is achieved by substituting the binding , , defined in Equation (4), by a normal node mimicking this hierarchical connection. For the sake of simplicity, the definition presented here will be restricted to organisations which hierarchies have at most two levels, that is organisations such that is a tree of just one or two levels, by omitting the final recursion step. If in Definition 2, there are in meta-variables that are not bound to any . Likewise for any , such that . Let denote respectively the set of all meta-variables, unbounded meta-variables and bounded meta-variables in γ. For , and, in general, . Furthermore, let denote the set and the set of all .
Definition 6.
The values of the connective network function, , under the restrictions above, are given by:
- 1.
- If , then , where
- 2.
- If , , , then , whereand the latter are modifications of and given, respectively, by:
is non-injective. A rough idea of the set can be obtained for organisations with a two-level hierarchy through the following reasoning. A given hyper-graph represent the channels of possible interactions in a phenomenon. Any hyper-graph (network) can be organised in several ways by partitioning the network into sub-networks and encapsulating these as aggregate units of interaction. Network-partitions interact with other nodes as composite nodes by means of collective aspects (like temperature or pressure) and channels of interaction encompassing all possible interactions of encapsulated nodes with nodes “external” to it, i.e., nodes in other partitions. Network partitions are obtained by partitioning their node-sets and rearranging its arcs accordingly. This stand will be used in Section 4.2 to estimate the size of .
Lastly, the following observation will be supportive of several examples and arguments. A process is a collection of ‘states’ (enchainments or entailments of natural events) that cohere the enchainments with ordered temporal moments when instantiated into a physical space [106]. When life phenomena is described in , life processes are a collection of enchainments in with a condition about their immersion in the time axis, e.g., biochemical pathways. Hence, life processes naturally include actual organisations as part of their states. That is, spaces Γ and allow for considering and handling organisations of processes which states can contain organisations as components [3]. This is of great relevance for biological phenomena since chemical processes, as entailments of chemical reactions and substrates, are processes in this sense. Organisations of (bio-)chemical processes arise by considering two chemical processes to be associated whenever they exchange substrates or influence each other. Processes can be considered as wholes whenever they present homeostasis, or any other form of permanence or recurrence, and have a distinctive functional character.
3.2.
Organisations convey information. This is clear when organisations are texts or pictures and their information-content is conveyed to humans. But even microorganisms have memory, process information, anticipate, coordinate tasks and make decisions [33,107,108,109]. Thus, information-driven interactions are a distinctive feature of biological systems at all scales, as discussed in [79,80], from distinct standpoints. They provoke changes in internal organisations and behaviour of a thing, that is, they provoke .
Existing information concepts focus on transmission of information, having a statistical nature and requiring a large enough set of messages, known in advance. Therefore, their application to scales or contexts where the number of intervening “things” and factors is at most moderate and far from homogeneous or isotropic is delicate, ad hoc, and can only provide hints about the phenomenon propensities. The widespread employ of the term information has introduced overload and bias in its meaning since long (see [30], footnote 1 on p. 194) and its utilisation is often inadequate [51].
From a basal point of view, information need not be quantifiable [80,110,111], despite the usefulness of measures in comparing things and in describing natural phenomena. A novel, non-quantitative, concept of is introduced in the sequel. It can be employed at all (biological) scales [2] in an integrated manner and is related to how organisations change. Being grounded on synexions, it is ontologically bounded to biological interpreters and observers. In this sense, is closer to molecular processes and changes at the molecular scale that instantiate signal processing, memory, reactiveness and anticipation in cells and tissues; and covers most relevant aspects of information in biological phenomena. It also accommodates information exchange at the sub- and supra-cellular levels, being useable in other domains as well. Exchanged in (biological) interactions is retrospectively observed and identified thanks to changes in organisations and behaviour.
Information in the sense to be presented is not a measure, measurable, or quantifiable. It targets the etymological roots of the word information: in-formare, or “to form inside.” From an organisational stand, it addresses information at Level B (meaning) and C (effectiveness) of Shannon’s Communication Theory more directly than at the commonly addressed Level A (transmission) ([112] chapter 1). Moreover, transmission of does not require identifiable senders nor a fixed number of messages. Nor is it constrained by the pre-definition of a set of signals and messages. Notwithstanding, usual measures of information-transmission can be recast from the concept below once there is a sender besides a receiver and the set of messages can be determined a priori. In the sequel it will be assumed that any biological entity is represented as a ‘synexion’ or organised set (volume), , for a properly chosen atom set and .
3.2.1. Perceptions
The concept of to be presented is grounded on an ontological hypothesis. Namely, that all biological entities ‘perceive’ and, by extension, so do organisations that represent them. The purpose of this section is to clarify the use of this term since it has here a rather specific meaning.
Perceptions are strongly intertwined with signals. Biological entities often have a living boundary that filters and transduces incoming signals. Let us call this kind of boundary skin. In individual organisms, ‘skins’ occupy a connected physical region and are part of its organisation, dividing the world in two regions: inside and outside the entity [8,113]. They also help the maintenance of particular homeostatic internal conditions. The “outside” region immediately around, together with anything it contains that may interact with the entity itself, is the entity’s environment. Cell membranes are straightforward examples of ‘skins’. Notwithstanding, there are organisations inside cells, in cellular matrices, in the mucosa-epidermis complex of multi-cellular organisms and in collective entities that are ‘skins’ in the above sense and not easily recognised as such.
Definition 7.
A is any perturbation (sudden variation) in the environment conditions, concentrated in time and space, that propagates eventually encountering a biological entity or another appropriate receiver.
Encounters have the usual meaning of two or more things being at the same neighbourhood in space at the same moment. This definition includes as signals: travelling molecules or bodies, local variations in pressure, temperature and concentrations, waves etc. Due to space-time continuity, whenever a signal encounters an entity it reaches its outer boundary (skin) first.
In biological and ‘organised complexity’ phenomena, signals are expected to provoke drastic and disruptive changes in the structure of the systems involved, since this is how their organisations are altered; changes that eventually rend them them unrecognisable. Indeed, biological systems are signal-amplifiers par excellence [74]. Perceptions are effects that a signal has upon the biological organisation or organised receiver it encounters.
Definition 8.
is a two stage process: it has an imprint moment and a recall moment:
- ▸
- Any signal encountering sensory apparatuses (in the skin) of a biological entity at time and transmitted into it provokes (localised) changes in its organisation.
- ▸
- Moreover, if another signal encounters the same biological entity at time and tends to provoke the same change in the organisation of as signal , is recognised as being the same signal as .
Hence, perception is an action rather than an entity, organisation or fact and results in imprints (see Figure 11). Imprints, that are organisational alterations, may decay over a short time, remain for longer periods, or become part of the organisation. Signals and need not be exactly the same perturbations of the environment, taking all attributes into account. With respect to the perception process of a synexion , however, and will be considered to be the “same” signal, whenever they provoke the same imprint. This is dependent on the complexity of and of the signals in the imprint class associated with the same alteration in .

Figure 11.
Perception: imprint (a), recall (b).
This sharpens the idea of similarity of signals (see Remark 2). Mistaking strictly different signals related to a sole imprint as the same signal is part of the perception process. Therefore, we say that a long lasting imprint is a model for and its similarity class.
3.2.2. and Interpretation
Imprints are changes in the organisation of a synexion but do not enforce alterations in behaviour. Modifications in may affect only the associated volumes , only its organisation , or both . Generally, signals provoke initial changes in volumes (physico-chemical processes) that eventually migrate to changes in its organisation . In brains, the first relate to the electro-magnetic activity and are likely to decay; while the second and third involve synaptic and organisational changes and are long lasting. Depending on the level of detail, changes in organisation imply changes in the organised volumes as well. Good examples of this are the cellular signalling system and the sensory-nervous-brain systems in multi-cellular organisms (see Section 4.1.1 and Section 4.1.2). Imprints that do not provoke changes cannot be detected.
Imprints that change the behaviour of biological entities will be called . The definition below employs observers, that are biological entities themselves. The role of observers, notwithstanding, is primarily to acknowledge that some change has happened. They are needed to detect and compare changes. Thus, any artefact that make special observations and compare them is an observer. Their role will be greatly clarified while refining our understanding of what is .
The definition of below relies on the following ontological hypothesis that is suggested by living phenomena and entities:
Hypothesis 1.
- A.
- Any biological entity or process may be represented in .
- B.
- All biological and life-related entities or processes .
- C.
- Perceptions are unique for a given biological entity or process—same signal, same imprint;
- D.
- Biological organisations emit signals that uniquely characterise them, that is, they may be recognised by means of the signals they emit.
Hypothesis 1C and 1D are not strictly necessary to define . They are relevant though while considering information-exchange in biological interactions, for rendering as a usable concept, and for clearly understanding its biological consequences.
Hypothesis 1A is the kernel of the organisation perspective. Notwithstanding, the representation of biological elements and processes as synexions is not unique, nor coerced in any manner. Any biological entity may be represented by synexions and reflecting different organisations, levels of detail or perspectives of study. Changing the synexions that retract enriches our perspective in the same way as seeing matter distinctly as a cloud of particles, a body, or a substance does. Proteins may be seen as two organisations at least: a sequence of amino-acids while studying folding or as an assemblage of secondary domains and docks while studying function. Even so, one representation as a synexion is enough to discuss about exchange and interpretation of signals.
The imprint, of a signal in may be different from its imprint in synexions or , even when all these synexions retract the same entity . In consequence, the similarity classes of may be distinct in each representation of a biological entity as a synexion. This multiplicity accommodates the representation of different levels of detail and different dynamical states. Our ability to understand information-driven interactions will depend on how coherently organisational changes ascribed to perceptions are represented and this can only be solved by reference to the signal itself or its source. The organisation framework makes this subtle point explicit but there will always exist a compromise between the complexity of synexions and their reliability as representations of things in natural phenomena.
Furthermore, it is well accepted nowadays that cells perceive and remember [107,108]. There is a clear association between conditions in the environment and activation-deactivation patterns of biochemical switches in the cell nucleus, maintained by the signalling cell system [15,114]. Hypothesis 1B, however, goes beyond that extending the perception concept down towards the sub-cellular scale and above towards the non-organismic entities and collectivities scale. The synexions framework accommodates other than that processed by neural systems or DNA transcription and intergeneration transmission, that are the only forms of biological information generally considered until recently [80].
Before advancing further, it is worth making the following observations:
- A signal reaching two different biological entities and , represented by and may provoke different imprints in their organisations, even if , and , are similar. However, if the collection of signals associated with imprints and is the same, that is, if any signal leading to the first imprint will also lead to the second imprint, the perception should be considered same for and ;
- Stabilised imprints are models for signals or collections of signals;
- A travelling molecule is a signal, because it is a localised variation in density, mass and other aspects concentrated in time and space;
- Pressure and concentration variations, being more diffuse and collective perturbations of environment attributes, may not be always taken as signals. This suggests that environmental variations depend on scale sensitivities as well as the complexity of the perturbation and the perceiver to be considered as signals. Signal and perception are thus relative concepts;
- Encountering is always due to relative motion. Either the signal propagates or the organism is moving and reaches a resting obstacle that acts as a perturbation in the perceived environment. What is important is that signal and organism approach each other in space and time for an encounter to occur.
Besides signals and perception, two or more special biological entities will be required to specify . One shall be termed interpreter, the others will be observers.
Definition 9.
Given a signal and at least two biological entities and in , will be termed an if the following occurs conjointly:
That is, if perceives changes in , after its encounter and interaction with .
Thence, says that is an for and that has interpreted . The observer acknowledges changes and the interpretation of by . The observer is not needed otherwise and may be the observer itself if it is complex enough to perceive its own perceptions, that is, create a model for them, and maintain a model of itself. Since is in , so are its perceptions (models) of and . Both perceptions are sub-synexions of and have extensions in time as much as it does. The perceptions of and , though, need to extend beyond for be able to detect differences between its anticipation of at and its new perception of at .
Figure 12 portrays the information-interpretation arrangement, highlighting observers (named in the figure), models of and , and its dependence in time. The fact that causes changes in organisations is crucial for the organisation perspective (see Section 5), that also relies on the following premiss:
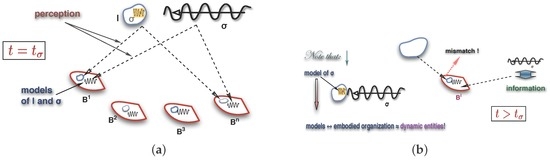
Figure 12.
: Observing a signal perception (a); recognizing as (b).
Hypothesis 2.
Every whole is an interpreter of its parts.
It should be noted though that Definition 9 does not, strictly speaking, describe information. Instead, it describes how to recognise that a signal is —through changes provoked by the signal in an organisation whose behaviour changes with respect to what was anticipated [8,115] with respect to it—suggesting a procedure to observe it.
This is somewhat analogous to energy which is a property attached to configurations in fields and can only be observed indirectly, through its effects in the components of a -phenomenon. , likewise, is essentially immaterial and intangible and can only be acknowledged through its effects in 4-dimensional (i.e., extending in ) -organisations.
4. Ontological Considerations
Subsections of this section contain arguments and discussions relative to employing the organisation-information theoretical framework to understand natural phenomena, as well as justifications for modelling decisions inspired by their observation as a collection of interacting organisations.
4.1. Space, Time and
The definition of provided above requires the immersion of organisations in space-time, and makes explicit use of space-time events and models immersed in space-time. In the sequel, arguments supporting the dependence of on space, time and are presented at several scales and domains. The examples below are simple and far from extensive. Their aim is to justify modelling decisions and clarify the constructs. Recently, though, a wealth of biological and biomedical scientific investigations provided many examples of the spatial and temporal nature and dependence of living components and phenomena (see, for instance, the last two sections of [51]).
4.1.1. Cognitive Domain
Talking about issues of cognition and models starts with humans; bringing the analysis closer to our usual sense of information. Cognition, seen as the acquisition and incorporation of and perceptions in the sense of previous definitions, is not restricted to humans. In multi-cellular organisms, a part of their organisation is specialised as a signal processing system. It also handles imprints resulting from perception process and any response or reaction to them. Complex signals, coming out of complex organisations, are processed by the sensorial-signal systems. Imprints are mostly associated with the brain and nervous system in multi-cellular organisms, although not being restricted to them. In cells, an elaborated and complex network of reactions centred around DNA, the chromatin, and nucleotides adapt and respond to variegated signals, recording them and changing gene-expression as well as behaviour [11,15,108,116]. The examples in this section nevertheless refer to human cognition.
Signals are apparently affected by the organisation of their sources. The organisation of sources seems to be reflected in imprints associated with signals emanating from them, at least partially. In the following discussion, association of parts is mainly given by topological proximity (neighbourhood), while the whole-part hierarchy by encapsulation of groups into unities.
In texts, that are sequential objects, the vicinity is given by collaterality, grouping letters into words. Words side by side form phrases. Phrases side by side, with punctuation marks interspersed, form sentences, and so on. However, parenthesis, notes and footnotes relief a bit texts from dependence on sequential vicinity, by enclosing their contents into units. Technical texts have other forms of encapsulating and naming text unities that may be referred later or earlier in the text, thus indirectly re-appearing at several points of the text.
The same coalescent mechanism appears in figures and pictures, although their inherent two or three dimensions (2D or 3D) make the idea of neighbourhood much richer from a practical point of view. To exemplify how neighbouring associate things, let’s consider two simple geometrical objects, here taken as wholes and as logical atoms in the universe : a circle (◯) and a line segment (∣). The circle can be rotated at will without appearing to be modified but the line segment will present different inclinations if rotated. Bringing the circle and line segment together (see Figure 13) in different manners will reveal important characteristics of information.

Figure 13.
Geometrical objects (a); joining them by contact into a whole (b).
The line segment may remain tangent to the circle after joining or may transect it and that the circumference of the circle may touch or cross the line segment at different distances form its centre. These different possibilities of contact will result in different tokens, symbols and signs that may convey different meanings, if they convey information at all. Each circle-glued-with-segment forms now a visual unit (Figure 14), a whole in the sense of both Γ and .
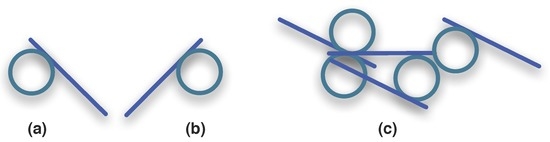
Figure 14.
Circle-line-segment visual unit (a); a mirrored circle-line-segment visual unit (b); and a collection of them (c).
Not all wholes convey information, but there are cases where a whole may not convey information, or convey a different information, due to the manner it relates to its surroundings. For instance, it may be difficult to recognise anything or associate a meaning to the circle-segment wholes as they appear in Figure 14. There is no easy clue in the visual units of images in Figure 14a and b or in those in the the heap of the image in Figure 14c that help us recognizing them. What, by Definition 8, means associating these units with previous imprints (known signals).
Considering Figure 14a and b together as unique image, it becomes possible to vaguely identify this new whole as a pair of (angry) eyes... if one has seen lots of animations. But what if we straight these units and line them up like in Figure 15?

Figure 15.
Letters.
Don’t they become immediately recognizable as letters {p,q,d,b}? Implicit subliminal visual references to the borders of the paper, that provide a sense of verticality, and from one whole to the other enforce their identification as letters. Anyhow, a group of letters like a syllable can be rotated to any degree and be severely distorted remaining recognizable, similarly to a group of chemical atoms forming a molecule. This observation and Hypothesis 2 suggest why molecules often have different functions in cells.
Relative distances between letters are strong topological clues enforcing the recognition of letter-groups as units or wholes, reducing the relevance of clues related to the environment. Similar phenomena occur in time when considering sounds and music. Groups of sounds or musical notes are more stable signals to our perception than scattered individual sounds or notes. It is more difficult to make known music or meaningful words unrecognizable than individual sounds or uncorrelated sound sequences.
Other visual units can be formed out of a circle and a line segment, as shown in Figure 16.
Figure 16.
Non-letter visual units out of a circle and line segment. See text for details.
Some will be easily recognised like those in Figure 16a–c; while others like Figure 16d will not, even if the sketches in (c) and (d) differ due to very small relative displacements of one part in relation to the other. Possible meanings for the resulting symbols are indicated by labels within the picture.
From another stand, there are visual units that defy interpretation no matter what is done with them, like the image of Figure 17a in two dimensions.
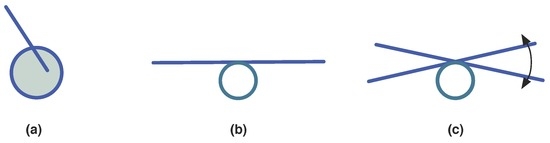
Figure 17.
Visual circle-segment wholes difficult to recognise. (a) non-sensical image; (b) purposeless thing; (c) swinging b. See text for details.
The image of Figure 17b may initially defy recognition. However, shaking or moving it a little bit, as indicated in Figure 17c will make them recognizable as the sketch of a see-saw. This recalls the importance of movement and the time-component in models and imprints, while recognizing many things and phenomena, particularly life-phenomena. Synexions are space-time objects and tubes resulting from the displacement of any spatially organised volume along time is a synexion. Hence, Definitions 8 and 9 contemplate cases where the recognition and interpretation of objects can only be made along time, while moving. The fact that certain changes are anticipated help the recognition of changes by observers in Definition 2.
Visual assemblages that form wholes and have been recognised may be associated in distinct forms resulting in new units or wholes, as suggested in Figure 18.
Figure 18.
More complex (made from previous units) visual wholes difficult to recognise. Images (a) and (b) are composed exclusively from units in Figure 14a–b, while (c) adds a rectangle-like visual unit.
This process may be carried out indefinitely resulting in visual units composed of other visual units which recognition helps the recognition of the whole unit. Abstracting from top-down or bottom-up stands, the explicit recursion in Definition 2 and Equation (9), inherited by Definitions 8 and 9, allows for the appearance of however complex organisations, signals, imprints and information. Cells present several space-time organisations that conform to these definitions, but their biological function, including molecules, motifs, or modules, may change depending on their localisation in the cell, their conformation and their motion.
4.1.2. Molecular Scale
Simple molecules are straightforwardly represented in Γ (Section 3.1.2) and (Section 3.1.3). Following the same procedure, proteins and other large molecules can also be represented in Γ and as huge, difficult to comprehend, plain hyper-graphs. Notwithstanding, using the now established hierarchical structure of proteins (see, for instance, [104] and the articles at http://www.proteinspotlight.org), proteins may be easily represented in Γ. Given that the basic constituent parts of a protein are amino-acids rather than atoms, a protein can be depicted as a sequence of the hyper-graphs representing amino-acids. Sequences of hyper-graphs belong to Γ [105]. Thus, proteins have at least two representations in Γ.
Yet, proteins are often depicted in terms of secondary structures and other familiar molecular components. Considering secondary and ancillary protein structures as nodes and the chemical bonds holding together these molecular structures as arcs, proteins may be represented Γ in several ways, where connectivity always reflects chemical bonds and hierarchy isolates identifiable protein domains and sub-units. The nodes of this hyper-graph are hyper-graphs themselves retracting (sub)molecules and by Definition 2 this assemblage belongs to Γ. Therefore, any assemblage of secondary protein structures is also straightforwardly modelled in Γ.
Their representation in is also immediate, since volumes occupied by atoms in space do not interpenetrate and their nearby positions unite the atom-volumes into molecule-volumes respecting constraint (9). Apart from that, protein components vibrate coherently, extending these volumes into space-time. The space-time volumes of the atoms of a protein and the partial union of them by amino-acids and subdomains provide the required volumes of letting synexions represent proteins. It is also clear from the previous discussion that any macro-molecule has many different representations , depending on which sub-units are being considered as wholes. Consequently, they also have multiple representations in . Anyway, protein folding is a transformation from Γ, or , into itself.
At any point in time, a protein may be activated or deactivated by a signal reaching it. Activation and deactivation are due to changes in protein organisation either in Γ, if new chemical bonds are formed or small molecules become attached to the protein, or just in , due to a re-arrangement in the tertiary structure caused by changes in the relative positions of its constituent parts. These organisational changes modify their “function”, that is, the manner a protein chemically behaves and reacts to external stimuli. Therefore, a perturbation in the environment, be it a travelling molecule or variations in distribution of energy or mass concentrations, may cause an alteration in the organisation of a protein and change its behaviour. Under Definitions 8 and 9, the (chemical) perturbation is a signal that provokes a re-organisation of the protein changing its chemical behaviour. Therefore, the perturbation is an for the protein.
An example of organisation alteration due sole to changes in space and not in Γ, is the pigmentation protein chameleonine. Chameleonine is the name given to the protein that change the colour of certain spots down the spine of Chamaeleo differensis individuals when temperature changes and is apparently present only on them [117]. Changes in environmental conditions, in this case temperature variations, provoke a change in the colour of the tissue where it resides. Temperature variations really change only the energy of its components that reside in the vibrations of the protein’s atoms. Chameleonine responds to this variation with a deformation in its spatial configuration due to structural stresses. As a result of this new (spatial) organisation, it changes behaviour reflecting, or re-emitting, a different light frequency. The signal in this case is temperature variation that, at the molecular scale, means a change in the mean number of molecules hitting a chameleonine molecule per time slice, and the consequent amplification of the vibration of its parts. This example conspicuously shows the need to include space and time in the conceptualisation of .
Chiral isomers further highlight spatial dependence of perception and information. This rich subject will not be discussed here, but a few observations illustrate important aspects of perception and information. Chiral isomers commonly respond equally to simple stimuli like temperature or to simple substrates in chemical reactions. However, depending on the complexity of molecules involved, on the environment where they are immersed or on the complexity of the signals reaching them, the effect in the organisation of wholes of which they are parts may be dramatically distinct, provoking deforming developmental diseases in humans, like the infamous thalidomide, particularly when chiral isomers are the signals [118,119]. Effective life phenomena is strongly dependent on essentially one of the chiral isomers.
From another stand, many biochemical organisations and structures in cells are not static. They are grounded on interacting homeostatic chemical processes, particularly when cycling. Hence, biological phenomena are really build on organisations of processes. A now canonic example of this character of theirs are the bacteria molecular motors. There are pictures of these ingenious engines available. However, these “structures” do not appear in engine snapshots at fixed moments in time. They arise from the superimposition of several snapshots at distinct times ([45] Figure 2), meaning that a flux of chemical substrates cooperate to give existence to these “motors”. In terms of organisations and synexions, notwithstanding, fluxes are naturally represented as process-organisations and may be considered either as observers or interpreters of signals that revert turning and “swimming” directions, for instance.
4.1.3. Cellular Scale
At the cellular scale, organisations are more conspicuous in eukaryotes than prokaryotes, despite the complex organisation of biochemical processes existing in the latter. The preferred reference to organisations in eukaryotes in the sequel, though, is just a matter of explanatory simplification.
Looking into the internal organisation of cells, there are organelles, special “tissues” like the endoplasmic reticulum and the variegated membranes, recurrent and stable molecular agglomerates, as well as a plethora of sustained biochemical processes that serve a variety of purposes in the cell. Among them we find transportation systems that using vacuoles protect substrates from reacting with chemicals existing in the cell before they reach a certain place or organelle in the cell. Signalling systems, extending from plasmatic to nucleus membranes, are responsible to transmit changes in the state of chemical switches in the cell membrane, that act as sensors, to the nucleus. There, the nucleotide-DNA-chromatin-nucleosome system retains the conditions of membrane-switches in the form of genetic inhibition-activation patterns [116] and more elaborated settings, like the CRIPSP-Cas systems, even alter DNA loci [108,120]. These nuclear reorganisations are imprints following Definition 8.
This travelling-wave system changes the switching status of a collection of molecules that surrounds the DNA and maintains some portions of the genes active while inhibiting others. Besides messenger RNA, that transports , there are also signalling pathways from the nucleus to the cytoplasm that activate or deactivate protein synthesis, that are determined by the inhibition-enhancing patterns of DNA sites. Therefore, it is not DNA that really controls cell activities but a complex formed by DNA and nucleotides that “switch” genes on and off. The messenger and transcriptions systems transmit these patterns of free DNA to appropriate sites in the cell. Also in this case, environmental perturbations that change the status of membrane sensors are signals as in Definition 7, while the DNA-nucleotide complex acts, in turn or concomitantly, as a memory (the organisation that concentrate imprints), a decider, or a transducer and may affect cell behaviour, that is, DNA-nucleotide-chromatin complex interprets (Definition 9) changes in the state of membrane-switches.
To see biotic-interactions as exchange of , we need to understand the effects of differences in time propagation between two wholes. In prokaryotes, transportation is due mainly to diffusion in heterogeneous media. Diffusion takes little time to bring molecules from one extreme of the cell to another, due to the small volume of the latter. Eukaryotes are about 10 times as lager as prokaryotes. To get equivalent transfer and reaction rates and characteristic times in larger cells, there must be some facility to accelerate and “direct” diffusion to the proper places. This “facility” is organisation. More precisely, dynamical organisations.
4.1.4. Physiology and Behaviour
Comments in this subsection refer to multi-cellular organisms. Toward larger scales, the organisation of multi-cellular organisms pretty resembles that of eukaryotes and prokaryotes from the right perspective. Following J.G. Miller [2], living systems are composed of nineteen systems, independently of scale. For instance, there is the nervous (electrical signaling) system, the immune (repair) system, the digestive system, the motor system, the boundary (environmental interface) system, the memory and learning systems. In spite of being analogous, they may however appear in completely distinct forms from one scale to another.
Anyhow, there are systems in larger organisms that are not as easily paired among scales as those enrolled above, like the endocrine system (chemical signaling?) or boundary systems in ecosystems and societies. Moreover, the categorisation in nineteen systems is somewhat arbitrary, since some of them may be considered as one single thing while others further subdivided. Also, focusing on the nervous and endocrine systems, we see that two signaling systems with remarkably different characteristic propagation times co-exist, one based on electro-chemical wave propagation and the other on diffusion and transport. Nonetheless, phenomena occurring within the limits of these systems are much richer and complex in multi-cellular organisms than in cells and, by extrapolation, in phenomena at larger scales.
In contrast to systems, multi-cellular and more complex organisms are better represented as a collection of superimposed organisations symbiotically cooperating by exchanging matter, energy and , particularly the latter. The characterisation of which organisations should be used in a representation will always be arbitrary to a certain extent, as in the case of systems. Finding (biologically) sound heuristics to support this task will greatly enhance our knowledge about phenomena involving living things. Clearly the macroscopic size of multi-cellular and more complex organisms impose a strong dependence on space and time for any exchange within and among the organisations representing living systems. Therefore, a mandatory guideline is the rendering of categorised organisations compatible in terms of characteristic times, volumes and frequencies while discovering organisations. Equation (9) proposes a possible guideline with respect to these concerns.
When we acknowledge exchange of information as a distinguishing characteristic of living matter, the necessity of this compatibility greatly promotes the use of space and time in the definition of .
4.1.5. Cultural Domain
Contrary to knowledge that may be individual, culture is a collective phenomenon. Notwithstanding, human purposeful collectivities (like enterprises, firms, industries, households, schools, communities etc), collective phenomena (like person-to-person contacts that propagate diseases, gossips, information and matter; or like cultural centers and cultural networks attracting and educating people) and human creations (like science, music, art and literature) are all founded on organisations and information as pictured in Section 3.1 and Section 3.2.
Purposeful human collectivities are formed by a collection of people that associate with each other with a (pre-)determined objective. Some organisations, like firms, industries, enterprises and (big) households, spontaneously or not organise themselves by having a hierarchy where smaller groups report to and/or are commanded by others. Such organisations are the basic action units in a cultural domain and have been extensively studied since long [121]. Most of them comply at least approximatively if not strictly to Definition 1.
Knowledge may be represented by classes of signals and their imprints organised as a consequence of associations and abstractions which are formed out of multiple interpretations and experiences, in the lines suggested by the discussion of Section 4.1.1. Culture also may be viewed as an organisation (in the sense of Definition 1) composed of concepts, ideas, taboos, individual knowledge, the imprints of a man or population etc. For instance, in a piece of music, art or literature there are often references, usually implicit, to another work in the same class or even in another class. There is music inspired by literature and folkloric wisdom, literature that refers to lyrics and so on. Literature is itself a web of veiled references to pieces of other literature.
In science, referencing is made explicitly; and this differentiates it from the other cultural expressions. Books, although sequentially arranged, may be as tortuous as any folding molecule due to backward and forward cross-referencing [8, Note to the reader]. In science, literature or music, one can only appreciate the beauty and depth of a piece of work if one has good acquaintance with a large number of other pieces of work, at least in the same cultural class or domain of discourse. Even within the explicit referential system of science, one has to know and understand the referred material to properly perceive and understand the work that referred to them.
Culture, however, differs from individual knowledge in two important ways. Instead of residing in the memory of a person, it is registered in books, writings, unspoken cultural premisses, “learning by seeing” or by “experiencing” and many other extra-organic media and conveyors. Furthermore, the interpretation of this collection of signals and imprints is not made by an individual in particular but, instead, by the whole community that produces and retains the culture. This means that many of the writings and extra-organic, non-individual, media contain ideas and discussion about elements of the culture itself, being self-reflexive and self-referential to a much higher extent than knowledge.
The nature of science is not much different from culture. Its findings (atoms in ) are also organised in the manner of Definition 1 and its interpretation is a collective enterprise. The distinctions reside primarily on rules, methods and underlying paradigms about how to collectively develop scientific knowledge and the relying on observation as a conflict resolver. The language of science, even when it doesn’t use mathematics, for instance, is developed in such a manner that differences in interpretation are minimised, and arguing procedures are standardised and their rules accessible to everyone. In this sense, the collective doing of science is more self-conscious than that of culture. Culture is produced more instinctively and intuitively than science. In Science, there are a more definite and explicit purpose to be followed and methodological rules to abide to.
4.2. Organisation and Complexity
The intertwining among living phenomena, organisation, information, language and complexity has been acknowledged since long [52,61,122,123,124,125,126], as well as the importance of innumerous concepts introduced by system thinking to deal with them [52,54,56,124]. Nevertheless, a consensual concept of complexity, whilst important, is still elusive [43]. Until now, there is no widely accepted definition of complexity, which meaning depends on the domain of inquiry. Those more widely used resort to concepts in computation, Shannon-Brillouin information or code-interpretation for their definition [41,51,127]; not to systems or organisation concepts. A few largely neglected efforts to connect information and organisation do, however, exist: the work of H. Atlan [40,62,65,77] and I. Walker [63]. This section discusses how complexity inserts itself in the organisation framework introduced above (Section 3).
The first thing to note is that there is no single way to represent things that compose phenomena in Γ or . Different representations may reflect different emphasis, different details, or distinct perspectives, standpoints, purpose, and questioning. To see this, consider again the example of the house (Section 3.1). A possible organisation for the house in Figure 1a, , is given by the graph in Figure 2a and belongs to Γ, since undirected graphs are special cases of hyper-graphs [97] and hyper-graphs belong to Γ (Definition 2, case 1). In , the house-rooms and entrance are atomic elements and are the only nodes, i.e., . The graph is such that:
The arcs of represent the access (doors) between the rooms and the external access.
Other organisations for this same house can be constructed by resourcing to meta-variables and different levels of detail, as indicated in Figure 19.
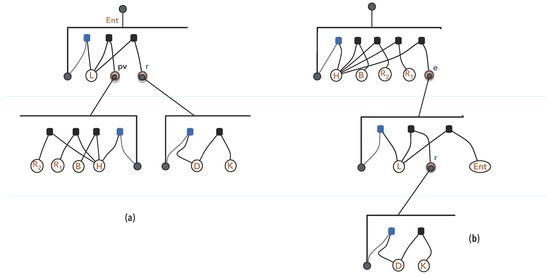
Figure 19.
Two alternative organisations of house in Figure 1a with different levels of detail: (a) has two levels and (b) has three. Objects represented are the same, but the perception of what is primary or secondary is different.
The first is a two-level organisation and the second a three-level organisation. Their expressions in Γ refer to the universal set of Equation (21) and are given in the sequel. The formal specification for organisation , pictured in Figure 19a, is:
while for organisation , of Figure 19b, it is:
Any of the three organisations and can be further detailed by adding more levels. For instance, any room ρ in can be turned into a meta-variable and bound to copies of the hyper-graph below that describes four connected walls, one of which has a door. Its nodes and arcs are given by:
where represent the walls, and the arcs represent their bindings at the corners. The wall with the door is associated with .
It is important to note that while creating this new level of detail the walls are duplicated in each room as part of the organisation. The concrete instances of organisations may collapse distinct parts of them in one object. This highlights the fact that there is a lot of freedom in the immersion of -graphs in the physical reality, or else, in . Otherwise, walls in Figure 1 may be double-walls, with an air cushion in-between, or even be separated by larger spaces, the organisation of the house remaining the same, although not its instantiation in .
Another important thing to note is that, except for the intermediate nodes and , which can be erased from without loss of connectivity,
This illustrates that the mapping (Definition 6) is non-injective and there is always more than one multi-level (hierarchical) organisation which can be associated with a given network under no matter which heuristics. Hence, network topology is largely insufficient to determine the organisation of bio-chemical networks or of connection-diagrams of other complex phenomena.
This observation leads to considering the question: “How many ways are there to hierarchically organise a network?”. Or else, what is the size of , for ? A more in-depth discussion of this problem is outside the scope of this text. However, a hint about its magnitude can be obtained by inspecting Definition 6, even if it is constrictive and considers only a two-level hierarchy.
From Equations (14)–(17), it is clear that and are constructed respectively as unions of the node-sets and arc-sets of and parts of γ and that neither nodes nor arcs can appear concomitantly at more than one level or more than one part of γ. If they do appear, like the walls in the example above, they are different instances of the same organisation and must be considered distinct in Γ; that is, a can never be equal to, or the same as, a copy of it that is part of another -graph . Furthermore, from the definition of hyper-graph [41,97],
and it is always possible to recover from , for all .
On that account and from an inspection of case 2 of Definition 6, it is clear that, to construct any : needs to be partitioned into subsets, , ; meta-variables assigned to the partition that will be ; and the assignments of Equation (4) established. Since is a parameter and, for each partition, there is a circular choice about which partition will be and which partition will be bound to which meta-variable,
where is the number of ways the integer κ can be partitioned into q distinct integers [128]. Considering that, for any reasonably representative network , is anywhere in the range from to , , is astronomically large for any useful set of nodes and arcs.
By weighting atoms and counting interrelations against maximum relevance and total number of possible interconnections under variegated guidelines, a wealth of complexity measures can be defined for organisations modelled in , that help comparing the many . The family of complexity measures presented in the sequel illustrates this. It has well defined, finite values for each , including self-referential ones. Self-referential -graphs can be obtained by, e.g., considering the nodes of as variables and binding copies of to them indefinitely but finitely [100].
The family of measures below is defined for organisations that may be constructed or observed. It is thus computed over the observed organisations rather than metered directly in phenomena. Despite being inherently non-determinant, it can be used to help choosing the best representation of constituents of phenomena in terms of organisations, as indicated in the end of this section. It is based on the following heuristics: (1) conformity to the whole-part relation (Definition 4), in the sense that the complexity of a part should be smaller than that of the whole, (2) possibility of gauging relative contributions of atoms and parts to the overall organisation without biases, and (3) sets of organisations (Definition 1) should also have a complexity assigned to them.
The relative contributions may be dependent on external factors like the ‘purpose’ or ‘function’ of the organisation as well as in the domain of application of the model and, thus, are subject to ontological guidelines. For instance, the rooms of the house in Section 3.1.1 may have different degrees of importance (weighting) depending if it is intended to become a residence, an office, or a restaurant. On top of that, organisations representing visual elements in Figure 17 may be used to discuss them from a cognitive perspective (Section 4.1.1) or a cultural perspective (Section 4.1.5). It is likely that the parts and atomic elements composing the visual units will have completely different degrees of importance in each analysis.
So, let be a given weight-function defined for the elements of , such that ():
where λ relates to the inter-level significance and is such that . Contributions are generally evaluated with respect to a single γ. Because of this, it is enough to consider since there exits several procedures to normalise ω in a way that it reflects only relative contributions of atoms within a γ, due to the finiteness of node-sets in γ.
The formulas for complexity measures in this family will be presented case by case for the three classes of organisations induced by Definitions 1 and 2:
It will help to use a few shorthands. For , let and be as above (Equation (25)) and
where is the cardinality of S and is the number of possible combinations of k elements in groups with p elements. The value is an estimate of the connectivity of h and of the influence of node n on its neighbours. Moreover, means that and γ is bound to meta-variable (see Equation (4)).
That stated,
If , , and its complexity is:
where . For , is a set of positive real values and any monotone statistics ( etc), for instance, provides a complexity measure that abide to the properties below. That is, if is monotone for each argument,
Each measure Ξ in this family is such that:
- The more associations there are in a -graph, higher its complexity is;
- ;
- The more detailed an organisation is, that is, deeper the hierarchical levels go or bigger the number of its parts is, higher its complexity is;
- The finiteness of and the existence of self-similar , require that the contribution of deeper levels in the hierarchy decays rapidly, e.g., .
Therefore, since the same natural object or entity may be associated with distinct organisations, it is a fortiori possible that natural objects have several (organisational) complexities. Thence, complexity in Γ is a concept associated with the description and representation of a phenomenon in terms of organisations rather than with the phenomenon itself [41,129]. Moreover, can be used as an indicator to find the “most convenient” representation (organisation) of some-thing in , where retract its raw connections.
Remark 2 (On the complexity of signals and imprints:).
The discussion about imprints just after Hypothesis 1 can be illuminated by the following observations
- 1.
- By definition, ;
- 2.
- If then , since ;
- 3.
- does not imply that ;
- 4.
- It is possible that and , or even that .
4.3. Organisation Perspective in Action: Re-Thinking Flows
Thinking in terms of organisations and organisational changes can bring new approaches to traditional scientific modelling. The investigation of cellular cytoskeleton, in particular flows and mechanical effects in plasma-membrane protrusions, is an active field of experimentation [36,46,130] and modelling [131]. These investigations focus on actin-polymerisation versus disassembly determining the retrograde flow of actin-filaments, its stability and force through actin-binding myosin motors. Mechanical and dynamical effects on the plasma-membrane result primarily from these phenomena. Notably, the cytosol is considered as a backcloth substrate, although actin diffusion, translation and sequestration in the cytosol may have important regulatory contributions to the actin network dynamics. Filopodia are long membrane protrusions containing parallel bundles of F-actin whose dynamics is governed by the regulation of polymerisation/disassembly processes. How the required amounts of G-actin are delivered within the challenging filopodial structure is a fascinating question. Existing models build on diffusion as the key actin-deliver mechanism [132,133].
In collaboration with A. Prokop and C.A. de Moura [134], we started to include a further hypothesis, where a mixture of cytosol and G-actin circulates changing organisation at the tip and bottom of the filopodium (Figure 20), from an incoming diluted solution into an outgoing tube formed by cytosol trapped into the actin filament bundle. The tip polymerisation drives a steady back-flow of the F-actin filament bundle at the core of the filopodium, with cytoplasm caught between the filaments (Phase B). This volume “outflow” is replaced by a compensatory influx of a diluted solution of G-actin molecules in the cytoplasm (Phase A) towards the tip of the filopodium, that occurs between the F-actin filament bundle and the cell membrane. At the tip of the filament the flow of the diluted solution blends conveniently towards the polymerisation points, guided by molecular organisations around it.
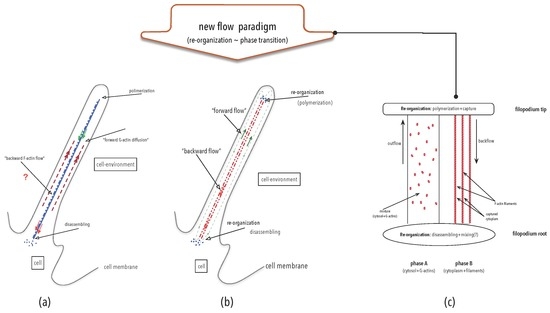
Figure 20.
Filopodia: (a) Diffusion Flow, (b) Mixture Flow, (c) Re-organisation Diagram.
The latter hypothesis is suggested by the organisation perspective. However, the models and observations needed to verify it are quite different from those under the diffusion view. Despite its hydrodynamical elegance, this alternative cannot arise under the diffusion perspective because cytosol displacements are disregarded, and only diffusive movements represented. A new starting point, the organisation perspective, is needed to suggest this alternative and the models and observations required. This perspective also suggests that, at a second stage, the actin flow together with the polymerisation and disassembling processes are to be considered as a biological organisation unit responding to cellular regulatory processes and signals coming from both the cell’s interior and exterior.
5. Interacting Organisations: Biological and Complex Phenomena
What makes living phenomena so singular? Things in physical phenomena move and exchange mass, momenta, and energy. When a cell phagocytes another cell, does it absorb just energy and mass, or does it intake as well, as suggested in several recent publications about cell immunology? When a firm “phagocytes” another (mergers & acquisitions), does it earns just material and monetary assets, or does it income know-how and knowledge as well? Presently, self-organisation and information help our understanding of systems out of equilibrium and sustain the perception that organisation is a natural phenomenon. They do not, however, help our understanding about the absorption of and knowledge suggested above, nor about the different effects of signals and imprints.
Also, neither organisation nor information is thought to affect/effect exchanges or cause specific behaviour [20,24,127]. Nor are they considered as possible and valid components of system states, promoting propensities and patterns of behaviour. In chemical phenomena, new substances are created when portions of molecules, organisations as well, are permuted among molecules, re-organising them. Chemical reactions re-organise substances into other substances, that appear and disappear. Molecular organisation determines which reactions are possible and which substances become which. Nevertheless, substrates and products remain the same, as well as their possible interactions, and organisation is but a parametric descriptive factor fixed once-for-all, as a natural law, in present day theoretical efforts towards living and complex phenomena.
Notwithstanding, outcomes in living phenomena require specific types of matter-energy in adequate amounts [32] that are properly interrelated and positioned [14]. Conformation states of molecules change, changing the reactions its substance can perform. Another distinctive aspect of living phenomena is that interactions exchange besides mass and energy [79,80]. Hence, the network of possible interactions is altered by the exchanged and by dynamics itself, as a consequence of conformal changes in molecules, changes protein activation states, the intensity of regulatory reactions, and so on. The present framework naturally allows for considering organisations as components of a system’s state by modelling a phenomenon in instead of .
Molecules are the simplest non-trivial organisations (see Section 3.1.2). At the molecular and intra-cellular scales, modules [96] may be interchanged with organisations, as here understood. At higher, more aggregate scales, it is not clear that observable aggregate modules [135] equate with organisations. Nor is it sharply clear that that near decomposability [121,136] will be applicable at smaller scales. Yet, organisations are aggregates of interrelated things that act as an unit at a higher, more aggregate scale and whose aggregation affects and effects behaviour at the higher scale.
The organisation perspective hypothesises that living and general complex phenomena result from a collection of interacting organisations, which interact exchanging , besides organised mass-energy assemblages. Moreover, interactions change the interacting organisations what, in turn, changes how organisations react as wholes. This is expressed by Definition 9. Hypothesis 2 realises emergence. The following arguments further illustrates the soundness of these conjectures.
The organisation framework straightforwardly allows for considering a dynamics of organisations, where changes in organisations (architectures) affect dynamical possibilities and propensities while dynamical stresses induce changes in organisations. Let’s consider the smallest living unit—a cell. An eukaryotic cell for explanatory reasons.
Figure 21 displays sketches of two cell-views as organisations. One is grounded on space-topology and A is hierarchically “below” B if A is contained or occurs inside B [104,137]. The other is based on (chemical) affinity. The conspicuous incompleteness of each is due to very different reasons. It is easy to grasp that the -graph in Figure 21a is indeed a synexion, since our perception of organelles include their volumes in space and the relation represented is induced by set inclusion. The number of molecules in each unit depicted grows from bottom to top. This representation clearly conforms to Equation (9) and is a synexion. Although there is no time, and things like vibrations, relative displacements etc, depicted in the figure, they do exist and may become part of the model. As suggested by this figure, synexions accommodate different levels of detail and some branches may reflect more detail than others.
Figure 21.
Cellular Organisation: topology bound (a) and re-action bound (b). Both are synexions (see Section 3.1.3).
Figure 21b displays biochemical networks of the cell schematically, the metabolic one with a little more detail. Cellular networks are hyper-graphs [99]. Although almost never made explicit, they are also synexions since molecules occupy volumes in space and chemical reactions require time. Or, at least, the sub-network that is “active” under any given conditions is a synexion, since they spread in space and time. First, because chemical reactions have characteristic times that may be regulated by interactions among cellular bio-chemical networks. Hence, each pathway or cycle has a tunable characteristic time. Second, because reactions and substrates are localised in specific regions of the cell. Unfortunately, information about frequencies, rates and localisation are still largely missing in biochemical data-bases nowadays, even though this has been changing in the last years [114,138].
Many essays in literature investigate the organisation of cellular networks [100]. Explicitly or not, they represent networks as bi-partite graphs [139] and their results are often in the form -graphs. But their results are not synexions since in general they do not use information about space-time localisation or characteristic times, distances and frequencies, even when available. Discarding this sort of information may hamper our understanding of life’s liveness due to several reasons. To cite one, molecules of substrates and residues of proteins need to fit the space-time niche where they are, e.g., by adjusting vibrations. Besides that, connections between chemical reactions occurring in distinct regions of a cell are often fake, since different molecules of the same substrate take part in each of them and there is no real connection between the reactions [100,140].
This indicates how important it is to map the many networks in Figure 21b onto one another and onto the organisation sketched in Figure 21a. The constraint given by Equation (9) is an important guide to obtain data that support more realistic biochemical networks. It is also an important designing clue towards a coherent representation of living entities in Γ and from the structural and dynamical stands.
From the organisation perspective, biological entities intake organisations, decompose them into simpler organisations (catabolism), storing the energy released, and rearrange these simpler organisations into parts of their own (anabolism) with little expenditure of energy, outputting matter and energy that is not usable. Instead of being completely catabolised, assimilated organisations may resonate with definite portions of the assimilating entity, being perceived and provoking imprints that may eventually induce behavioural alterations [141]. A good example is the assimilation of viruses by prokaryotic cells as immunological memories [108]. However, absorbed nucleic-acids may distort the cell’s behaviour [142,143,144], often making it destroy itself. In both cases, which may occur alongside, biological interactions entail organisational changes and exchange. Hypotheses 1, particularly C and D, are necessary to make this description sound. This kind of reasoning allows for addressing questions about the input of and knowledge during phagocytosis or the merging of firms and other human organisations, as long as the complexity of catabolic results is large enough to retain meaningful signals.
Two concepts in physics and chemistry are important to understand dynamics, stasis and stable dynamical regimes—fluctuations and perturbation by virtual changes. The first is ontological and effectively occur in phenomena inducing swiftness and ability to change. The second is methodological and enables ideal inspection of alternative virtual behaviours, not observed but possible, and the subsequent questioning about why Nature has chosen the path we observe. Fluctuations are ubiquitous in living phenomena. Lively proteins flop between stable conformal states, altering interaction possibilities [50]. Changes in population density are identified by quorum-sensing set-ups that oscillate around chemical equilibrium points [33]. Complex oscillations between cellular modules and processes that occur during cell division determine form and localisation [14], and so on. That is, they manifest organisation-fluctuations.
Spaces Γ and provide tools that allow for defining organisational difference and distance between as well as neighbourhoods around organisations in Γ, that may be processes like the TCA cycle. Two points in Γ are close together if they differ only near the leaves of their recursive skeletons; that is, near the bottom of the diagrams in Figure 9 or Figure 10. Elements of present two forms of fluctuation. They can oscillate around a state in , which means that their instantiations (the “phase-space” retract) oscillate around a stable “set-value” in or γ cyclicly exchanges some of its lower-complexity parts with elements available in its Γ-neighbourhood, like when a protein docking-site is constantly being flip-flopped. One of the main features of the formalism underlying this perspective is the ability to enforce distinguishing organisational fluctuations in a complex from dynamics [134]. Nevertheless, a would-be methodology of virtual behaviours (counterpart of virtual displacements) needs to encompass evolutionary perspectives to be effective in explaining how the actual organisation of ‘problems of complexity’ came to be.
6. Conclusions
This work addresses the description of organisation and suggests means to use organisation as a tool to understand and explain living as well as other complex phenomena. Concomitantly, it enlarges the general systems approach, sharpening systemic terms like: interacting parts, communication, wholeness, modular organisation, whole-part relationship, hierarchical systems and so on. The elements in may be time-functions or space-time-functions that chronicle observable aspects of phenomena [145] and compose system states ([8] chapter 4), offering other means to obtain immersions in , that do not necessarily comply with Equation (9). Thus, it also supports considering organisations that are permanently being rebuilt by steady substitution of their hierarchically lower parts, as if the walls of Equation (24) were a collection of dynamical systems permanently substituting their bricks.
It introduces mathematical spaces for modelling organisations and defines grounded on the organisation concept and the tools provided by these spaces. These spaces accommodate organisational fluctuations and the concept extends the usual Shannon-Brillouin information; addressing the Shannon-Weaver levels of meaning and effectiveness [112] with no a fortiori reference to senders, the pre-established set of messages or “intention”. Organisations and augments our tools to address questions about the acquisition of imprints, know-how and knowledge during interaction of organisations, phagocytosis and mergers included.
The complexity of organisation is central in these enquiries since an imprint, being part of an organisation, is bound to have a complexity smaller than that of the organisation. Being dynamical (like all elements in ), it is prone to oscillation and destabilisation. On the other side, any imprint is a candidate to become a model and a model, knowledge; depending on relative complexities, stasis, and dynamical relations.
Definition 9 looks anticipatory. Although only observers are required to anticipate the interpreter’s behaviour to acknowledge change, the basic property enabling anticipation is the immersion of Γ in , i.e., the space , and thus any synexion with internal models may anticipate. Imprints in an interpreter may be rock-stable, hampering change. Organisation fluctuations distort perceptions, destabilises and fluidises imprints and models, perturbing interpretations. The organisation perspective thus support investigations about the role of imprecision, fuzziness, adaptation, anticipation, emotion and moods in adaptation, learning and cognition [115,146,147,148] during transfer of information at any scale and encompasses all three Shannon-Weaver levels [112].
The introduced framework is able to represent (biological) organisations from a relational stand. To define information in this context, organisations were immersed in a physical fabric endowed with space-time, resulting in “concrete” organisations that associate structural organisation with the dynamics of usual changes (Section 3.1.3). It presents examples and arguments supporting and justifying this immersion and a statement about using the organisational perspective for studying living phenomena. One advantage is that a mathematics can be developed in Γ (and ) that contemplates transformations of organisations, relations among organisations and other mathematical tools, where theorems can be proven and relational reasoning sharpened. It enables reasoning about organisations independently of any ontological references or immersions in , unveiling properties intrinsic to the relational aspects of organisations (Section 3.1.4 and Section 4.2). Furthermore, it supports the distinction and identification of organisation changes amid dynamics in . This text extends and improves previous presentations of the above definitions [106,149,150].
Although inspired on life phenomena, the present concept of biological organisation extends beyond life and may be used at the molecular and supra-organismic scales [10,14,28,66,121]. General systems researchers think organisations as a collection of interacting components, alluding only informally to hierarchy and treating organisation components as deciders [30]. On the other hand, the definition of systems is meant to capture organisation [23,30,124]. General systems are special cases of organisations as above, since a set of interacting components can be formalised as mathematical relations [23], that are associated with hyper-graphs without isolated nodes [98] and, thus, are exactly the organisations delineated in case 1 of Definition 2. Hence, the term organisation may be used instead of the term system wherever it appears, providing a solid ground to handle systems of varying structure [4], even with unknown bounds and domains for the structural variations.
Definition 2 formalises the hierarchy inherent in organisations, extending the former definitions in many ways. It gives a formal meaning to terms like “system within a system” and “system composed of systems”, so common in the literature. Mappings and relations can be defined in Γ, allowing for comparing organisations, transforming them into one another, and considering organisations as the same while portions of theirs vary (Section 3.1.4 and Section 4.2). The concepts and tools here introduced, therefore, add to the system thinking framework [124].
There are many ways of building bio-mathematical worlds, life-like universes and theoretical explanations to study and understand life and richer phenomena, that develop along variegated reasoning directives [10,41,53,55,57,60,65,82,151,152,153,154]. Some consider singular characteristics of life, like organisation or reproduction, trying to explain them in terms of Shannon-Brillouin information, computational metaphors, bio-semiotics, or self-organising dynamical systems [8,41,60,84,155]. They draw on partial analogies, focus on the onset of life, or target the definition of life and organisation. Many go beyond and aim towards a theoretical biology [8,56,152,156,157]. Of the latter, two stand out: Rosen’s (M,R)-systems and Gánti’s Chemoton Theory [55].
It is not yet known whether these approaches relate to or can be rephrased in Γ and . Probing into possibilities, the following can be said about the last two. The Chemoton represents a would-be pre-biotic living entity, being formed by three interconnected cyclic processes, represented by symbols standing for unspecified molecules, enclosed by a boundary molecular circle plus a hypothesis about diffusion of food and waste through the enclosing circle. Cyclic processes and the molecular circle can be straightforwardly represented in Γ (see Section 3.1). The diffusion of waste and nutrients through the boundary circle can be approximatively represented in , even without the instantiation of the Chemoton in . Representing the Chemoton in , however, requires the specification of physical and chemical properties for each (molecular) node. Chemical properties will affect the relative position of would-be molecules in the cycles, as well as the length and stabililty of the cycles. Physical properties should determine which molecules compose the cycles. Thence, expressing the Chemoton in amounts to solving Gánti’s riddle.
Rosen addressed different questions. His formalism [8,68] is directed to express features that could unquestionably identify and distinguish life, independently of how living entities are instantiated. Despite both approaches being relational, it is unclear how to map his categorical diagrams into Γ or . Without resource to mathematical underpinnings, the following provides a possible interpretation of Rosen’s ideas under the organisation perspective.
Block diagrams unite components into wholes. Their homologies in the organisation perspective are thus the hyper-graphs at the root and forks of a γ’s recursive skeleton, , the root acting also as a closure. Rosen’s basic components , , or [8, Section 5K]. may be atoms in Γ or , depending on how the sets A and B are constructed. The functors Φ and are not in Γ but could be transformations from Γ into Γ.
From a formalistic standpoint, Γ and are closer to Rashevsky’s (1954) initial ideas, grounded on graphs and discrete topology. Notwithstanding, the present approach departs from the majority of existing approaches, perhaps radically, in what it abstracts from “biological function” or “origins of life” phenomena and questions, instead of attempting to represent and answering them. Arcs and bindings in whole-part graphs represent more directly observable and identifiable relations: spacial contiguity, chemical bonds or affinity, preferences, channels of interaction, and so on; instead of function which is a difficult observable [158], often only recognised a posteriori and in relation to external interactions with elements of the organisation’s environment. Biological function is a many-to-many relationship. This means that a biological entity may play several functional roles, while the same biological function may be fulfilled by different entities. Having hyper-graphs as a fundamental building block, the present perspective does support the organisation of pathways [100] and the identification functional modules in a many-to-many fashion [34,35].
Complexity science, see [11, Chap 10] and [49], adaptive systems [153,159] and systems biology [160] are revivals of the “general systems theory”, initiated by von Bertalanffy around the 1930s [52] to address problems in biology [22,52], that were boosted by cybernetics concepts [136]. Organisations may be employed in these fields in substitution of the systems concept, considering interactions among them and adding the benefit of reasoning about systems while organisations globally, instead of in a case by case, ad hoc, and behaviour centred manner.
For instance, with the tools offered by general systems theory, it is not possible to describe a collection of interacting (eco)systems, maintaining their identity and internal dynamics while considering interactions and dynamics among them, as it is not possible to consider a collection of interacting organelles (biochemical systems) in a cell, maintaining their identities in the same way. It is necessary to smash these phenomena into a huge and all-encompassing (dynamical) system and be compelled to analyse the behaviour of the latter, loosing sight of the inherent hierarchy in the phenomena and the fact that organelles are often standalone entities encapsulated by permeable membranes that modulate their interactions with rest of the cell. Organisations, synexions in particular, help to keep behavioural levels separate and subsystems as units, studying each separately or in conjunction in several ways.
It allows also for discussing organisations as such, independently of any dynamics, as when discussing the relation of organisation and complexity in Section 4.2. Furthermore, techniques already developed for composite systems can support the disentanglement of dynamics between the various hierarchy levels of organisations and creation of aggregated states for sub-organisations. Moreover, mathematical investigations about properties of Γ and , may develop a coherent and encompassing platform to address Weaver’s ‘problems of organised complexity’. It allows for extending formalisations to chemistry [56], aligning with Gánti’s proposal of addressing questions of how can raw materials and simple organisations assemble into larger organisations [161], maintain themselves and evolve, addressing also more complex entities like communities, societies and beyond [2], by using the concept of modules [135].
It is worth emphasising that the framework presented above adds but does not supersedes or substitute any of the previous essays. Instead, it offers a formalism where they can possibly be rephrased and brought together providing richer pictures of complex features and traits. Even communication channels and Shannon-Brillouin information can be regained in Γ or , if the sender is made explicit and the set of exchangeable messages is known and fixed in advance. Yuri Lazebnik in 2002 [162] ingeniously claimed for an unambiguous biological language. Such language is vital for biotechnology [109] and for constructing (more fundamental) biological theories [17]. In physical and chemical phenomena, only the attributes of ‘things’ change. Interactions and the possibilities of interaction do not change, or change mildly and are considered fixed. In living and complex phenomena, on the contrary, ‘interactions’ (or relations) change wildly, even more often than ‘things’. It is my hope that the organisation framework and the perspective it introduces, bringing interactions and things to the same level of attention, will serve as a basis for developing such language and help boosting the description of inter-level relations [163] and the concomitant addressing of proximal and ultimate explanations [164] by allowing the consideration of a dynamics of relations associated with the usual dynamics, as discussed in [26]. If it comes to assist the establishment of a philosophy and basic principles for the living and complex sciences [16,165] will be an added bonus. The importance of having a consensual philosophy and well-established principles is addressed in [22,23,124] in an unsurpassable manner.
Acknowledgments
These ideas and framework have been being developed and polished during almost three decades. During this time, I had the pleasure to be assisted by a great number of colleagues and people that supported the long standing process of consolidating and testing them. They helped through discussions, advice, key questions, and aids of variegated kind. The list presented in the sequel is certainly incomplete, due to misfirings in synapses of my memory-brain. My heartfelt apologies to those not in the list but who should be, many of whom read previous versions and drafts of this paper, providing criticisms that greatly improved the text. I would like to express my deepest gratitude to the whole group of Mathematical Modelling of Knowledge Diffusion, headquartered at LNCC and the Federal University of Bahia, to the Faculty of Life Sciences of the University of Manchester, as well as, to W. Aber, D.S. Alves, P. Antonelli, J.-Y. Béziau, L.H. Coutinho, A.C. Gadelha Vieira, M. Grinfeld, D.C. Krakauer, R. Lins de Carvalho (In Memoriam), F. Lopes, C. Menezes, S. McKee, A.C. Olinto, N. Papavero, M.M. Peixoto, A.A. Pinto, J.C. Portinari, A. Prokop, D.L. Robertson, M. Trindade dos Santos, J.-M. Schwartz, Iain W. Stewart, C.M.M. Vianna, S. Webb, my present and former students, and persons who attended the many talks, short-courses, and normal courses I presented on this topic or on “modelling techniques” posing wise questions. Last but not least, M.C.F. Bittencourt, M. Butturini and I.U. Cavalcanti invaluable suggestions improved the style, expressiveness, organisation, and quality of this writing. Any remaining flaws are, nevertheless, my sole responsibility.
I would also like to acknowledge the financial support of the following agencies and programs, through several grants and financial support along the development of this work: CAPES (specially fellowship No. 10313/88-2), PCI/LNCC, Wellcome Trust (specially grant 097820/Z/11/B) and FAPERJ (specially grant No. 101.261/2014).
Conflicts of Interest
The authors declare no conflict of interest. Financial sponsors had no word whatsoever in any phase of this study.
References
- Weaver, W. Science and Complexity. Am. Sci. 1948, 36, 536–544. [Google Scholar] [PubMed]
- Miller, J.G. Living Systems; McGraw-Hill Book Co., Inc.: New York, NY, USA, 1978. [Google Scholar]
- Kritz, M.V. Boundaries, Interactions and Environmental Systems. Mec. Comput. 2010, 29, 2673–2687. [Google Scholar]
- Mohler, R.R.; Ruberti, A. (Eds.) Recent Developments in Variable Structure Systems, Economics and Biology; Lecture Notes in Economics and Mathematical Systems; Springer: Berlin/Heidelberg, Germany, 1978; Volume 162.
- Junk, W.J. (Ed.) The Central Amazon Floodplain: Ecology of a Pulsating System; Springer: New York, NY, USA, 1997.
- De Ruiter, P.C.; Wolters, V.; Moore, J.C. (Eds.) Dynamic Food Webs : Multi-species Assemblages, Ecosystem Development, and Environmental Change; Theoretical Ecology Series; Academic Press: Boston, MA, USA; Elsevier: Amsterdam, The Netherlands, 2005.
- Pascual, M.; Dunne, J.A. Ecological Networks: Linking Structure to Dynamics in Food Webs; Studies in the Sciences of Complexity, Santa Fe Institute, Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Rosen, R. Life Itself: A Comprehesive Inquiry into the Nature, Origin, and Fabrication of Life; Complexity in Ecological Systems Series; Columbia University Press: New York, NY, USA, 1991. [Google Scholar]
- Hofmeyr, J.H.S. The biochemical factory that autonomously fabricates itself: A systems biological view of the living cell. In Systems Biology: Philosophical Foundations; Elsevier B.V.: Amsterdam, The Netherlands, 2007; pp. 217–242. [Google Scholar]
- Miller, J.G. Living Systems: The Organization. Behav. Sci. 1972, 17, 1–182. [Google Scholar] [CrossRef] [PubMed]
- Harold, F.M. The Way of the Cell: Molecules, Organisms and the Order of Life; Oxford University Press: Oxford, UK, 2001. [Google Scholar]
- Lillie, R.S. Living Systems and Non-living Systems. Philos. Sci. 1942, 9, 307–323. [Google Scholar] [CrossRef]
- Ulanowicz, R.E. On the nature of ecodynamics. Ecol. Complex. 2004, 1, 341–354. [Google Scholar] [CrossRef]
- Harold, F.M. Molecules into Cells: Specifying Spatial Architecture. Microbiol. Mole. Biol. Rev. 2005, 69, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits; Mathematical and Computational Biology, Chapman & Hall/CRC: London, UK, 2007. [Google Scholar]
- Boogerd, F.C.; Bruggeman, F.J.; Hofmeyr, J.H.S.; Westerhoff, H.V. Systems Biology: Philosophical Foundations; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Krakauer, D.C.; Collins, J.P.; Erwin, D.; Flack, J.C.; Fontana, W.; Laubichler, M.D.; Prohaska, S.J.; West, G.B.; Stadler, P.F. The challenges and scope of theoretical biology. J. Theor. Biol. 2011, 276, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Jeldtoft-Jensen, H.; Oliveira, L.P.; Sibani, P. Evolution in complex systems. Complexity 2004, 10, 49. [Google Scholar] [CrossRef]
- Roberts, A.J. Model Emergent Dynamics in Complex Systems; Mathematical Modeling and Computation, SIAM, Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2015. [Google Scholar]
- Bailly, F.; Longo, G. Extended Critical Situations: The Physical Singularity of Life Phenomena. J. Biol. Syst. 2008, 16, 309–336. [Google Scholar] [CrossRef]
- Watson, D.L. Biological Organization. Q. Rev. Biol. 1931, 6, 143–166. [Google Scholar] [CrossRef]
- Mesarović, M.D. (Ed.) System Theory and Biology; Springer: New York, NY, USA, 1968.
- Klir, G.J. Facets of Systems Science, 2nd ed.; Plenum Press: New York, NY, USA, 2001. [Google Scholar]
- Balescu, R. Equilibrium and Non-Equilibrium Statistical Mechanics; A Wiley-Interscience Publication, John Wiley & Sons: New York, NY, USA, 1975. [Google Scholar]
- Rosen, R. On interactions between dynamical systems. Math. Biosci. 1975, 27, 299–307. [Google Scholar] [CrossRef]
- Kritz, M.V.; dos Santos, M.T. Dynamics, Systems, Dynamical Systems and Interaction Graphs. In Dynamics, Games and Science II; Peixoto, M.M., Rand, D., Pinto, A.A., Eds.; Springer: Berlin, Germany, 2011; pp. 507–541. [Google Scholar]
- Ashby, W.R. Principles of the Self-Organizing System. In Principles of Self-Organization: Transactions of the University of Illinois Symposium; von Foster, H., Zopf, G.W., Jr., Eds.; University of Illinois, Pergamon Press: London, UK, 1962; pp. 255–278. [Google Scholar]
- Rosen, R. Biological Systems as Organizational Paradigms. Int. J. Gen. Syst. 1974, 1, 165–174. [Google Scholar] [CrossRef]
- Varela, F.G.; Maturana, H.R.; Uribe, R. Autopoiesis: The Organization of Living Systems, Its Characterization and a Model. Biosystems 1974, 5, 187. [Google Scholar] [CrossRef]
- Miller, J.G. Living Systems: Basic Concepts. Behav. Sci. 1965, 10, 193–237. [Google Scholar] [CrossRef] [PubMed]
- Auger, P. The Methods and Limits of Scientific Knowledge. In On Modern Physics; Clarkson N. Potter, Inc. Publisher: New York, NY, USA, 1961; pp. 79–108. [Google Scholar]
- Miller, J.G. Living systems. Curr. Mod. Biol. 1971, 4, 55–256. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing In Bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Stoney, R.A.; Ames, R.M.; Nenadic, G.; Robertson, D.L.; Schwartz, J.M. Disentangling the multigenic and pleiotropic nature of molecular function. BMC Syst. Biol. 2015, 9, S3. [Google Scholar] [CrossRef] [PubMed]
- Oyeyemi, O.J. Modelling HIV-1 Interaction with the Host System. Ph.D. Thesis, University of Manchester, Manchester, UK, 2016. [Google Scholar]
- Prokop, A.; Beaven, R.; Qu, Y.; Sánchez-Soriano, N. Using fly genetics to dissect the cytoskeletal machinery of neurons during axonal growth and maintenance. J. Cell Sci. 2013, 126, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zheng, H. Organized modularity in the interactome: Evidence from the analysis of dynamic organization in the cell cycle. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Brownridge, P.; Lawless, C.; Payapilly, A.B.; Lanthaler, K.; Holman, S.W.; Harman, V.M.; Grant, C.M.; Beynon, R.J.; Hubbard, S.J. Quantitative analysis of chaperone network throughput in budding yeast. Proteomics 2013, 13, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Peacocke, A.R. An Introduction to the Physical Chemistry of Biological Organization; Clarendon Press: Oxford, UK, 1983. [Google Scholar]
- Atlan, H. Entre le Cristal et la Fumée. Essay sur l’Organization du Vivant; Éditions du Seuil: Paris, France, 1986. [Google Scholar]
- Kritz, M.V. Creating Bio-Mathematical Worlds. P&D Report 29/95, LNCC/MCT, Petrópolis, 1995. In Proceedings of the 13th European Meeting on Cybernetics and Systems Research, Vienna, Austria, 9–12 April 1996.
- Bizzarri, M.; Palombo, A.; Cucina, A. Theoretical aspects of Systems Biology. Prog. Biophys. Mol. Biol. 2013, 112, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Kitto, K. High end complexity. Int. J. Gen. Syst. 2008, 37, 689–714. [Google Scholar] [CrossRef]
- Waddington, C.H. (Ed.) Biological Organization, Cellular and Sub-cellular; Pergamon Press: London, UK, 1959.
- Berg, H. Motile behavior of bacteria. Phys. Today 2000, 53, 24–29. [Google Scholar] [CrossRef]
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin Dynamics, Architecture, and Mechanics in Cell Motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Needham, J. On the dissociability of the fundamental processes in ontogenesis. Biol. Rev. 1933, 8, 180–233. [Google Scholar] [CrossRef]
- Rosen, R. Structural And Functional Considerations in the Modelling of Biological Organization; Technical Report 77 25; The Center for Theoretical Biology, SUNY: Buffalo, NY, USA, 1977. [Google Scholar]
- Schweitzer, F. (Ed.) Self-Organization of Complex Structures: From Individual to Collective Dynamics; CRC Press, Taylor and Francis Group, LLC.: Boca Raton, FL, USA, 1997.
- Vinson, V.J. Proteins in Motion. Science 2009, 324, 197. [Google Scholar] [CrossRef] [PubMed]
- Longo, G.; Miquel, P.A.; Sonnenschein, C.; Soto, A.M. Is information a proper observable for biological organization? Prog. Biophys. Mol. Biol. 2012, 109, 108–114. [Google Scholar] [CrossRef] [PubMed]
- von Bertalanffy, L. General Systems Theory; Allen Lane The Penguin Press: London, UK, 1968. [Google Scholar]
- Fontana, W.; Buss, L. The Barrier of Objects: From Dynamical Systems to Bounded Organizations. In Boundaries and Barriers; Casti, J.L., Karlqvist, A., Eds.; Addison-Wesley Publishing Company, Inc.: Reading, MA, USA, 1996; pp. 55–115. [Google Scholar]
- Letelier, J.C.; Soto-Andrade, J.; Guíñez Abarzúa, F.; Cornish-Bowden, A.; Luz Cárdenas, M. Organizational invariance and metabolic closure: Analysis in terms of systems. J. Theor. Biol. 2006, 238, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A. Tibor Gánti and Robert Rosen: Contrasting approaches to the same problem. J. Theor. Biol. 2015, 381, 6–10. [Google Scholar] [CrossRef] [PubMed]
- de la Escosura, A.; Briones, C.; Ruiz-Mirazo, K. The systems perspective at the crossroads between chemistry and biology. J. Theor. Biol. 2015, 381, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Montévil, M.; Mossio, M. Biological Organization as Closure of Constraints. J. Theor. Biol. 2015, 372, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.; di Fenizio, P.S. Chemical Organisation Theory. Bull. Math. Biol. 2007, 69, 1199–1231. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A. At Home in the Universe: The Search for Laws of Self-Organization and Complexity; Oxford University Press: New York, NY, USA; Oxford, UK, 1995. [Google Scholar]
- Kauffman, S.A. The Origins of Order: Self-Organization and Selection in Evolution; Oxford University Press: New York, NY, USA; Oxford, UK, 1993. [Google Scholar]
- Rashevsky, N. Life, information theory, and topology. Bull. Math. Biol. 1955, 17, 229–235. [Google Scholar] [CrossRef]
- Atlan, H. Application of information theory to the study of the stimulating effects of ionizing radiation, thermal energy, and other environmental factors. Preliminary ideas for a theory of organization. J. Theor. Biol. 1968, 21, 45–70. [Google Scholar] [CrossRef]
- Walker, I. The Evolution of Biological Organization as a Function of Information; Editora INPA: Manaus, Brazil, 2005. [Google Scholar]
- Rashevsky, N. Topology and life: In search of general mathematical principles in biology and sociology. Bull. Math. Biol. 1954, 16, 317–348. [Google Scholar] [CrossRef]
- Atlan, H. On a formal definition of organization. J. Theor. Biol. 1974, 45, 295–304. [Google Scholar] [CrossRef]
- Pahl-Wostl, C. The Dynamic Nature of Ecosystems, Chaos and Order Entwined; John Wiley & Sons: Chichester, UK, 1995. [Google Scholar]
- Maturana, H. The Organization of the Living: A Theory of the Living Organization. Int. J. Hum. Comput. Stud. 1999, 51, 149–168. [Google Scholar] [CrossRef]
- Louie, A.H. (M,R)-Systems and their Realizations. Axiomathes 2006, 16, 35–64. [Google Scholar] [CrossRef]
- Kineman, J.J. Relational Science: A Synthesis. Axiomathes 2011, 21, 393–437. [Google Scholar] [CrossRef]
- Baas, N.A. On structure and organization: An organizing principle. Int. J. Gen. Syst. 2013, 42, 170–196. [Google Scholar] [CrossRef]
- Baas, N.A. On higher structures. Int. J. Gen. Syst. 2016, 45, 747–762. [Google Scholar] [CrossRef]
- Hellerman, L. The Animate—Inanimate Relationship. Int. J. Gen. Syst. 2016, 45, 734–746. [Google Scholar] [CrossRef]
- Baas, N.A. Self-organisation and Higher Order Structures. In Self-organisation of Complex Structures: From Individual to Collective Dynamics; Schweitzer, F., Ed.; CRC Press: Boca Raton, FL, USA, 1997; pp. 71–81. [Google Scholar]
- Bohr, N.H.D. Light and Life. Nature 1933, 131, 457–459. [Google Scholar] [CrossRef]
- Maynard Smith, J. The Concept of Information in Biology. Philos. Sci. 2000, 67, 177–194. [Google Scholar] [CrossRef]
- Adami, C. Information theory in molecular biology. Phys. Life Rev. 2004, 1, 3–22. [Google Scholar] [CrossRef]
- Atlan, H.; Cohen, I.R. Self-organization and meaning in immunology. In Self-Organization and Emergence in Life Sciences; Feltz, B., Crommelinck, M., Goujon, P., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 121–139. [Google Scholar]
- Hauhs, M.; Lange, H. Ecosystem dynamics viewed from an endoperspective. Sci. Total Environ. 1996, 183, 125–136. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variaton in the History of Life. In Life and Mind: Philosophical Issues in Biology and Psycology, A Bradford Book; The MIT Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Roederer, J.G. Information and its Role in Nature; The Frontiers Collection; Springer: Berlin, Germany, 2005. [Google Scholar]
- Adamatzky, A.; Armstrong, R.; Jones, J.; Gunji, Y.P. On creativity of slime mould. Int. J. Gen. Syst. 2013, 42, 441–457. [Google Scholar] [CrossRef]
- Chaitin, G. To a Mathematical Definition of ‘LIFE’. SIGACT News, 1970; 4, 12–18. [Google Scholar] [CrossRef]
- Chaitin, G.J. Toward a Mathematical Definition of “Life”. In Maximum Entropy Formalism; Levine, R.D., Tribus, M., Eds.; M.I.T. Press: Cambridge, MA, USA, 1979; pp. 477–498. [Google Scholar]
- Pattee, H.H. Simulations, Realizations, and Theories of Life. In Artificial Life; Number VI in SFI Series in the Sciences of Complexity; Langton, C., Ed.; Addison-Wesley Publishing Company, Inc.: Redwood City, CA, USA, 1989; pp. 63–77. [Google Scholar]
- Atlan, H.; Koppel, M. The cellular computer DNA: Program or data. Bull. Math. Biol. 1990, 52, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Emmeche, C. The Computational Notion of Life. Theoria 1994, 9, 1–30. [Google Scholar]
- Griffiths, P.E. Genetic Information: A Metaphor in Search of a Theory. Philos. Sci. 2001, 68, 394–412. [Google Scholar] [CrossRef]
- Jablonka, E. Information: Its interpretation, its inheritance, and its sharing. Philos. Sci. 2002, 69, 578–605. [Google Scholar] [CrossRef]
- Thaller, B. Advanced Visual Quantum Mechanics; Springer Science+Business Media Inc.: New York, NY, USA, 2005. [Google Scholar]
- Scott Kelso, J.A. Dynamic Patterns: The Self-Organization of Brain and Behaviour; The MIT Press, A Bradford Book: Cambridge, MA, USA, 1999. [Google Scholar]
- Bruggeman, F.J.; Westerhoff, H.V.; Boogerd, F.C. BioComplexity: A pluralist research strategy is necessary for a mechanistic explanation of the “live” state. Philos. Psychol. 2002, 15, 411–440. [Google Scholar] [CrossRef]
- Lloyd, E.K. Counting Isomers and Isomerizations. In Graph Theory and Its Applications: East and West, Proceedings of the First China_USA International Graph Theory Conference, Jinan, China, June 9–20, 1986; Annals of the New York Academy of Sciences; Capobianco, M.F., Guan, M., Hsu, D.F., Tian, F., Eds.; The New York Academy of Sciences: New York, NY, USA, 1989; Volume 576, pp. 377–384. [Google Scholar]
- Dobrowolski, J.C. The chiral graph theory. MATCH Commun. Math. Comput. Chem. 2015, 73, 347–374. [Google Scholar]
- Barwise, J. Admissible Sets and Structures; Springer: Berlin, Germany, 1975. [Google Scholar]
- Kritz, M.V. On Biology and Information; P&D Report 25/91; LNCC/MCTI: Petrópolis, Brazil, 1991. [Google Scholar]
- Hartwell, L.H.; Hopfield, J.J.; Leibler, S.; Murray, A.W. From molecular to modular cell biology. Nature 1999, 402, C47–C52. [Google Scholar] [CrossRef] [PubMed]
- Berge, C. Graphs and Hypergraphs; North-Holland: Amsterdam, The Netherlands, 1973. [Google Scholar]
- Schmidt, G.; Ströhlein, T. Relations and Graphs: Discrete Mathematics for Computer Scientists; EACTS Monagraphs on Theoretical Computer Science; Springer: Berlin, Germany, 1993. [Google Scholar]
- Klamt, S.; Haus, U.U.; Theis, F. Hypergraphs and Cellular Networks. PLoS Comput. Biol. 2009, 5, e1000385. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, M.T.; Kritz, M.V. On the Hierarchical Organization of Metabolic Networks: An Underlying Mathematical Model. In Proceedings of the International Symposium on Mathematical and Computational Biology (BIOMAT 2005); Mondaini, R., Dilão, R., Eds.; E-papers Serviços Editoriais Ltda.: Rio de Janeiro, Brazil, 2006; pp. 221–241. [Google Scholar]
- Manna, Z. The Mathematical Theory of Computation; McGraw-Hill Co.: New York, NY, USA, 1974. [Google Scholar]
- Bailly, A. Dictionnaire Grec Français, 26th ed.; Librairie Hachette: Paris, France, 1963; (1e Édition, 1894). [Google Scholar]
- Arnol’d, V.I. Mathematical Methods of Classical Mechanics; Vol. 60, Graduate Texts in Mathematics; Springer: Berlin, Germany, 1978. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Kritz, M.V. On Relations between i-graphs and Data Structures. Logique Analyse 1996, 39, 153–164. [Google Scholar]
- Kritz, M.V. Biological Organizations. In Proceedings of the IV Brazilian Symposium on Mathematical and Computational Biology (BIOMAT IV); Mondaini, R., Ed.; BIOMAT Consortium, E-papers Serviços Editoriais Ltda.: Rio de Janeiro, Brazil, 2005; Volume 2, pp. 89–103. [Google Scholar]
- Mitchell, A.; Romano, G.H.; Groisman, B.; Yona, A.; Dekel, E.; Kupiec, M.; Dahan, O.; Pilpel, Y. Adaptive prediction of environmental changes by microorganisms. Nature 2009, 460, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, J.K.; Lee, A.S.Y.; Engelman, A.; Doudna, J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 2015, 519, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef] [PubMed]
- Stonier, T. Information and the Internal Structure of the Universe; Springer: London, UK, 1990. [Google Scholar]
- Fenzl, N.; Hofkirchner, W. Information Processing in Evolutionary Systems. In Self-organisation of Complex Structures: From Individual to Collective Dynamics; Schweitzer, F., Ed.; CRC Press: Boca Raton, FL, USA, 1997; pp. 59–70. [Google Scholar]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Hoath, S.B. Considerations on the Role of the Skin as the Boundary of an Autopoietic System. In Proceedings of the Biology, Language, Cognition and Society: International Symposium on Autopoiesis, Belo Horizonte, Brazil, 18–21 November 1997.
- Santra, T.; Kolch, W.; Kholodenko, B.N. Navigating the multilayered organization of eukaryotic signaling: A new trend in data integration. PLoS Comput. Biol. 2014, 10, e1003385. [Google Scholar] [CrossRef] [PubMed]
- Nadin, M. Anticipation and dynamics: Rosen’s anticipation in the perspective of time. Int. J. Gen. Syst. 2010, 39, 3–33. [Google Scholar] [CrossRef]
- Iyer, K.V.; Pulford, S.; Mogilner, A.; Shivashankar, G.V. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys. J. 2012, 103, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, V.B. Moody wallpaper. Protein Spotlight 2003, 33. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.616.8546&rep=rep1&type=pdf (accessed on 1 March 2017). [Google Scholar]
- Ai, C.; Li, Y.; Wang, Y.; Chen, Y.; Yang, L. Insight into the effects of chiral isomers quinidine and quinine on CYP2D6 inhibition. Bioorg. Med. Chem. Lett. 2009, 19, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Levinson, H.; Mori, K. The Pheromone Activity of Chiral Isomers of Trogodermal for Male Khapra Beetles. Naturwissenschaften 1980, 67, 148–149. [Google Scholar] [CrossRef]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.A. The Sciences of the Artificial, 3rd ed.; The MIT Press: Cambridge, MA, USA, 1996. [Google Scholar]
- Lenneberg, E.H. Biological Foundations of Language, reprint ed.; Robert E. Krieger Publ. Co.: Malabar, FL, USA, 1984; (Original edition and copyright: John Wiley & Sons, Inc., 1967.). [Google Scholar]
- Casti, J.L.; Karlqvist, A. (Eds.) Complexity, Language, and Life: Mathematical Approaches; Vol. 16, Lecture Notes in Biomathematics; Springer: Berlin, Germany, 1986.
- Weinberg, G.M. An Introduction to General Systems Thinking; Dorset Hause Publishing: New York, NY, USA, 2001. [Google Scholar]
- Bosch, O.; Maani, K.; Smith, C. Systems Thinking—Language of Complexity for Scientists and Managers; The University of Queensland: Queensland, Australia, 2007; pp. 57–66. [Google Scholar]
- Corning, P.A.; Szathmáry, E. “Synergistic selection”: A Darwinian frame for the evolution of complexity. J. Theor. Biol. 2015, 371, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Zurek, W.H. Complexity, Entropy, and the Physics of Information. In Santa Fe Institute Studies in the Sciences of Complexity; Perseus Publishing: Cambridge, MA, USA, 1990. [Google Scholar]
- Watkins, J.J. Number Theory: A Historical Approach; Princeton University Press: Princeton, NJ, USA; Oxford, UK, 2014. [Google Scholar]
- Rosen, R. Complexity and System Descriptions. In Systems, Approaches, Theories, Applications; Hartnett, W., Ed.; D. Reidel Publishing Co.: Dordrecht-Holland, The Netherlands, 1977; pp. 169–178. [Google Scholar]
- Shao, X.; Li, Q.; Mogilner, A.; Bershadsky, A.D.; Shivashankar, G.V. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl. Acad. Sci. USA 2015, 112, E2595–E2601. [Google Scholar] [CrossRef] [PubMed]
- Ditlev, J.A.; Mayer, B.J.; Loew, L.M. There is More Than One Way to Model an Elephant. Experiment-DrivenModeling of the Actin Cytoskeleton. Biophys. J. 2013, 104, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Erban, R.; Flegg, M.B.; Papoian, G.A. Multiscale Stochastic Reaction–Diffusion Modeling: Application to Actin Dynamics in Filopodia. Bull. Math. Biol. 2013, 76, 799–818. [Google Scholar] [CrossRef] [PubMed]
- Mogilner, A.; Rubinstein, B. The physics of filopodial protrusion. Biophys. J. 2005, 89, 782–795. [Google Scholar] [CrossRef] [PubMed]
- De Moura, C.A.; Kritz, M.V.; Leal, T.F.; Prokop, A. Mathematical-computational Simulation of Cytoskeletal Dynamics. In Modeling and Computational Intelligence in Engineering Applications; Santiago, O.L., da Silva-Neto, A.J., Silva, G., Eds.; Springer: Berlin, Germany, 2016. [Google Scholar]
- Callebaut, W.; Rasskin-Gutman, D. Modularity: Understanding the Development and Evolution of Natural Complex Systems; The Viena Series in Theoretical Biology; The MIT Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Simon, H.A. The Architecture of Complexity. Proc. Am. Philos. Soc. 1962, 106, 467–485. [Google Scholar]
- Ehret, C.F. Organelle Systems and Biological Organization. Science 1960, 132, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Herrgård, M.J.; Swainston, N.; Dobson, P.; Dunn, W.B.; Arga, K.Y.; Arvas, M.; Büthgen, N.; Borger, S.; Costenoble, R.; Heinemann, M.; et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 2008, 26, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.; Dubois, J.; Bourqui, R. Pathway Preserving Representation of Metabolic Networks. Comput. Graph. Forum 2011, 30, 1021–1030. [Google Scholar] [CrossRef]
- Kritz, M.V.; dos Santos, M.T.; Urrutia, S.; Schwartz, J.M. Organizing Metabolic Networks: Cycles in Flux Distribution. J. Theor. Biol. 2010, 265, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Keung, A.J.; Khalil, A.S. A unifying model of epigenetic regulation. Science 2016, 351, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Breinig, F.; Sendzik, T.; Eisfeld, K.; Schmitt, M.J. Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc. Natl. Acad. Sci. USA 2006, 103, 3810–3815. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Breinig, F. Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded rna virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, L.A.; Polak, E. System Theory, Vol. 8, Inter-University Electronics Series; TATA McGraw-Hill Publishing Co. Ltd.: Bombay/New Delhi, India, 1969. [Google Scholar]
- Louie, A.H. Anticipation in (M,R)-systems. Int. J. Gen. Syst. 2012, 41, 5–22. [Google Scholar] [CrossRef]
- Burstein, G.; Negoita, C.; Kranz, M. Postmodern Fuzzy System Theory: A Deconstruction Approach Based on Kabbalah. Systems 2014, 2, 590–605. [Google Scholar] [CrossRef]
- Igamberdiev, A.U. Anticipatory dynamics of biological systems: From molecular quantum states to evolution. Int. J. Gen. Syst. 2015, 44, 631–641. [Google Scholar] [CrossRef]
- Kritz, M.V. Biological Information and Knowledge; Relatório de P&D 23/2009; LNCC/MCT: Petrópolis, Brazil, 2009. [Google Scholar]
- Kritz, M.V. Biological Organization, Biological Information, and Knowledge. bioRxiv 2014, 2014, 012617. [Google Scholar]
- Johnson, J. A Theory of Stars in Complex Systems. In Complexity, Language, and Life: Mathematical Approaches; Lecture Notes in Biomathematics; Casti, J.L., Karlqvist, A., Eds.; Springer: New York, NY, USA, 1986; Volume 16, pp. 21–61. [Google Scholar]
- Bergareche, A.M.; Ruiz-Mirazo, K. Metabolism and the problem of its universalization. BioSystems 1999, 49, 45–61. [Google Scholar] [CrossRef]
- Holland, J.H. Studying complex adaptive systems. J. Syst. Sci. Complex. 2006, 19, 1–8. [Google Scholar] [CrossRef]
- Letelier, J.C.; Cárdenas, M.L.; Cornish-Bowden, A. From L’Homme Machine to metabolic closure: Steps towards understanding life. J. Theor. Biol. 2011, 286, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Emmeche, C. A Bio-semiotic Note on Organisms, animals, machines, Cyborgs, and the Quasi-autonomy of Robots. Pragmat. Cogn. 2007, 15, 455–483. [Google Scholar]
- Luz Cárdenas, M.; Letelier, J.C.; Gutiérrez, C.; Cornish-Bowden, A.; Soto-Andrade, J. Closure to efficient causation, computability and artificial life. J. Theor. Biol. 2010, 263, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Villani, M.; Filisetti, A.; Graudenzi, A.; Damiani, C.; Carletti, T.; Serra, R. Growth and Division in a Dynamic Protocell Model. Life 2014, 4, 837–864. [Google Scholar] [CrossRef] [PubMed]
- Shrager, J. The fiction of function. Bioinformatics 2003, 19, 1934–1936. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, J. Complex Adaptive Systems; CIS Technical Report 070302A; Complex Intelligent Systems Laboratory, Centre for Information Technology Research: Melbourne, Australia, 2007. [Google Scholar]
- Wolkenhauer, O. Systems biology: The reincarnation of systems theory applied in biology? Brief. Bioinform. 2001, 2, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E. Founder of systems chemistry and foundational theoretical biologist: Tibor Gánti (1933-2009). J. Theor. Biol. 2015, 381, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Lazebnik, Y. Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell 2002, 2, 179–182. [Google Scholar] [CrossRef]
- Boogerd, F.; Bruggeman, F.; Jonker, C.; de Jong, H.L.; Tamminga, A.; Treur, J.; Westerhoff, H.; Wijngaards, W. Inter-level relations in computer science, biology, and psychology. Philos. Psychol. 2002, 15, 463–471. [Google Scholar] [CrossRef]
- Mayr, E. This Is Biology: The Science of the Living World; Belknap Press/Havard University Press: Cambridge, MA, USA, 1997. [Google Scholar]
- Kitto, K. A Contextualised General Systems Theory. Systems 2014, 2, 541–565. [Google Scholar] [CrossRef]
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).