Fluxomics of the Eastern Oyster for Environmental Stress Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Comparison of the Perchloric Acid versus Methanol Extraction Methods Applied to Oyster Tissues
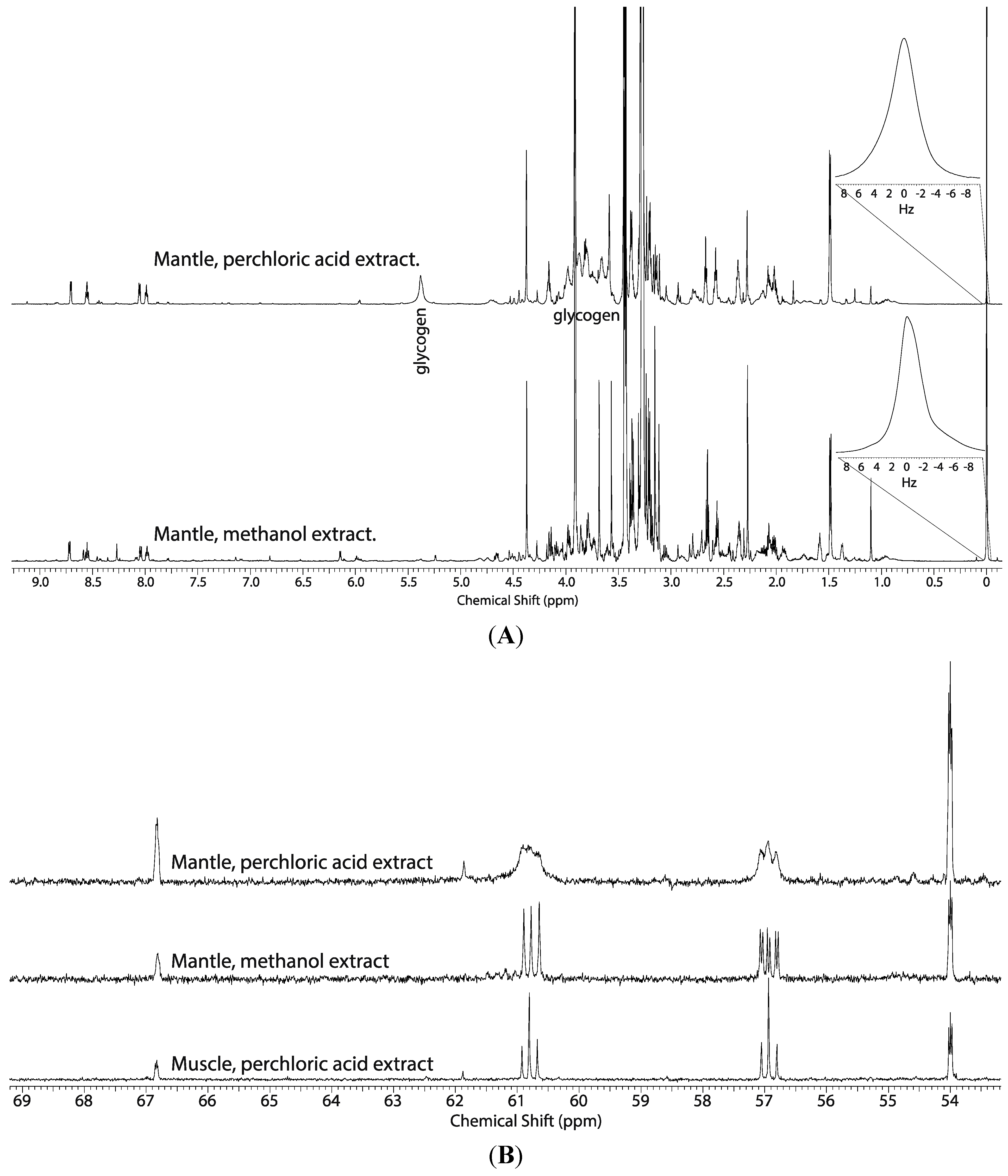
2.2. Tissue Distribution of 2-13C/15N-Glycine and U-13C-Glucose in the Four Tissues and Treatments
| Treatment | N | Total * | Muscle | Gills | Stomach | Mantle |
|---|---|---|---|---|---|---|
| Weight ± SD, g (%13C Ala ± SD) | Weight ± SD, g (%13C Ala ± SD) | Weight ± SD, g (%13C Ala ± SD) | Weight ± SD, g (%13C Ala ± SD) | Weight ± SD, g (%13C Ala ± SD) | ||
| Control | 4 | 56.3 ± 13.2 | 0.43 ± 0.12 | 0.67 ± 0.10 | 1.26 ± 0.64 | 1.60 ± 0.59 |
| - | (ND) | (ND) | (ND) | (ND) | ||
| 2-13C/15N-Glycine | 3 | 53.5 ± 13.5 | 0.45 ± 0.18 | 1.03 ± 0.35 | 1.16 ± 0.11 | 1.39 ± 0.69 |
| - | (ND) | (ND) | (ND) | (ND) | ||
| U-13C-Glucose | 4 | 55.5 ± 21.5 | 0.42 ± 0.23 | 0.63 ± 0.26 | 1.00 ± 0.65 | 0.89 ± 0.31 |
| - | (23% ± 3%) | (20% ± 3%) | (23% ± 10%) | (15% ± 2%) | ||
| 2-13C/15N-Glycine + U-13C-Glucose | 4 | 56 ± 10.8 | 0.48 ± 0.15 | 0.95 ± 0.12 | 1.23 ± 0.39 | ± 0.42 |
| - | (18% ± 2%) | (16% ± 2%) | (25% ± 12%) | (13% ± 2%) |
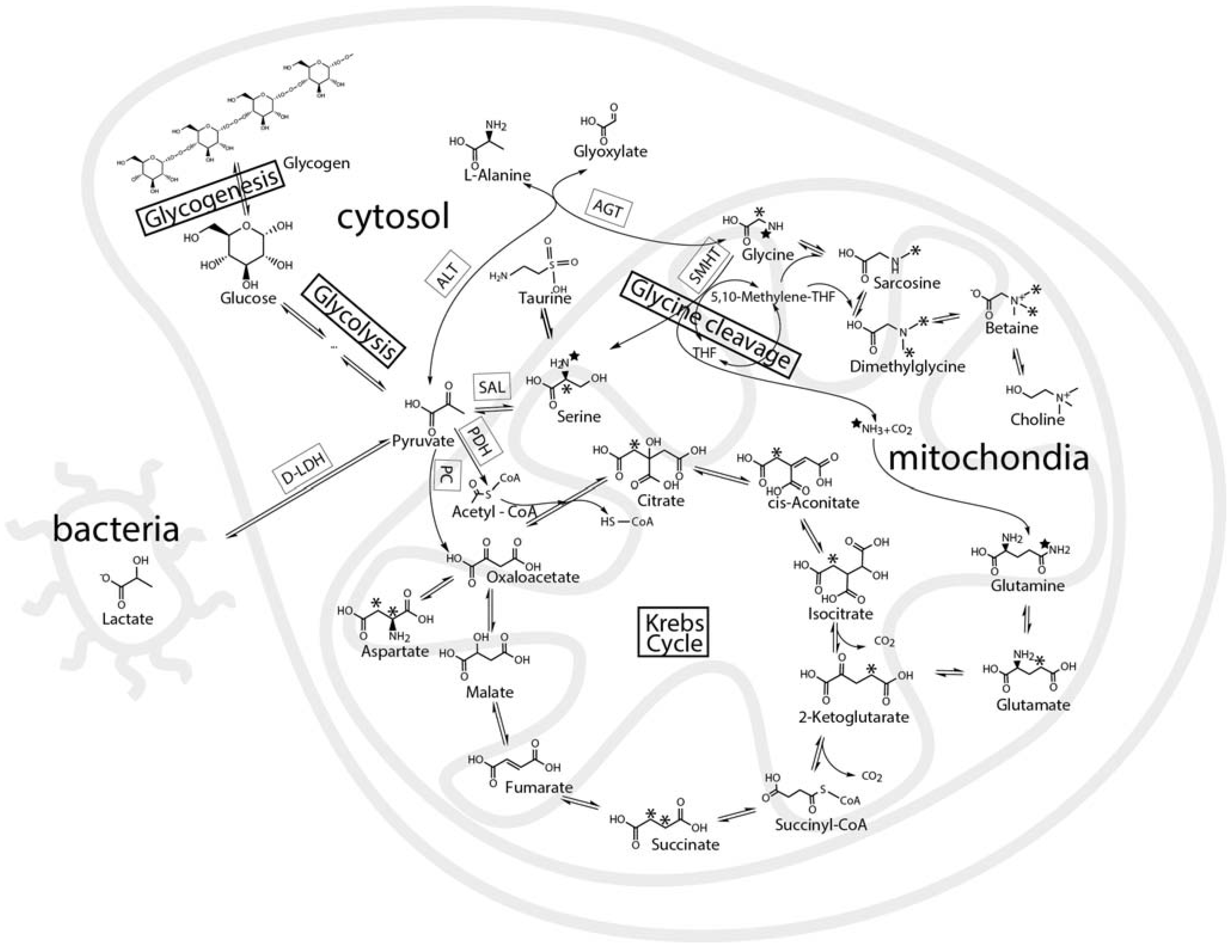
2.3. Mass Balance of 13C-labeled Metabolites within Each of the Tissue Blocks
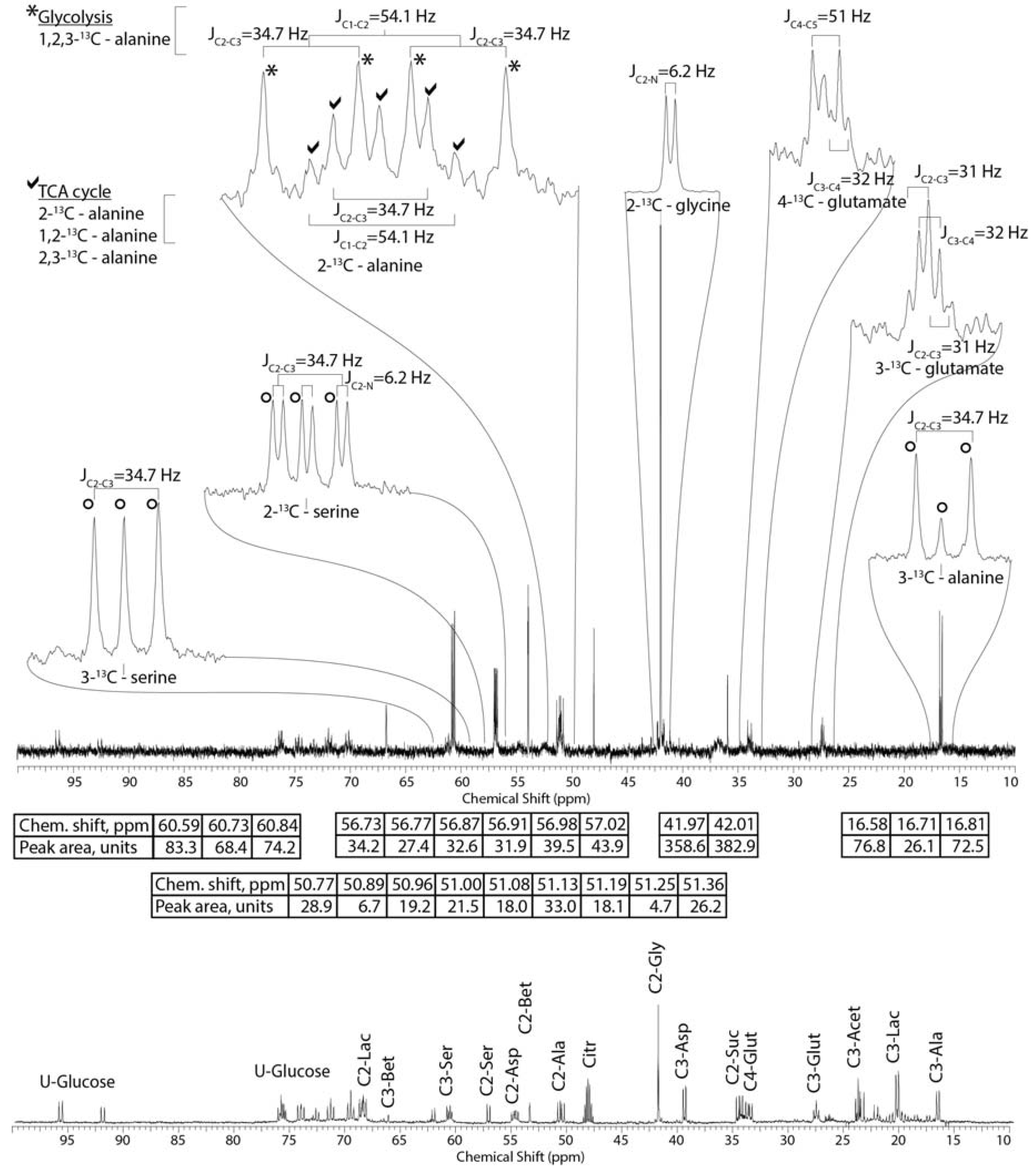
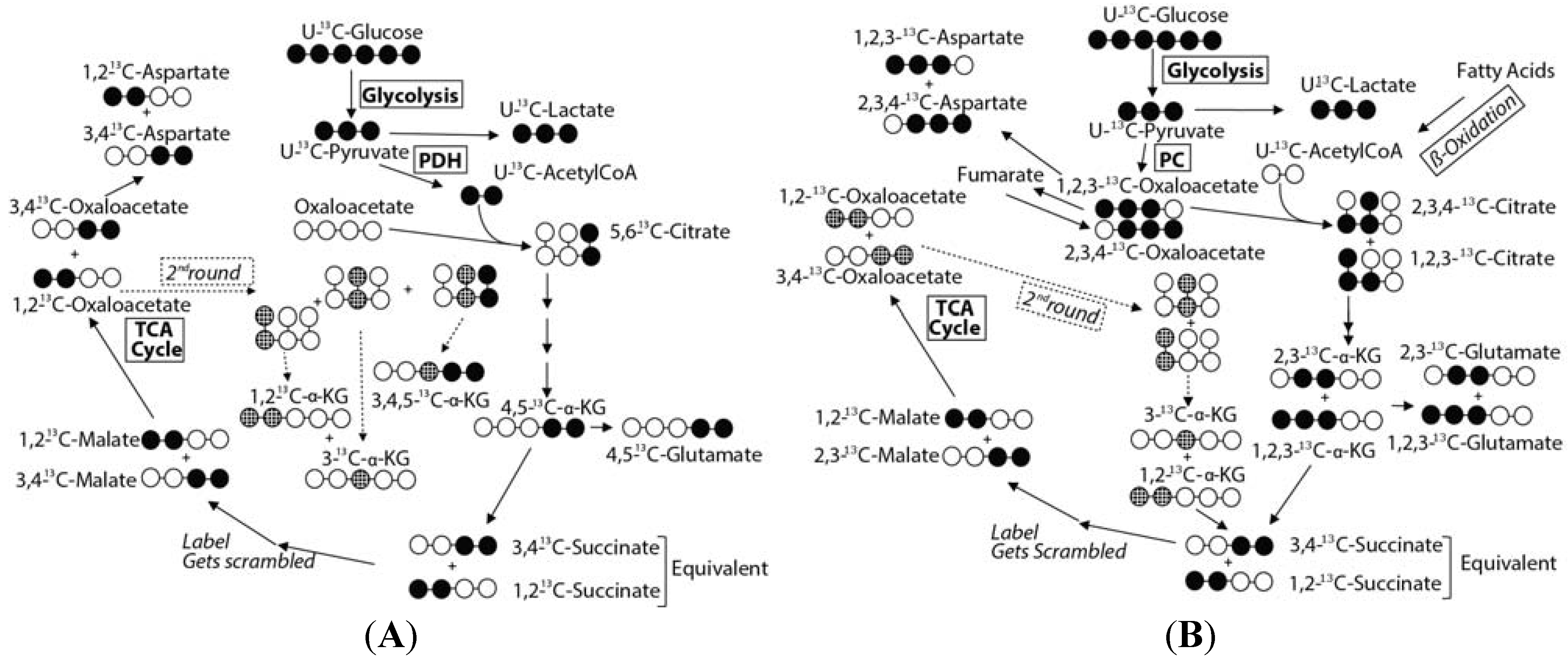
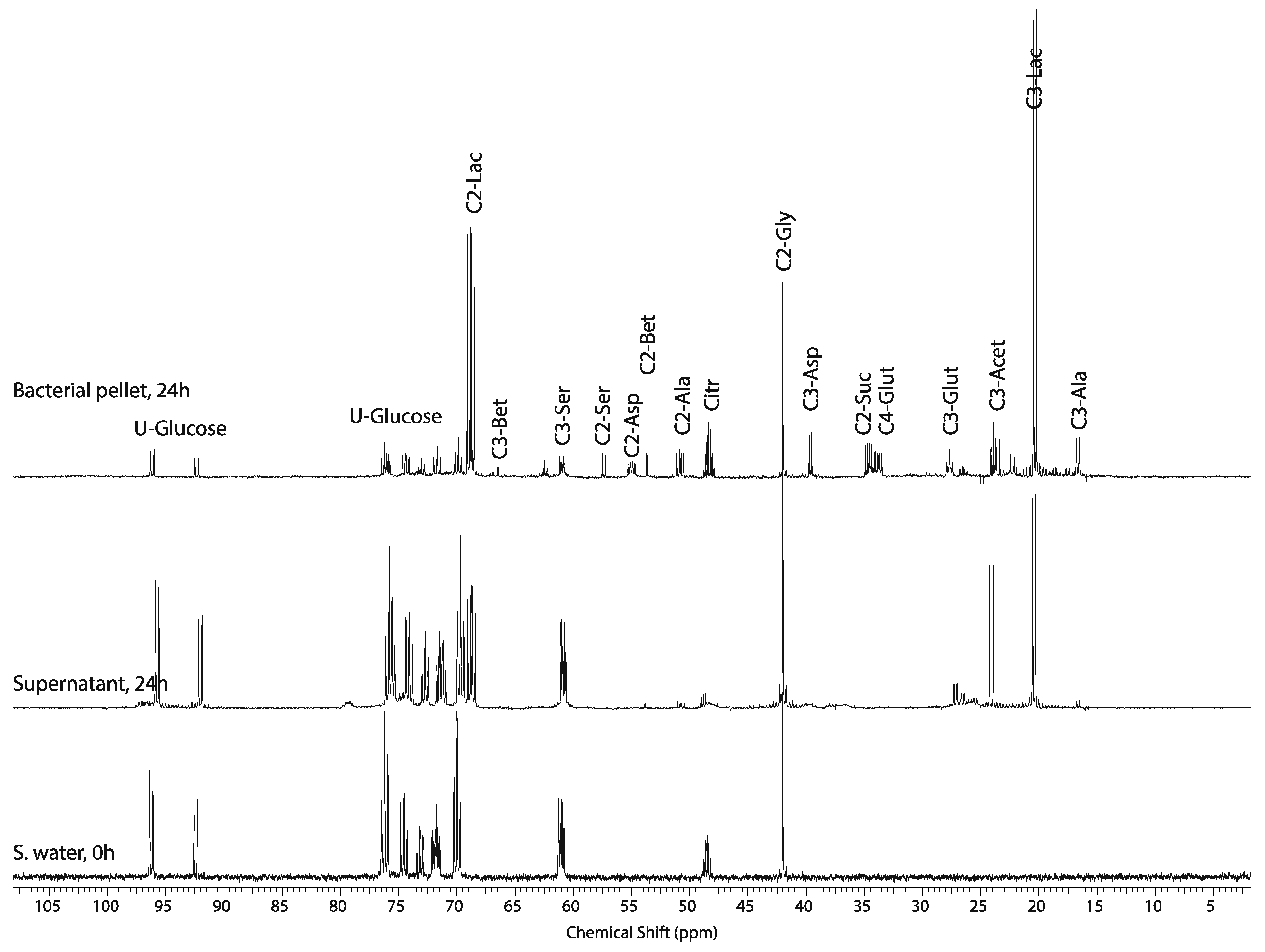
2.4. Effect of Hypoxic Stress on the Oyster Mass Balance

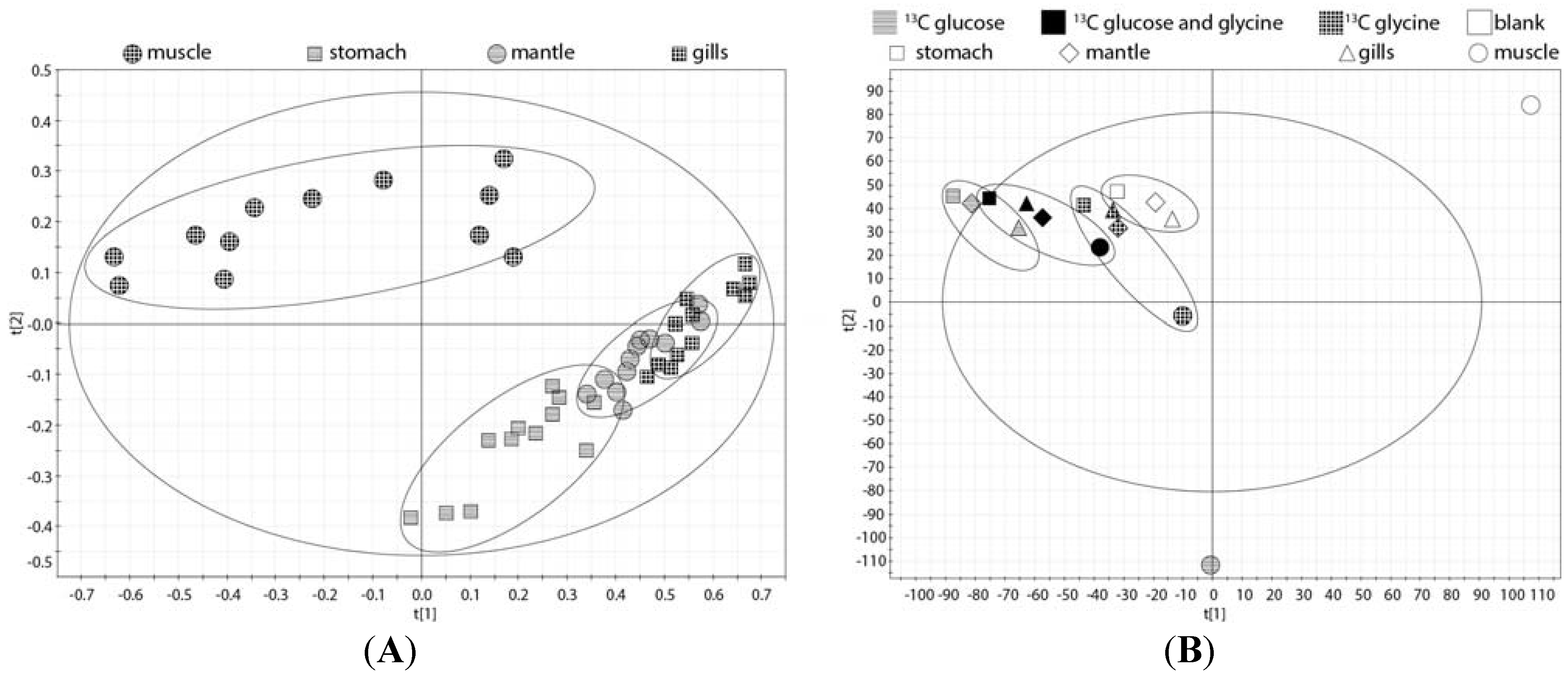
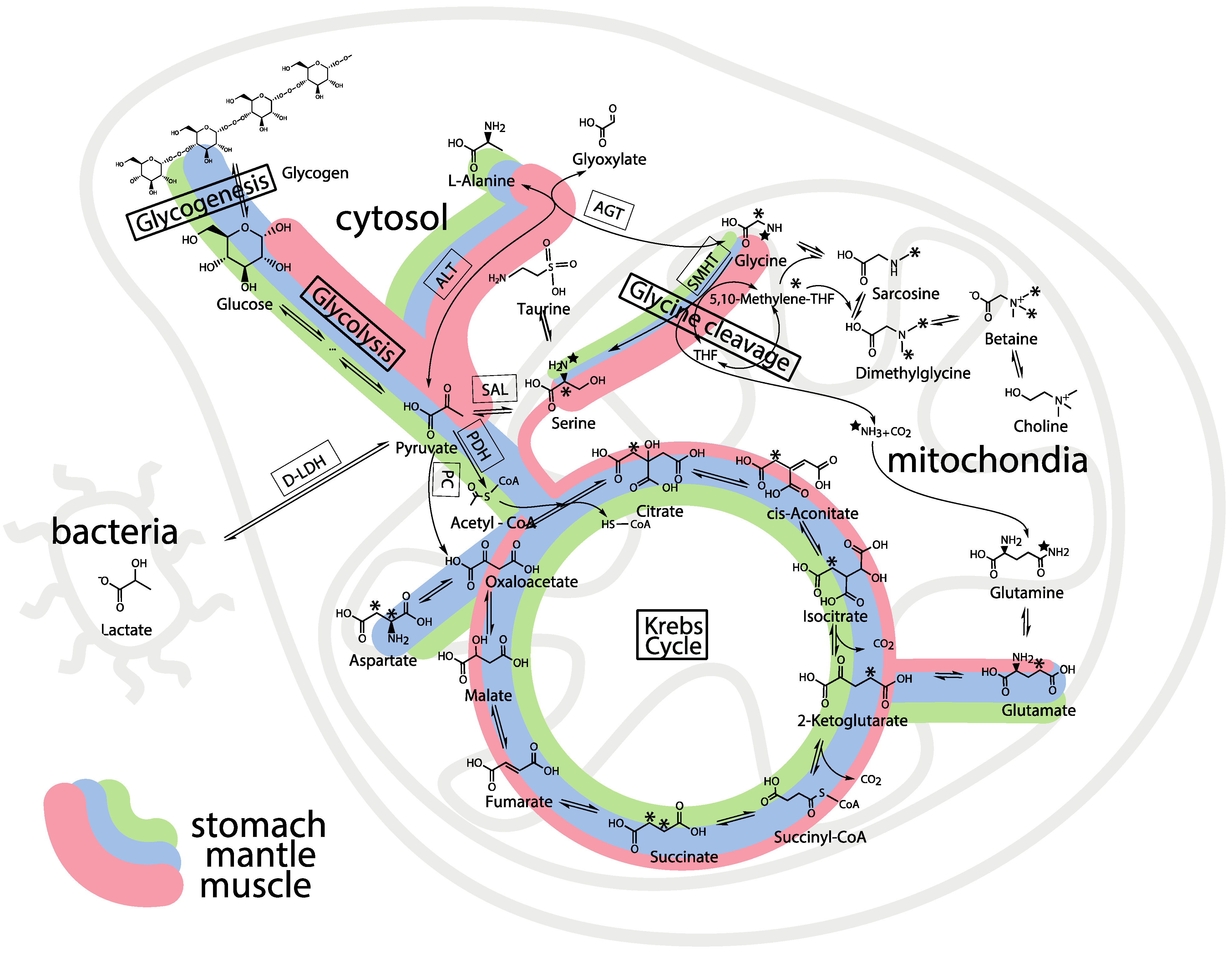
2.5. Whole Body Mass Balance Phenotyping (WoMBaP) Summary Plot
3. Experimental Section
3.1. Acquisition and Husbandry of Oysters
3.2. WoMBaP Experimental Procedure
3.3. Dissection and Extraction of Oyster Tissue Blocks
3.4. Emergence and Crushed Oyster Experimental Procedure
3.5. 1H and 13C NMR Spectroscopy of Extracted Tissue Blocks
3.6. Analysis of 1H and 13C NMR Spectra and Generation of WoMBaP Summary Plots
4. Conclusions
Acknowledgments
References
- Smart, R.C.; Hodgeson, E. Molecular and Biochemical Toxicology, 4th ed.; Wiley, John & Sons, Incorporated: Hoboken, NJ, USA, 2008; p. 948. [Google Scholar]
- Klaassen, C.D. Casarett & Doull’s Toxicology: The Basic Science of Poisons, 7th ed.; McGraw-Hill Professional: New York, NY, USA, 2007; p. 1280. [Google Scholar]
- Lee, H.; Tikunov, A.; Stoskopf, M.K.; Macdonald, J.M. Applications of chemical shift imaging to marine sciences. Mar. Drugs 2010, 8, 2369–2383. [Google Scholar] [CrossRef]
- Tikunov, A.P.; Winnike, J.H.; Tech, K.; Jeffries, R.E.; Semelka, C.T.; Martin, J.; McClelland, R.; Graves, L.M.; Macdonald, J.M. Fluxomics by NMR spectroscopy from cells to organisms focusing on liver. Curr. Metabolomics 2013, 1, 128–159. [Google Scholar] [CrossRef]
- Tikunov, A.; Johnson, C.B.; Lee, H.; Stoskopf, M.K.; Macdonald, J.M. Metabolomic investigations of American oysters Using 1H-NMR spectroscopy. Mar. Drugs 2010, 8, 2578–2596. [Google Scholar] [CrossRef]
- Fan, T.W.; Higashi, R.M.; Macdonald, J.M. Emergence and recovery response of phosphate metabolites and intracellular pH in intact Mytilus edulis as examined in situ by in vivo 31P-NMR. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 1991, 1092, 39–47. [Google Scholar] [CrossRef]
- Higashi, R.M.; Fan, T.W.-M.; Macdonald, J.M. Monitoring of metabolic responses of intact Haliotis (abalones) under salinity stress by 31P surface probe localized NMR. J. Exp. Zool. 1989, 249, 350–356. [Google Scholar] [CrossRef]
- Hines, A.; Oladiran, G.S.; Bignell, J.P.; Stentiford, G.D.; Viant, M.R. Direct sampling of organisms from the field and knowledge of their phenotype: Key recommendations for environmental metabolomics. Environ. Sci. Technol. 2007, 41, 3375–3381. [Google Scholar] [CrossRef]
- Jones, O.A.H.; Spurgeon, D.J.; Svendsen, C.; Griffin, J.L. A metabolomics based approach to assessing the toxicity of the polyaromatic hydrocarbon pyrene to the earthworm Lumbricus rubellus. Chemosphere 2008, 71, 601–609. [Google Scholar] [CrossRef]
- Tuffnail, W.; Mills, G.A.; Cary, P.; Greenwood, R. An environmental 1H NMR metabolomic study of the exposure of the marine mussel Mytilus edulis to atrazine, lindane, hypoxia and starvation. Metabolomics 2009, 5, 33–43. [Google Scholar] [CrossRef]
- Viant, M.R.; Bundy, J.G.; Pincetich, C.A.; de Ropp, J.S.; Tjeerdema, R.S. NMR-derived developmental metabolic trajectories: An approach for visualizing the toxic actions of trichloroethylene during embryogenesis. Metabolomics 2005, 1, 149–158. [Google Scholar] [CrossRef]
- Macdonald, J.M.; Schmidlin, O.; James, T.L. In vivo monitoring of hepatic glutathione in anesthetized rats by 13C NMR. Magn. Reson. Med. 2002, 48, 430–439. [Google Scholar] [CrossRef]
- Jo, P.G.; Choi, Y.K.; Choi, C.Y. Cloning and mRNA expression of antioxidant enzymes in the Pacific oyster, Crassostrea gigas in response to cadmium exposure. Comp. Biochem. Physiol. Part C 2008, 147, 460–469. [Google Scholar]
- Ivanina, A.V.; Sokolov, E.P.; Sokolova, I.M. Effects of cadmium on anaerobic energy metabolism and mRNA expression during air exposure and recovery of an intertidal mollusk Crassostrea virginica. Aquat. Toxicol. 2010, 99, 330–342. [Google Scholar] [CrossRef]
- Johnson, C.B.; Tikunov, A.P.; Lee, H.; Wolak, J.E.; Pediaditakis, P.; Romney, D.; Holmuhamedov, E.; Gamcsik, M.P.; Macdonald, J.M. 13C MRS detection of changes in serine isotopomers reflects changes in mitochondrial redox status. Magn. Reson. Med. 2012, 68, 671–679. [Google Scholar] [CrossRef]
- Meng, J.; Zhu, Q.; Zhan, L.; Li, C.; Li, L.; She, Z.; Huang, B.; Zhang, G. Genome and transcriptome analyses provide insight into the euryhaline adaptation mechanism of Crassostrea gigas. PLoS One 2013, 8, e58563. [Google Scholar]
- Cunningham, P.A.; Tripp, M.R. Accumulation, tissue distribution and elimination of 203HgCl2 and CH3 203HgCl in the tissues of the American oyster Crassostrea virginica. Mar. Biol. 1975, 31, 321–334. [Google Scholar] [CrossRef]
- Denton, G.R.W.; Burdon-Jones, C. Influence of temperature and salinity on the uptake, distribution and depuration of mercury, cadmium and lead by the black-lip oyster Saccostrea echinata. Mar. Biol. 1981, 64, 317–326. [Google Scholar]
- Martincié, D.; Nürnberg, N.W.; Stoeppler, M.; Branica, M. Bioaccumulation of heavy metals by bivalves from Lim Fjord (North Adriatic Sea). Mar. Biol. 1984, 81, 177–188. [Google Scholar] [CrossRef]
- Bryan, G.W.; Langston, W.J. Bioavailability, accumulation and effects of heavy metals in sediments with special reference to United Kingdom estuaries: A review. Environ. Pollut. 1992, 76, 89–131. [Google Scholar] [CrossRef]
- Kurochkin, I.O.; Ivanina, A.V.; Eilers, S.; Downs, C.A.; May, L.A.; Sokolova, I.M. Cadmium affects metabolic responses to prolonged anoxia and reoxygenation in eastern oysters (Crassostrea virginica). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, 1262–1272. [Google Scholar] [CrossRef]
- Paynter, K.T.; Karam, G.A.; Ellis, L.L.; Bishop, S.H. Subcellular distribution of aminotransferases, and pyruvate branch point enzymes in gill tissue from four bivalves. Comp. Biochem. Physiol. Part B: Comp. Biochem. 1985, 82, 129–132. [Google Scholar] [CrossRef]
- Donald, K.M.; Hawkins, A.J.S.; Smerdon, G.R. Transcript analysis of the genes encoding aminopeptidase N and alanine aminotransferase, two enzymes involved in protein turnover, in the pacific oyster, Crassotrea gigas. Comp. Biochem. Physiol. 2001, 128, 459–467. [Google Scholar] [CrossRef]
- Katz, J.; Lee, W.-N.; Wals, P.A.; Bergner, E.A. Studies of glycogen synthesis and the Krebs cycle by mass isotopomer analysis with [U-13C]Glucose in rats. J. Biol. Chem. 1989, 264, 12994–13001. [Google Scholar]
- Uehara, T.; Kosyk, O.; Jeannot, E.; Bradford, B.U.; Tech, K.; Macdonald, J.M.; Boorman, G.A.; Chatterjee, S.; Mason, R.P.; Melnyk, S.B.; et al. Acetaminophen-induced acute liver injury in HCV transgenic mice. Toxicol. Appl. Pharmacol. 2013, 266, 224–232. [Google Scholar] [CrossRef]
- Greenway, S.C.; Storey, K.B. Seasonal change and prolonged anoxia affect the kinetic properties of phosphofructokinase and pyruvate kinase in oysters. J. Comp. Physiol. 2000, 170, 285–293. [Google Scholar] [CrossRef]
- Le Moulla, G.; Bacca, H.; Huvet, A.; Moal, J.; Pouvreau, S.; van Wormhoudt, A. Transcriptional regulation of pyruvate kinase and phosphoenolpyruvate carboxykinase in the adductor muscle of the oyster Crassostrea gigas during prolonged hypoxia. J. Exp. Zool. 2007, 307, 371–382. [Google Scholar]
- Hammen, C.S. Metabolism of the oyster, Crassostrea virginica. Am. Zool. 1969, 9, 309–318. [Google Scholar]
- Lannig, G.; Eilers, S.; Pörtner, H.O.; Sokolova, I.M.; Bock, C. Impact of ocean acidification on energy metabolism of oyster, Crassostrea gigas-changes in metabolic pathways and thermal response. Mar. Drugs 2010, 8, 2318–2339. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, G.H.; Ivanina, A.V.; Matoo, O.B.; Pörtner, H.O.; Lannig, G.; Bock, C.; Beniash, E.; Sokolova, I.M. Interactive effects of salinity and elevated CO2 levels on juvenile eastern oysters, Crassostrea virginica. J. Exp. Biol. 2012, 215, 29–43. [Google Scholar] [CrossRef]
- Sussarellu, R.; Fabioux, C.; le Moullac, G.; Fleury, E.; Moraga, D. Transcriptomic response of the Pacific oyster Crassostrea gigas to hypoxia. Mar. Genomics 2010, 3, 133–143. [Google Scholar] [CrossRef]
- Macey, B.M.; Achilihu, I.O.; Burnett, K.G.; Burnett, L.E. Effects of hypercapnic hypoxia on inactivation and elimination of Vibrio campbellii in the Eastern oyster, Crassostrea virginica. Appl. Environ. Microbiol. 2008, 74, 6077–6084. [Google Scholar] [CrossRef]
- Chapman, R.W.; Mancia, A.; Beal, M.; Veloso, A.; Rathburn, C.; Blair, A.; Holland, A.F.; Warr, G.W.; Didinato, G.; Sokolova, I.M.; et al. The transcriptomic responses of the eastern oyster, Crassostrea virginica, to environmental conditions. Mol. Ecol. 2011, 20, 1431–1449. [Google Scholar] [CrossRef]
- David, E.; Tanguy, A.; Pichavant, K.; Moraga, D. Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 2005, 272, 5635–5652. [Google Scholar] [CrossRef]
- Dewar, B.J.; Keshari, K.; Jeffries, R.; Dzeja, P.; Graves, L.M.; Macdonald, J.M. Metabolic assessment of a novel chronic myelogenous leukemic cell line and an imatinib resistant subline by 1H NMR spectroscopy. Metabolomics 2010, 6, 439–450. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Colmer, T.D.; Lane, A.N.; Higashi, R.M. Determination of metabolites by 1H NMR and GC: Analysis for organic osmolytes in crude tissue extracts. Anal. Biochem. 1993, 214, 260–271. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Higashi, R.M.; Lane, A.N.; Jardetzky, O. Combined use of 1H-NMR and GC-MS for metabolite monitoring and in vivo 1H-NMR assignments. Biochim. Biophys. Acta 1986, 882, 154–167. [Google Scholar] [CrossRef]
- Szyperski, T. 13C-NMR, MS and metabolic flux balancing in biotechnology research. Q. Rev. Biophys. 1998, 31, 41–106. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tikunov, A.P.; Stoskopf, M.K.; Macdonald, J.M. Fluxomics of the Eastern Oyster for Environmental Stress Studies. Metabolites 2014, 4, 53-70. https://doi.org/10.3390/metabo4010053
Tikunov AP, Stoskopf MK, Macdonald JM. Fluxomics of the Eastern Oyster for Environmental Stress Studies. Metabolites. 2014; 4(1):53-70. https://doi.org/10.3390/metabo4010053
Chicago/Turabian StyleTikunov, Andrey P., Michael K. Stoskopf, and Jeffrey M. Macdonald. 2014. "Fluxomics of the Eastern Oyster for Environmental Stress Studies" Metabolites 4, no. 1: 53-70. https://doi.org/10.3390/metabo4010053




