A Comparative Analysis of MicroRNA Expression in Mild, Moderate, and Severe COVID-19: Insights from Urine, Serum, and Nasopharyngeal Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects and Study Design
2.2. Definitions
2.3. Data Collection and Sample Processing
2.4. Total RNA Extraction
2.5. SARS-CoV-2 Real-Time RT-PCR
2.6. MicroRNA Profile Expression
2.7. Data Analysis
3. Results
3.1. Demographic, Laboratory Markers and Clinical Symptoms
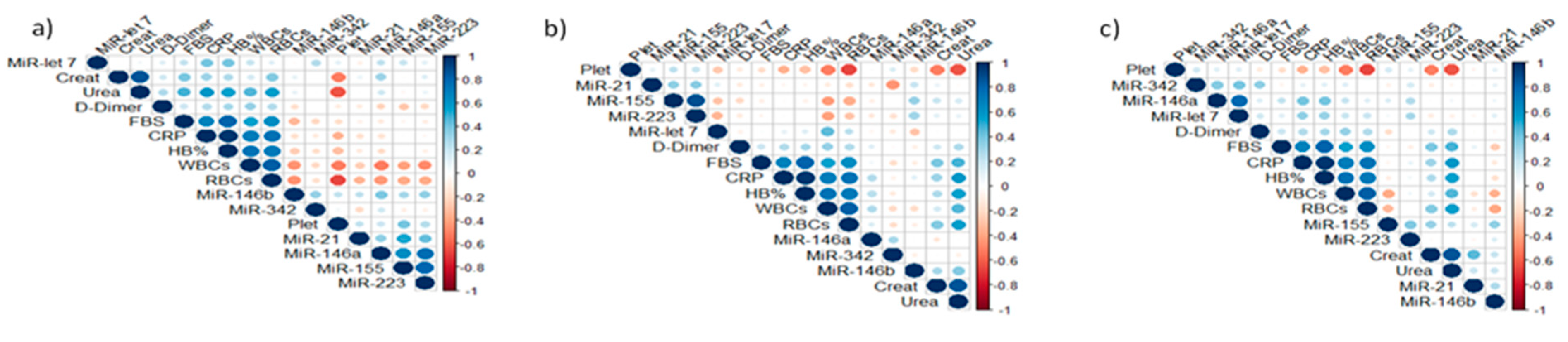
3.2. Expression Analysis of Tested miRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maranini, B.; Ciancio, G.; Ferracin, M.; Cultrera, R.; Negrini, M.; Sabbioni, S.; Govoni, M. microRNAs and Inflammatory Immune Response in SARS-CoV-2 Infection: A Narrative Review. Life 2022, 12, 288. [Google Scholar] [CrossRef]
- Ramos-Casals, M.; Brito-Zeron, P.; Mariette, X. Systemic and organ-specific immune-related manifestations of COVID-19. Nat. Rev. Rheumatol. 2021, 17, 315–332. [Google Scholar] [CrossRef]
- Cron, R.Q. COVID-19 cytokine storm: Targeting the appropriate cytokine. Lancet Rheumatol. 2021, 3, e236–e237. [Google Scholar] [CrossRef]
- Jin, Y.; Ji, W.; Yang, H.; Chen, S.; Zhang, W.; Duan, G. Endothelial activation and dysfunction in COVID-19: From basic mechanisms to potential therapeutic approaches. Signal Transduct. Target. Ther. 2020, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Kessel, C.; Vollenberg, R.; Masjosthusmann, K.; Hinze, C.; Wittkowski, H.; Debaugnies, F.; Nagant, C.; Corazza, F.; Vély, F.; Kaplanski, G.; et al. Discrimination of COVID-19 from inflammation-induced cytokine storm syndromes by disease-related blood biomarkers. Arthritis Rheumatol. 2021, 73, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.; Mallea, J.; Zubair, A.C. Insights into the use of mesenchymal stem cells in COVID-19 mediated acute respiratory failure. NPJ Regen. Med. 2020, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020, 11, 509–522. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug. Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Hardin, L.T.; Xiao, N. miRNAs: The Key Regulator of COVID-19 Disease. Int. J. Cell Biol. 2022, 2022, 1645366. [Google Scholar] [CrossRef] [PubMed]
- Ryu, G.; Shin, H.W. SARS-CoV-2 infection of airway epithelial cells. Immune. Netw. 2021, 21, e3. [Google Scholar] [CrossRef]
- Nik Mohamed Kamal, N.N.S.B.; Shahidan, W.N.S. Non-exosomal and exosomal circulatory microRNAs: Which are more valid as biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef]
- Formosa, A.; Turgeon, P.; Dos Santos, C.C. Role of miRNA dysregulation in sepsis. Mol. Med. 2022, 28, 99. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Svegliati Baroni, S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, A.S.; Visacri, M.B.; da Ronda, C.R.D.S.C.; Vasconcelos, P.E.D.N.S.; Quintanilha, J.C.F.; de Souza, R.N.; Ventura, D.S.; Eguti, A.; Silva, L.F.S.; Perroud Junior, M.W.; et al. Differentially expressed plasmatic microRNAs in Brazilian patients with Coronavirus disease 2019 (COVID-19): Preliminary results. Mol. Biol. Rep. 2022, 49, 6931–6943. [Google Scholar] [CrossRef]
- Kim, W.R.; Park, E.G.; Kang, K.W.; Lee, S.M.; Kim, B.; Kim, H.S. Expression analyses of microRNAs in hamster lung tissues infected by SARS-CoV-2. Mol. Cells 2020, 43, 953–963. [Google Scholar] [CrossRef]
- De Gonzalo-Calvo, D.; Benitez, I.D.; Pinilla, L.; Carratala, A.; Moncusi-Moix, A.; Gort-Paniello, C.; Molinero, M.; Gonzalez, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Letafati, A.; Najafi, S.; Mottahedi, M.; Karimzadeh, M.; Shahini, A.; Garousi, S.; Abbasi-Kolli, M.; Sadri Nahand, J.; Tamehri Zadeh, S.S.; Hamblin, M.R.; et al. MicroRNA let-7 and viral infections: Focus on mechanisms of action. Cell Mol. Biol. Lett. 2022, 27, 14. [Google Scholar] [CrossRef]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J.; et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011, 208, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yu, L.; Zhang, R.; Zhu, L.; Shen, W. Correlation of microRNA-146a/b with disease risk, biochemical indices, inflammatory cytokines, overall disease severity, and prognosis of sepsis. Medicine 2020, 99, e19754. [Google Scholar] [CrossRef] [PubMed]
- Kassif-Lerner, R.; Zloto, K.; Rubin, N.; Asraf, K.; Doolman, R.; Paret, G.; Nevo-Caspi, Y. miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. J. Pers. Med. 2022, 12, 324. [Google Scholar] [CrossRef] [PubMed]
- Houshmandfar, S.; Saeedi-Boroujeni, A.; Rashno, M.; Khodadadi, A.; Mahmoudian-Sani, M.R. miRNA-223 as a regulator of inflammation and NLRP3 inflammasome, the main fragments in the puzzle of immunopathogenesis of different inflammatory diseases and COVID-19. Naunyn. Schmiedebergs Arch. Pharmacol. 2021, 394, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Li, Y.; Wang, P.; Yu, P.; Lang, L. Circulating miR-342-5p serves as a diagnostic biomarker in patients with carotid artery stenosis and predicts the occurrence of the cerebral ischemic event. Ir. J. Med. Sci. 2022, 191, 713–718. [Google Scholar] [CrossRef]
- World Health Organization (WHO); Working Group on the Clinical Characterisation and Management of COVID-19 Infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020, 20, e192–e197, Erratum in Lancet Infect Dis. 2020, 20, e250. [Google Scholar] [CrossRef]
- Swaminathan, G.; Martin-Garcia, J.; Navas-Martin, S. RNA viruses and microRNAs: Challenging discoveries for the 21st century. Physiol. Genom. 2013, 45, 1035–1048. [Google Scholar] [CrossRef]
- Chow, J.T.S.; Salmena, L. Prediction and analysis of SARS-CoV-2-targeting microRNA in human lung epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Fani, M.; Zandi, M.; Ebrahimi, S.; Soltani, S.; Abbasi, S. The role of miRNAs in COVID-19 disease. Future Virol. 2021, 16, 301–306. [Google Scholar] [CrossRef]
- Jiang, Z.; Ma, J.; Wang, Q.; Wu, F.; Ping, J.; Ming, L. Combination of Circulating miRNA-320a/b and D-Dimer Improves Diagnostic Accuracy in Deep Vein Thrombosis Patients. Med. Sci. Monit. 2018, 24, 2031–2037. [Google Scholar] [CrossRef]
- Gaytán-Pacheco, N.; Ibáñez-Salazar, A.; Herrera-Van Oostdam, A.S.; Oropeza-Valdez, J.J.; Magaña-Aquino, M.; Adrián López, J.; Monárrez-Espino, J.; López-Hernández, Y. miR-146a, miR-221, and miR-155 are involved in Inflammatory Immune Response in Severe COVID-19 Patients. Diagnostics 2022, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.K.; Cabrera-Luque, J.; Kar, S.; Sen, C.; Devaney, J.; Biswas, R. Suppression of miR-155 attenuates lung cytokine storm induced by SARS-CoV-2 infection in human ACE2-transgenic mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, F.; Sun, X.; Zhu, Y.; Qin, W. Downregulation of miR-146a inhibits influenza A virus replication by enhancing the type I interferon response in vitro and in vivo. Biomed. Pharmacother. 2019, 111, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Gilyazova, I.; Asadullina, D.; Kagirova, E.; Sikka, R.; Mustafin, A.; Ivanova, E.; Bakhtiyarova, K.; Gilyazova, G.; Gupta, S.; Khusnutdinova, E.; et al. MiRNA-146a-A Key Player in Immunity and Diseases. Int. J. Mol. Sci. 2023, 24, 12767. [Google Scholar] [CrossRef]
- Nersisyan, S.; Engibaryan, N.; Gorbonos, A.; Kirdey, K.; Makhonin, A.; Tonevitsky, A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ 2020, 8, 9994. [Google Scholar] [CrossRef]
- Pessôa, R.L.; da Rosa Abreu, G.; de Oliveira, R.B. MicroRNA Let-7 Plays an Important Role in the Immunopathology of COVID-19: A Systematic Review. Immuno 2023, 3, 112–121. [Google Scholar] [CrossRef]
- Neudecker, V.; Brodsky, K.S.; Clambey, E.T.; Schmidt, E.P.; Packard, T.A.; Davenport, B.; Standiford, T.J.; Weng, T.; Fletcher, A.A.; Barthel, L.; et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci. Transl. Med. 2017, 9, eaah5360. [Google Scholar] [CrossRef]
- Curcio, R.; Poli, G.; Fabi, C.; Sugoni, C.; Pasticci, M.B.; Ferranti, R.; Rossi, M.; Folletti, I.; Sanesi, L.; Santoni, E.; et al. Exosomal miR-17-5p, miR-146a-3p, and miR-223-3p Correlate with Radiologic Sequelae in Survivors of COVID-19-Related Acute Respiratory Distress Syndrome. Int. J. Mol. Sci. 2023, 24, 13037. [Google Scholar] [CrossRef]
- Lan, L.; Cao, H.; Zhao, L.; Cui, W.; Wang, B. PTPN12 down-regulated by miR-146b-3p gene affects the malignant progression of laryngeal squamous cell carcinoma. Open Med. 2023, 18, 20230727. [Google Scholar] [CrossRef]
- Seddiki, N.; Brezar, V.; Ruffin, N.; Lévy, Y.; Swaminathan, S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 2014, 142, 32–38. [Google Scholar] [CrossRef]
- Haroun, R.A.; Osman, W.H.; Amin, R.E.; Hassan, A.K.; Abo-Shanab, W.S.; Eessa, A.M. Circulating plasma miR-155 is a potential biomarker for the detection of SARS-CoV-2 infection. Pathology 2022, 54, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Widiasta, A.; Sribudiani, Y.; Nugrahapraja, H.; Hilmanto, D.; Sekarwana, N.; Rachmadi, D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Noncoding RNA Res. 2020, 5, 153–166. [Google Scholar] [CrossRef] [PubMed]



| Groups | Age (Mean & SD) | Male n (%) | Female n (%) | Total n (%) |
|---|---|---|---|---|
| Healthy | 36.7 ± 11.4 | 43 (75.4) | 14 (24.6) | 57 (100) |
| Mild | 38.5 ± 10.6 | 77 (72.6) | 29 (27.4) | 106 (100) |
| Moderate | 45.2 ± 9.3 | 39 (54.9) | 32 (45.1) | 71 (100) |
| Sever | 61.8 ± 9.7 | 26 (66.6) | 13 (33.4) | 39 (100) |
| Test | Healthy Mean (Range) | Mild Mean (Range) | Moderate Mean (Range) | Severe Mean (Range) |
| HB%, g/dL | 13.4 (12.2–14.5) | 13.2 (11.5–14.2) | 12.8 (12–13.9) | 11.6 (10.0–13.2) |
| WBCs, ×103/μL | 7.8 (5.9–9.5) | 8.5 (6.3–10.8) | 9.6 (7.1–12.7) | 10.0 (8.6–14.9) |
| RBCs, ×103/μL | 4.7 (3.6–5.5) | 4.2 (3.3–5.3) | 4.0 (2.8–5.6) | 3.9 (2.4–5.8) |
| PLT, ×103/μL | 223 (160–285) | 191 (166–273) | 168 (110–235) | 173.4 (107–240) |
| FBS, mg/dL | 85.6 (73–110) | 93 (88–120) | 139 (92–271) | 207 (112–405) |
| B. urea, mg/dL | 29.5 (22–45) | 37 (27–45) | 49 (30–84) | 69 (33–123) |
| Creatinine, mg/dL | 0.86 (0.5–1.1) | 0.95 (0.5–1.1) | 1.0 (0.7–1.4) | 3.8 (1.1–7.5) |
| CRP, mg/mL | 36 (12–96) | 72 (18–108) | 290 (60–560) | 810 (400–1200) |
| D-dimer, ng/mL | 210 (170–400) | 225 (180–550) | 366.2 (242–693) | 939.5 (892–1340) |
| Test | Healthy (n = 57) | Mild (n = 106) | Moderate (n = 71) | Severe (n = 39) |
|---|---|---|---|---|
| Fever; n, % | 0 (0) | 24 (22.6) | 48 (67.6) | 30 (76.9) |
| Cough | 5 (8.7) | 81 (76.4) | 66 (92.9) | 35 (89.7) |
| Dyspnea | 0 (0) | 15 (14.2) | 40 (56.3) | 33 (84.6) |
| Sore throat | 3 (5.2) | 19 (17.9) | 65 (91.5) | 27 (69.2) |
| Diarrhea | 6 (10.5) | 22 (20.7) | 39 (54.9) | 23 (58.9) |
| Fatigue | 10 (17.5) | 69 (65) | 62 (87.4) | 37 (94.8) |
| Abdominal pain | 5 (8.7) | 29 (27.3) | 34 (47.8) | 17 (44) |
| Myalgia | 0 (0) | 12 (11.3) | 36 (50.7) | 30 (77) |
| Headache | 17 (29.8) | 92 (86.8) | 51 (71.8) | 21 (54) |
| Group | Target | AUC | p Value | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Severe Group | MiR-21 | 0.838 | 0.006 | 100 | 85.7 |
| MiR-155 | 0.927 | 0.001 | 96.6 | 87.3 | |
| MiR-146a | 0.853 | 0.045 | 100 | 86.7 | |
| MiR-146b | 0.857 | 0.093 | 100 | 94.3 | |
| MiR-let-7 | 0.903 | 0.017 | 100 | 94.4 | |
| MiR-223 | 0.800 | 0.033 | 100 | 90.6 | |
| MiR-342 | 0.731 | 0.028 | 91 | 100 | |
| Moderate Group | MiR-21 | 0.851 | 0.004 | 86.7 | 71.4 |
| MiR-155 | 0.942 | 0.001 | 100 | 75 | |
| MiR-146a | 0.843 | 0.051 | 97.1 | 86.7 | |
| MiR-146b | 0.614 | 0.591 | 100 | 97.2 | |
| MiR-let-7 | 0.903 | 0.014 | 100 | 90.6 | |
| MiR-223 | 0.906 | 0.004 | 100 | 87.5 | |
| MiR-342 | 0.764 | 0.012 | 90.9 | 100 | |
| Mild Group | MiR-21 | 0.836 | 0.006 | 93.3 | 85.7 |
| MiR-155 | 0.929 | 0.001 | 86.6 | 75 | |
| MiR-146a | 0.775 | 0.119 | 97.1 | 100 | |
| MiR-146b | 0.757 | 0.227 | 100 | 97.3 | |
| MiR-let-7 | 0.972 | 0.011 | 100 | 96.9 | |
| MiR-223 | 0.984 | 0.001 | 100 | 87.5 | |
| MiR-342 | 0.863 | 0.001 | 91 | 100 |
| Group | Target | AUC | p Value | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Severe Group | MiR-21 | 0.981 | 0.001 | 100 | 81.2 |
| MiR-155 | 0.936 | 0.001 | 100 | 90.9 | |
| MiR-146a | 0.829 | 0.122 | 100 | 80 | |
| MiR-146b | 0.866 | 0.006 | 95 | 82.5 | |
| MiR-let-7 | 0.893 | 0.007 | 100 | 88.9 | |
| MiR-223 | 0.884 | 0.011 | 100 | 81.5 | |
| MiR-342 | 0.861 | 0.001 | 95.5 | 100 | |
| Moderate Group | MiR-21 | 0.810 | 0.001 | 100 | 87.5 |
| MiR-155 | 0.800 | 0.002 | 100 | 95.5 | |
| MiR-146a | 0.821 | 0.131 | 100 | 97 | |
| MiR-146b | 0.678 | 0.065 | 100 | 94.2 | |
| MiR-let-7 | 0.873 | 0.049 | 100 | 96.3 | |
| MiR-223 | 0.825 | 0.004 | 100 | 92.9 | |
| MiR-342 | 0.727 | 0.020 | 94.8 | 100 | |
| Mild Group | MiR-21 | 0.866 | 0.001 | 100 | 75 |
| MiR-155 | 0.855 | 0.009 | 100 | 90.9 | |
| MiR-146a | 0.929 | 0.044 | 100 | 91 | |
| MiR-146b | 0.819 | 0.001 | 100 | 94.1 | |
| MiR-let-7 | 0.919 | 0.001 | 100 | 96.3 | |
| MiR-223 | 0.913 | 0.001 | 100 | 96.4 | |
| MiR-342 | 0.876 | 0.001 | 100 | 93.2 |
| Group | Target | AUC | p Value | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Severe Group | MiR-21 | 0.779 | 0.19 | 100 | 94.3 |
| MiR-155 | 0862 | 0.003 | 100 | 71.5 | |
| MiR-146a | 0.869 | 0.002 | 100 | 82.4 | |
| MiR-146b | 0.766 | 0.100 | 100 | 80.6 | |
| MiR-let-7 | 0.806 | 0.074 | 100 | 87.8 | |
| MiR-223 | 0.569 | 0.574 | 100 | 96.7 | |
| MiR-342 | 0.784 | 0.017 | 87.5 | 100 | |
| Moderate Group | MiR-21 | 0.886 | 0.07 | 100 | 91.4 |
| MiR-155 | 0.879 | 0.002 | 90 | 78.6 | |
| MiR-146a | 0.767 | 0.022 | 100 | 87.3 | |
| MiR-146b | 0.839 | 0.116 | 100 | 98 | |
| MiR-let-7 | 0.800 | 0.095 | 100 | 88.9 | |
| MiR-223 | 0.788 | 0.019 | 100 | 90 | |
| MiR-342 | 0.897 | 0.001 | 100 | 96.4 | |
| Mild Group | MiR-21 | 0.921 | 0.08 | 100 | 97.1 |
| MiR-155 | 0.769 | 0.028 | 100 | 71.4 | |
| MiR-146a | 0.677 | 0.130 | 100 | 87.5 | |
| MiR-146b | 0.962 | 0.033 | 100 | 96.8 | |
| MiR-let-7 | 0.950 | 0.035 | 100 | 96.3 | |
| MiR-223 | 0.667 | 0.175 | 100 | 93.3 | |
| MiR-342 | 0.784 | 0.015 | 87.5 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltane, R.; Almulla, N.; Alasiri, A.; Elashmawy, N.F.; Qumsani, A.T.; Alshehrei, F.M.; Keshek, D.E.-G.; Alqadi, T.; AL-Ghamdi, S.B.; Allayeh, A.K. A Comparative Analysis of MicroRNA Expression in Mild, Moderate, and Severe COVID-19: Insights from Urine, Serum, and Nasopharyngeal Samples. Biomolecules 2023, 13, 1681. https://doi.org/10.3390/biom13121681
Soltane R, Almulla N, Alasiri A, Elashmawy NF, Qumsani AT, Alshehrei FM, Keshek DE-G, Alqadi T, AL-Ghamdi SB, Allayeh AK. A Comparative Analysis of MicroRNA Expression in Mild, Moderate, and Severe COVID-19: Insights from Urine, Serum, and Nasopharyngeal Samples. Biomolecules. 2023; 13(12):1681. https://doi.org/10.3390/biom13121681
Chicago/Turabian StyleSoltane, Raya, Nuha Almulla, Ahlam Alasiri, Nabila F. Elashmawy, Alaa T. Qumsani, Fatimah M. Alshehrei, Doaa El-Ghareeb Keshek, Taha Alqadi, Saleh Bakheet AL-Ghamdi, and Abdou Kamal Allayeh. 2023. "A Comparative Analysis of MicroRNA Expression in Mild, Moderate, and Severe COVID-19: Insights from Urine, Serum, and Nasopharyngeal Samples" Biomolecules 13, no. 12: 1681. https://doi.org/10.3390/biom13121681







