The Osteocyte with SB216763-Activated Canonical Wnt Signaling Constructs a Multifunctional 4D Intelligent Osteogenic Module
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Cell Culture
2.3. Activation and Inhibition of Wnt Signaling in Osteocyte MLO-Y4
2.4. Cell Proliferation Analysis
2.5. Immunofluorescence
2.6. Extraction of RNA and Analysis of Gene Expression
2.7. AP Staining and Quantitative Analysis
2.8. Staining with Alizarin Red S and Its Quantitative Assessment
2.9. Adipogenic Differentiation and Oil Red O Staining
2.10. Cell Migration Analysis
2.11. HUVEC Tube Formation Assay
2.12. Live/Dead Assay
2.13. Integrated 3D Printing
2.14. Statistical Analysis
3. Results
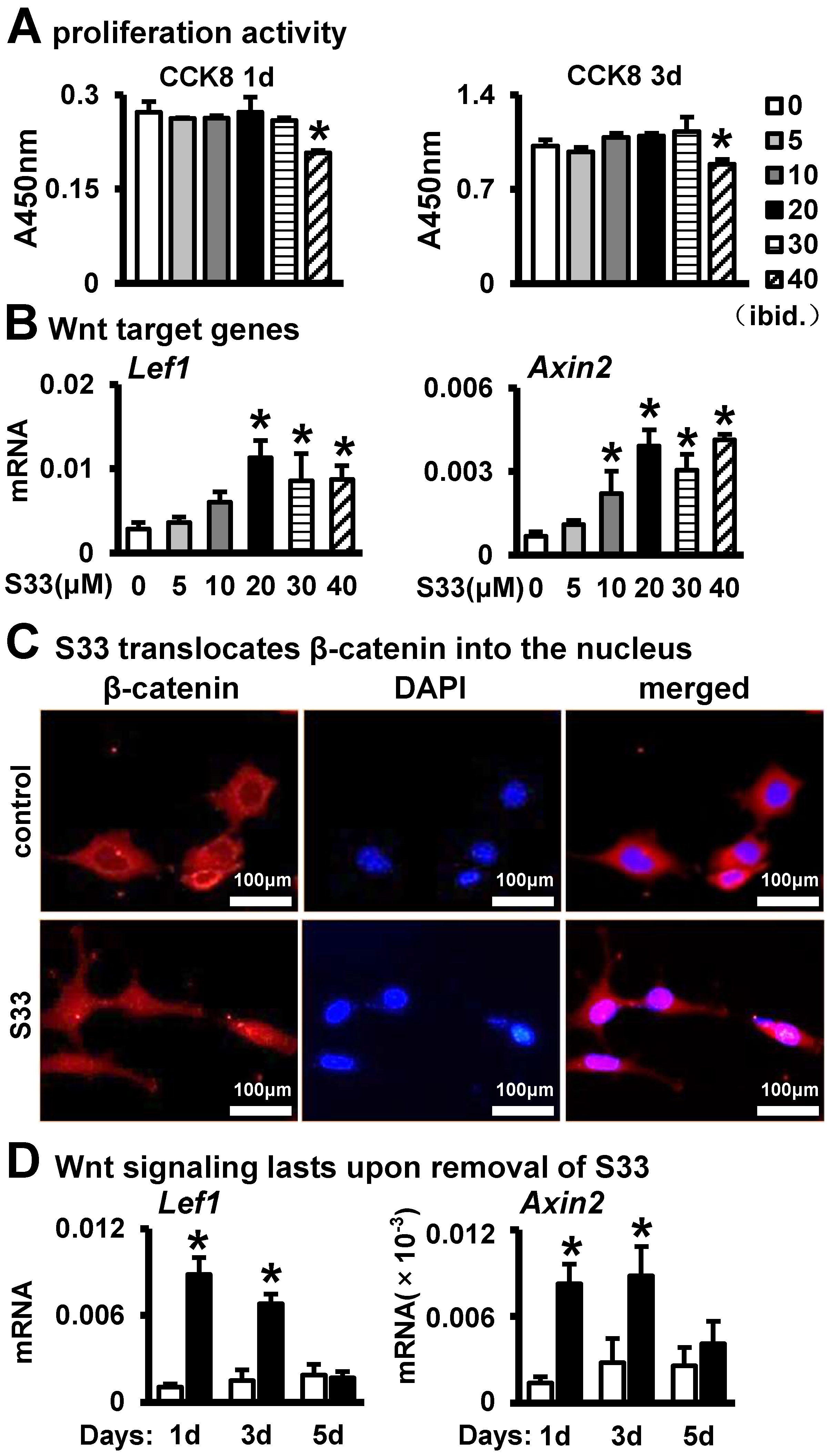
3.1. Activation of Wnt Signaling in Osteocytes by S33
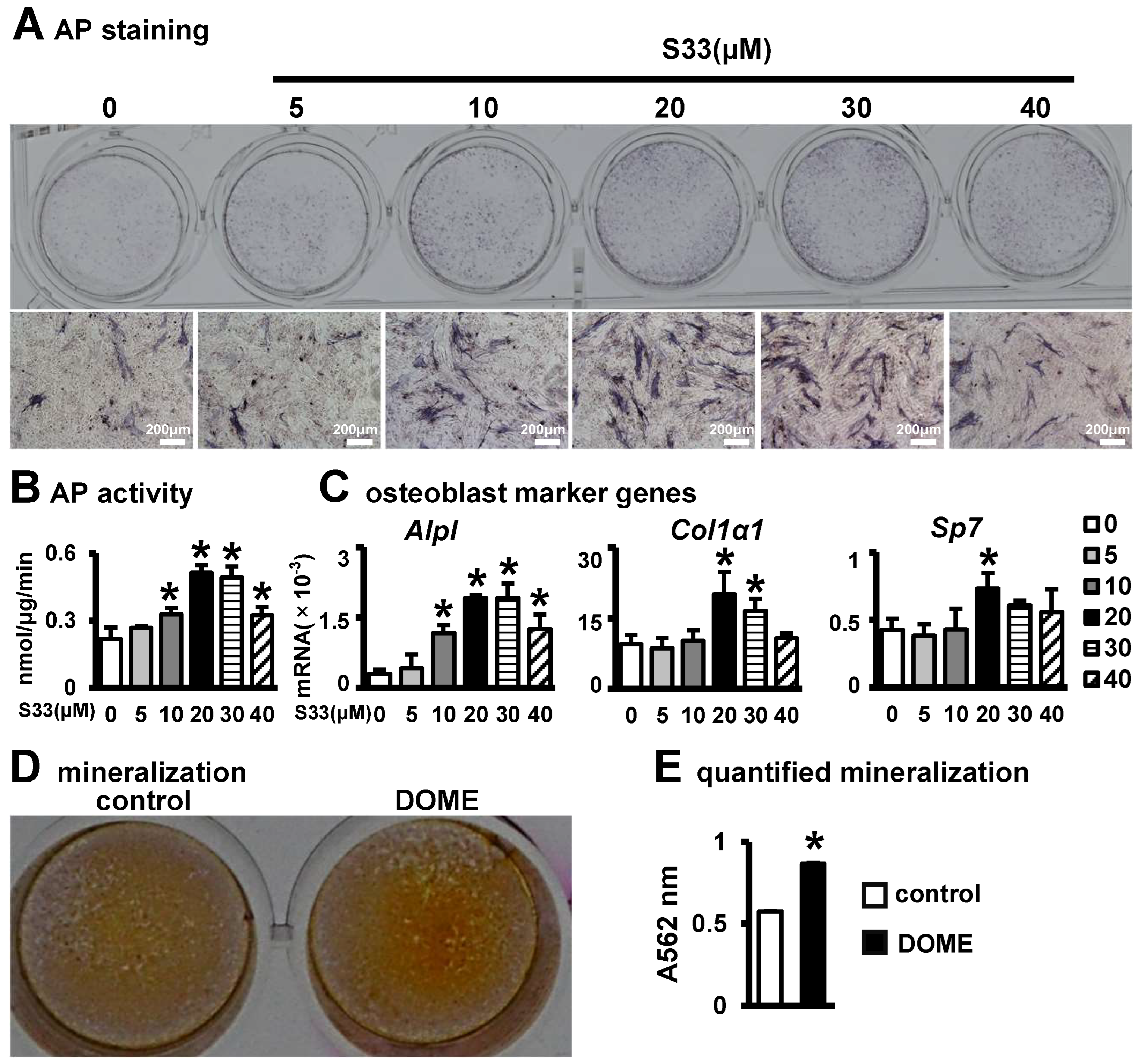
3.2. The Effect of S33-Stimulated Osteocytes on Osteoblast Differentiation
3.3. Inhibition of Adipogenic Differentiation in ST2 Cells by DOME
3.4. Promotion of Angiogenesis by DOME
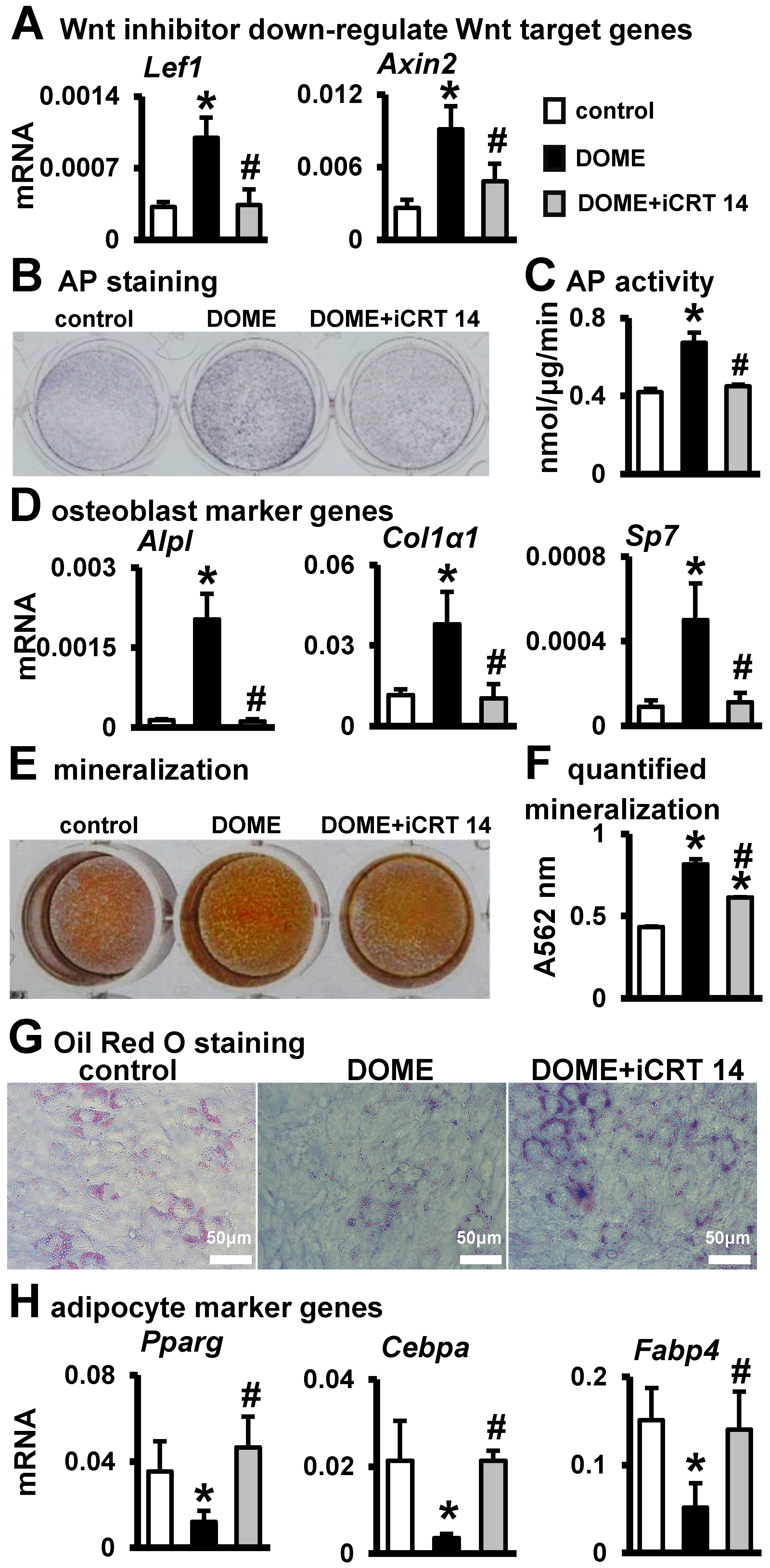
3.5. The Key Role of Wnt/β-Catenin Signaling in DOME for Promoting Osteoblast Differentiation and Inhibiting Adipogenic Differentiation
3.6. DOME Promotes Cell Proliferation in the PCI3D Scaffold
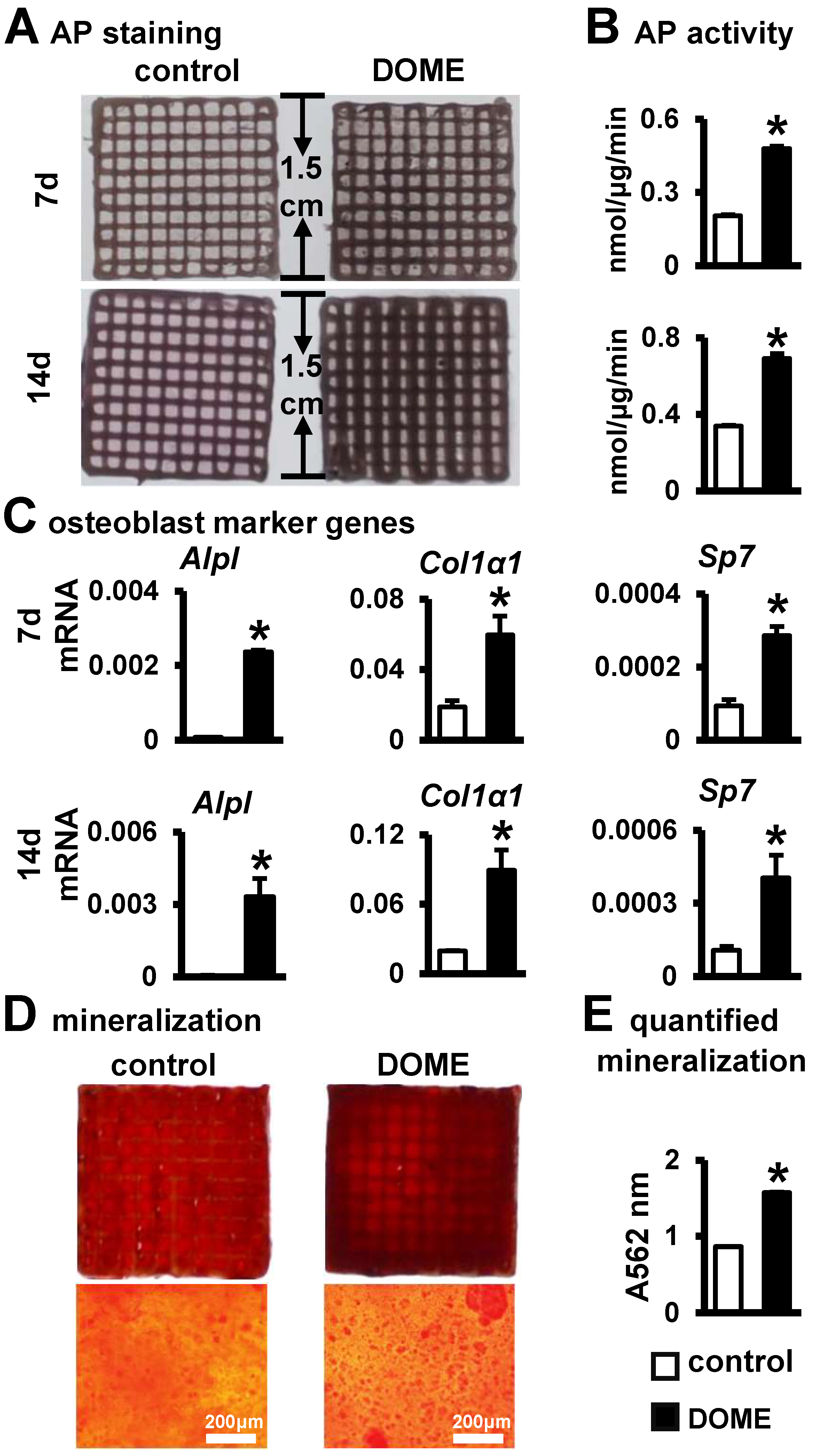
3.7. Promotion of Osteoblast Differentiation and Mineralization in the PCI3D Module by DOME
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaharwar, A.K.; Singh, I.; Khademhosseini, A. Engineered biomaterials for in situ tissue regeneration. Nat. Rev. Mater. 2020, 5, 686–705. [Google Scholar] [CrossRef]
- Zhu, G.; Zhang, T.; Chen, M.; Yao, K.; Huang, X.; Zhang, B.; Li, Y.; Liu, J.; Wang, Y.; Zhao, Z. Bone physiological microenvironment and healing mechanism: Basis for future bone-tissue engineering scaffolds. Bioact. Mater. 2021, 6, 4110–4140. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.B.; Koh, J.T.; Song, S.C. Tuning physical properties and BMP-2 release rates of injectable hydrogel systems for an optimal bone regeneration effect. Biomaterials 2017, 122, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Koons, G.L.; Diba, M.; Mikos, A.G. Materials design for bone-tissue engineering. Nat. Rev. Mater. 2020, 5, 584–603. [Google Scholar] [CrossRef]
- James, A.W.; LaChaud, G.; Shen, J.; Asatrian, G.; Nguyen, V.; Zhang, X.; Ting, K.; Soo, C. A Review of the Clinical Side Effects of Bone Morphogenetic Protein-2. Tissue Eng. Part B Rev. 2016, 22, 284–297. [Google Scholar] [CrossRef]
- Tibbits, S. 4D Printing: Multi-Material Shape Change. Archit. Des. 2014, 84, 116–121. [Google Scholar] [CrossRef]
- Wan, Z.; Zhang, P.; Liu, Y.; Lv, L.; Zhou, Y. Four-dimensional bioprinting: Current developments and applications in bone tissue engineering. Acta Biomater. 2020, 101, 26–42. [Google Scholar] [CrossRef]
- Gao, B.; Yang, Q.; Zhao, X.; Jin, G.; Ma, Y.; Xu, F. 4D Bioprinting for Biomedical Applications. Trends Biotechnol. 2016, 34, 746–756. [Google Scholar] [CrossRef]
- Wang, P.; Wang, X.; Wang, B.; Li, X.; Xie, Z.; Chen, J.; Honjo, T.; Tu, X. 3D printing of osteocytic Dll4 integrated with PCL for cell fate determination towards osteoblasts in vitro. Bio-Des. Manuf. 2022, 5, 497–511. [Google Scholar] [CrossRef]
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef]
- Azbazdar, Y.; Karabicici, M.; Erdal, E.; Ozhan, G. Regulation of Wnt Signaling Pathways at the Plasma Membrane and Their Misregulation in Cancer. Front. Cell Dev. Biol. 2021, 9, 631623. [Google Scholar] [CrossRef]
- Mikels, A.J.; Nusse, R. Wnts as ligands: Processing, secretion and reception. Oncogene 2006, 25, 7461–7468. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/beta-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4, a007906. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Delgado-Calle, J.; Condon, K.W.; Maycas, M.; Zhang, H.; Carlesso, N.; Taketo, M.M.; Burr, D.B.; Plotkin, L.I.; Bellido, T. Osteocytes mediate the anabolic actions of canonical Wnt/beta-catenin signaling in bone. Proc. Natl. Acad. Sci. USA 2015, 112, E478–E486. [Google Scholar] [CrossRef] [PubMed]
- Narcisi, R.; Arikan, O.H.; Lehmann, J.; Ten Berge, D.; van Osch, G.J. Differential Effects of Small Molecule WNT Agonists on the Multilineage Differentiation Capacity of Human Mesenchymal Stem Cells. Tissue Eng. Part A 2016, 22, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.; Hwang, S.G.; Park, H.S.; Choi, S.R.; Park, S.H.; Kim, H.; Ha, N.C.; Bae, S.J.; Han, J.K.; Kim, D.E.; et al. Small molecule-based disruption of the Axin/beta-catenin protein complex regulates mesenchymal stem cell differentiation. Cell Res. 2012, 22, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, M.P.; Culbert, A.A.; Cross, D.A.; Corcoran, S.L.; Yates, J.W.; Pearce, N.J.; Rausch, O.L.; Murphy, G.J.; Carter, P.S.; Roxbee Cox, L.; et al. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 2000, 7, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef]
- Liu, Y.; Ruan, X.; Li, J.; Wang, B.; Chen, J.; Wang, X.; Wang, P.; Tu, X. The Osteocyte Stimulated by Wnt Agonist SKL2001 Is a Safe Osteogenic Niche Improving Bioactivities in a Polycaprolactone and Cell Integrated 3D Module. Cells 2022, 11, 831. [Google Scholar] [CrossRef]
- Gonsalves, F.C.; Klein, K.; Carson, B.B.; Katz, S.; Ekas, L.A.; Evans, S.; Nagourney, R.; Cardozo, T.; Brown, A.M.; DasGupta, R. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 5954–5963. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Xie, Z.; Li, M.; Liu, Y.; Tu, X. Activating Wnt/beta-Catenin Signaling in Osteocytes Promotes Osteogenic Differentiation of BMSCs through BMP-7. Int. J. Mol. Sci. 2022, 23, 16045. [Google Scholar] [CrossRef]
- Wang, B.; Khan, S.; Wang, P.; Wang, X.; Liu, Y.; Chen, J.; Tu, X. A Highly Selective GSK-3beta Inhibitor CHIR99021 Promotes Osteogenesis by Activating Canonical and Autophagy-Mediated Wnt Signaling. Front. Endocrinol. 2022, 13, 926622. [Google Scholar] [CrossRef]
- Billiet, T.; Gevaert, E.; De Schryver, T.; Cornelissen, M.; Dubruel, P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 2014, 35, 49–62. [Google Scholar] [CrossRef]
- Moseti, D.; Regassa, A.; Kim, W.K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef]
- Ahmad, B.; Serpell, C.J.; Fong, I.L.; Wong, E.H. Molecular Mechanisms of Adipogenesis: The Anti-adipogenic Role of AMP-Activated Protein Kinase. Front. Mol. Biosci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Saran, U.; Gemini Piperni, S.; Chatterjee, S. Role of angiogenesis in bone repair. Arch. Biochem. Biophys. 2014, 561, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, A.; Mazloomnejad, R.; Kasravi, M.; Gholamine, B.; Bahrami, S.; Sarzaeem, M.M.; Niknejad, H. Recent advances on small molecules in osteogenic differentiation of stem cells and the underlying signaling pathways. Stem Cell Res. Ther. 2022, 13, 518. [Google Scholar] [CrossRef] [PubMed]
- Awale, G.M.; Barajaa, M.A.; Kan, H.M.; Seyedsalehi, A.; Nam, G.H.; Hosseini, F.S.; Ude, C.C.; Schmidt, T.A.; Lo, K.W.; Laurencin, C.T. Regenerative engineering of long bones using the small molecule forskolin. Proc. Natl. Acad. Sci. USA 2023, 120, e2219756120. [Google Scholar] [CrossRef]
- Ding, L.; Madamsetty, V.S.; Kiers, S.; Alekhina, O.; Ugolkov, A.; Dube, J.; Zhang, Y.; Zhang, J.S.; Wang, E.; Dutta, S.K.; et al. Glycogen Synthase Kinase-3 Inhibition Sensitizes Pancreatic Cancer Cells to Chemotherapy by Abrogating the TopBP1/ATR-Mediated DNA Damage Response. Clin. Cancer Res. 2019, 25, 6452–6462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lai, Z.-P.; Chen, P.; Ying, Y.; Zhuang, J.; Yu, K.-M. Glycogen synthase kinase-3β inhibitor SB216763 promotes DNA repair in ischemic retinal neurons. Neural Regen. Res. 2020, 16, 394–400. [Google Scholar] [CrossRef]
- Luo, G.; Chen, L.; Burton, C.R.; Xiao, H.; Sivaprakasam, P.; Krause, C.M.; Cao, Y.; Liu, N.; Lippy, J.; Clarke, W.J.; et al. Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors. J. Med. Chem. 2016, 59, 1041–1051. [Google Scholar] [CrossRef]
- Henriksen, E.J.; Kinnick, T.R.; Teachey, M.K.; O’Keefe, M.P.; Ring, D.; Johnson, K.W.; Harrison, S.D. Modulation of muscle insulin resistance by selective inhibition of GSK-3 in Zucker diabetic fatty rats. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E892–E900. [Google Scholar] [CrossRef]
- Rodriguez-Jimenez, F.J.; Vilches, A.; Perez-Arago, M.A.; Clemente, E.; Roman, R.; Leal, J.; Castro, A.A.; Fustero, S.; Moreno-Manzano, V.; Jendelova, P.; et al. Activation of Neurogenesis in Multipotent Stem Cells Cultured In Vitro and in the Spinal Cord Tissue After Severe Injury by Inhibition of Glycogen Synthase Kinase-3. Neurotherapeutics 2021, 18, 515–533. [Google Scholar] [CrossRef]
- Zhang, Y.; Zu, D.; Chen, Z.; Ying, G. An update on Wnt signaling pathway in cancer. Transl. Cancer Res. 2020, 9, 1246–1252. [Google Scholar] [CrossRef]
- Malhotra, A.; Habibovic, P. Calcium Phosphates and Angiogenesis: Implications and Advances for Bone Regeneration. Trends Biotechnol. 2016, 34, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fukushima, Y.; Nozaki, K.; Nakanishi, H.; Deng, J.; Wakabayashi, N.; Itaka, K. Enhancement of bone regeneration by coadministration of angiogenic and osteogenic factors using messenger RNA. Inflamm. Regen. 2023, 43, 32. [Google Scholar] [CrossRef] [PubMed]
- Zisch, A.H.; Lutolf, M.P.; Hubbell, J.A. Biopolymeric delivery matrices for angiogenic growth factors. Cardiovasc. Pathol. 2003, 12, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Zhang, X.-B.; Gao, X.-D.; Hao, D.-J.; Li, T.; Xu, Z.-W. Bioprinting for bone tissue engineering. Front. Bioeng. Biotechnol. 2022, 10, 1036375. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Chen, G.; Liu, C.; Ye, X.; Wang, S.; Dong, M.; Sun, M.; He, J.; Yu, X.; Ye, G.; et al. 4D Printing of Multi-Responsive Membrane for Accelerated In Vivo Bone Healing Via Remote Regulation of Stem Cell Fate. Adv. Funct. Mater. 2021, 31, 2103920. [Google Scholar] [CrossRef]
- Liu, X.; Chen, W.; Shao, B.; Zhang, X.; Wang, Y.; Zhang, S.; Wu, W. Mussel patterned with 4D biodegrading elastomer durably recruits regenerative macrophages to promote regeneration of craniofacial bone. Biomaterials 2021, 276, 120998. [Google Scholar] [CrossRef] [PubMed]



| Primer | Forward | Reverse |
|---|---|---|
| Gapdh | GCACAGTCAAGGCCGAGAAT | GCCTTCTCCATGGTGGTGAA |
| Lef1 | CCTACAGCGACGAGCACTTTT | CCTTGCTTGGAGTTGACATCTG |
| Axin2 | TGAGCGGCAGAGCAAGTCCAA | GGCAGACTCCAATGGGTAGCT |
| Alpl | TTCGCTATCTGCCTTGCCTG | AGTCTGTGTCTTGCCTGCC |
| Col1α1 | TCAACCCCGTCTACTTCCCT | TTCAACAGTCCAAGAACCCCAT |
| Sp7 | GCCCCCTGGTGTTCTTCATT | CTTCCCCCTTCTTGGCACTC |
| Pparg | AGCCCTTTACCACAGTTGATTTCTCC | GCAGGTTCTACTTTGATCGCACTTTG |
| Cebpa | TGGACAAGAACAGCAACGAG | TCACTGGTCAACTCCAGCAC |
| Fabp4 | GACGACAGGAAGGTGAAGAGCATC | GGAAGTCACGCCTTTCATAACACATTC |
| Vegfa | TTCGAGGAGCACTTTGGGTC | GTGGGTGGGTGTGTCTACAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, Y.; Chen, J.; Gong, W.; Tu, X. The Osteocyte with SB216763-Activated Canonical Wnt Signaling Constructs a Multifunctional 4D Intelligent Osteogenic Module. Biomolecules 2024, 14, 354. https://doi.org/10.3390/biom14030354
Zhang J, Zhang Y, Chen J, Gong W, Tu X. The Osteocyte with SB216763-Activated Canonical Wnt Signaling Constructs a Multifunctional 4D Intelligent Osteogenic Module. Biomolecules. 2024; 14(3):354. https://doi.org/10.3390/biom14030354
Chicago/Turabian StyleZhang, Jinling, Ying Zhang, Jiafeng Chen, Weimin Gong, and Xiaolin Tu. 2024. "The Osteocyte with SB216763-Activated Canonical Wnt Signaling Constructs a Multifunctional 4D Intelligent Osteogenic Module" Biomolecules 14, no. 3: 354. https://doi.org/10.3390/biom14030354







