Proteome-Wide Mendelian Randomization and Colocalization Analysis Identify Therapeutic Targets for Knee and Hip Osteoarthritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. MR and SMR Analysis
2.2.1. MR Analysis
2.2.2. SMR Analysis
2.3. Bayesian Colocalization Analysis and Steiger Filtering Analysis
2.4. Phenotype Scanning
2.5. Protein–Protein Interaction (PPI) Network and Drug Identification
2.6. Classification Hierarchy of Proteins as Potential Drug Targets
2.7. MR Phenome-Wide Association Study
3. Results
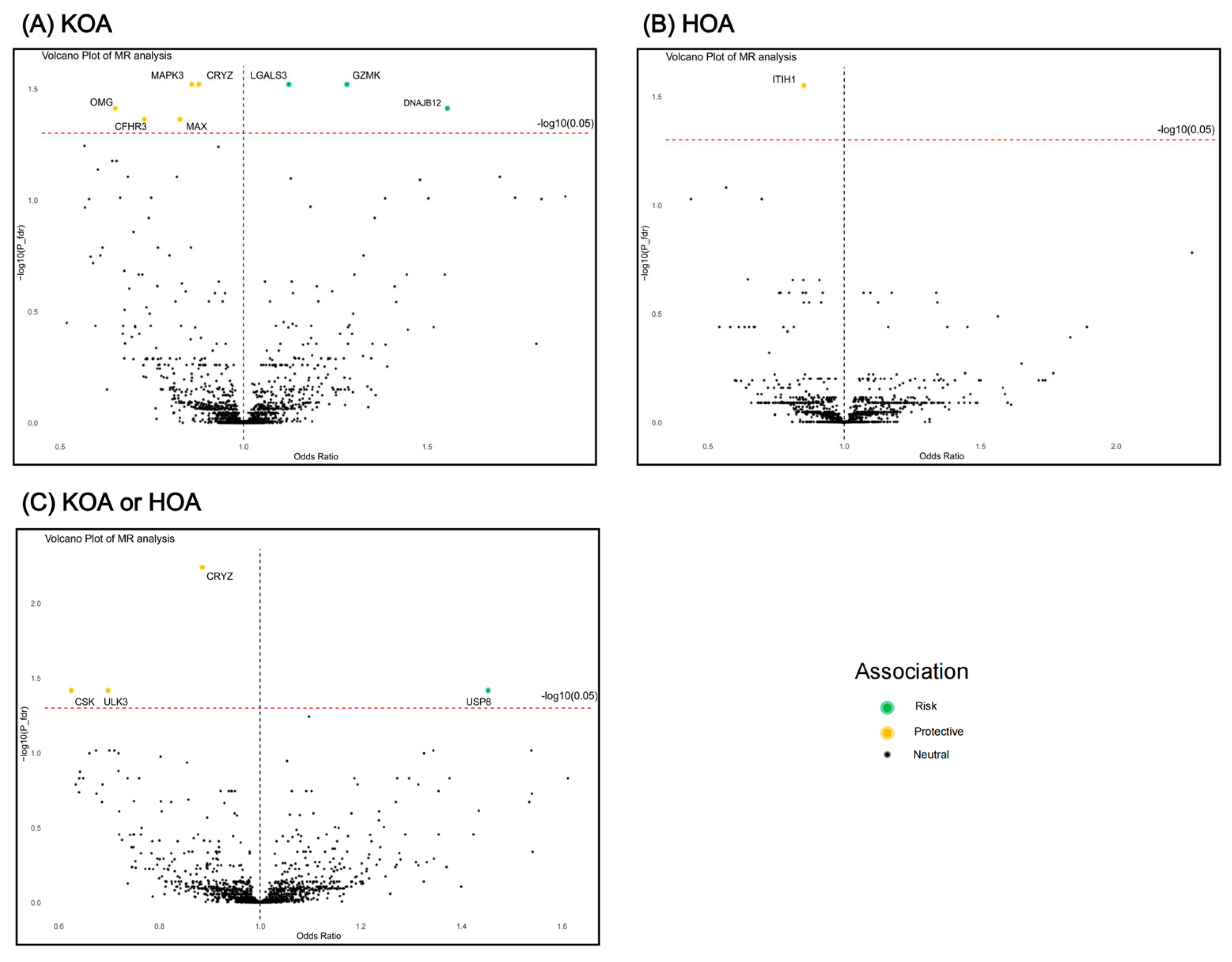
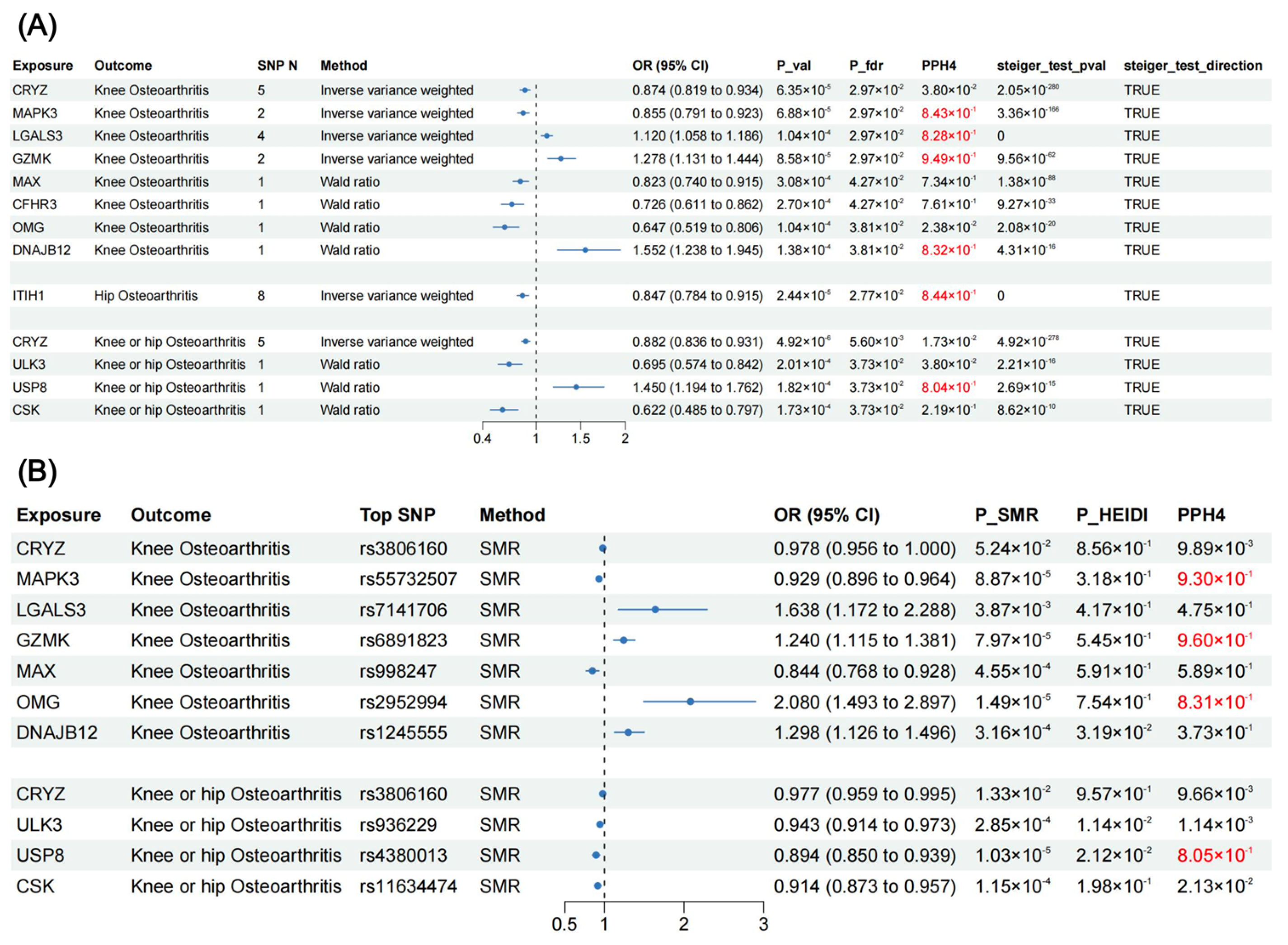
3.1. MR Results
3.2. SMR Results (Additional Validation)
3.3. Colocalization Analysis and Phenotype Scanning for Causal Proteins
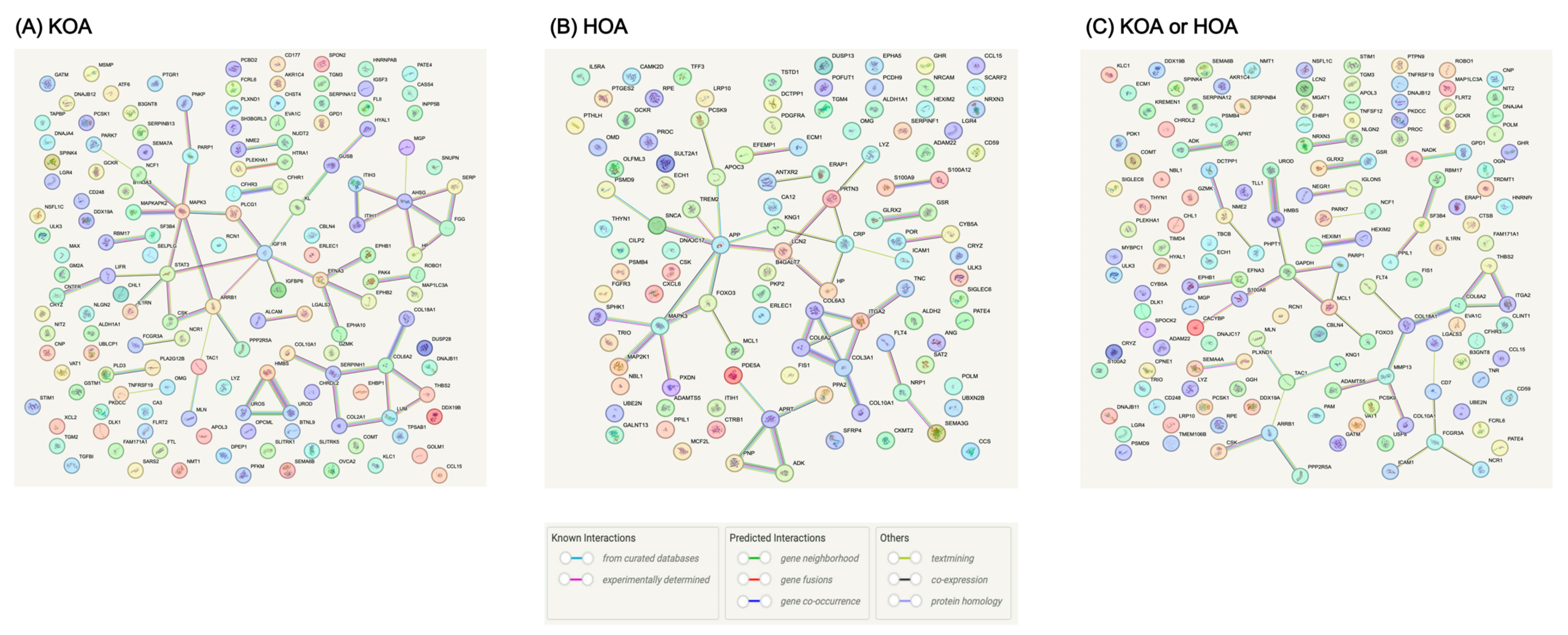
3.4. Interactions among MR-Prioritized Proteins and Their Association with Current Medication
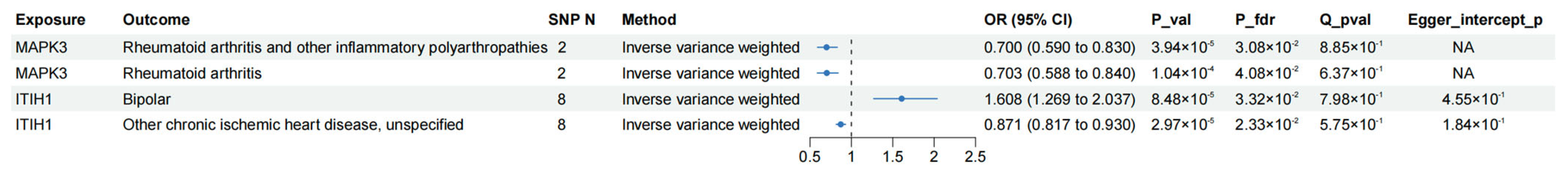
3.5. MR Phenome-Wide Association Studies of Core Therapeutic Targets of OA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martel-Pelletier, J.; Barr, A.J.; Cicuttini, F.M.; Conaghan, P.G.; Cooper, C.; Goldring, M.B.; Goldring, S.R.; Jones, G.; Teichtahl, A.J.; Pelletier, J.P. Osteoarthritis. Nat. Rev. Dis. Primers 2016, 2, 16072. [Google Scholar] [CrossRef]
- Turkiewicz, A.; Petersson, I.F.; Björk, J.; Hawker, G.; Dahlberg, L.E.; Lohmander, L.S.; Englund, M. Current and future impact of osteoarthritis on health care: A population-based study with projections to year 2032. Osteoarthr. Cartil. 2014, 22, 1826–1832. [Google Scholar] [CrossRef]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Aresti, N.; Kassam, J.; Nicholas, N.; Achan, P. Hip osteoarthritis. BMJ 2016, 354, i3405. [Google Scholar] [CrossRef]
- Latourte, A.; Kloppenburg, M.; Richette, P. Emerging pharmaceutical therapies for osteoarthritis. Nat. Rev. Rheumatol. 2020, 16, 673–688. [Google Scholar] [CrossRef]
- Leopoldino, A.O.; Machado, G.C.; Ferreira, P.H.; Pinheiro, M.B.; Day, R.; McLachlan, A.J.; Hunter, D.J.; Ferreira, M.L. Paracetamol versus placebo for knee and hip osteoarthritis. Cochrane Database Syst. Rev. 2019, 2, Cd013273. [Google Scholar] [CrossRef]
- Jüni, P.; Hari, R.; Rutjes, A.W.; Fischer, R.; Silletta, M.G.; Reichenbach, S.; da Costa, B.R. Intra-articular corticosteroid for knee osteoarthritis. Cochrane Database Syst. Rev. 2015, 2015, Cd005328. [Google Scholar] [CrossRef] [PubMed]
- Vasudeva, A.; Yadav, S.L.; Nanda, S.; Sahu, S. Assessment of pain and structure after an intra-articular injection of adalimumab in osteoarthritis of the knee: A case report. Medicine 2020, 99, e21131. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, X.; Goupille, P.; Beaulieu, A.D.; Burch, F.X.; Bensen, W.G.; Conrozier, T.; Loeuille, D.; Kivitz, A.J.; Silver, D.; Appleton, B.E. Intraarticular injection of anakinra in osteoarthritis of the knee: A multicenter, randomized, double-blind, placebo-controlled study. Arthritis Rheum. 2009, 61, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Grothe, K.; Flechsenhar, K.; Paehler, T.; Ritzeler, O.; Beninga, J.; Saas, J.; Herrmann, M.; Rudolphi, K. IκB kinase inhibition as a potential treatment of osteoarthritis—Results of a clinical proof-of-concept study. Osteoarthr. Cartil. 2017, 25, 46–52. [Google Scholar] [CrossRef]
- Assi, R.; Quintiens, J.; Monteagudo, S.; Lories, R.J. Innovation in Targeted Intra-articular Therapies for Osteoarthritis. Drugs 2023, 83, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; DeLisle, R.K.; et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, I.A.; Vallerga, C.L.; Boer, C.G.; Schiphof, D.; Ikram, M.A.; Bierma-Zeinstra, S.M.A.; van Meurs, J.B.J. Plasma proteomics identifies CRTAC1 as a biomarker for osteoarthritis severity and progression. Rheumatology 2023, 62, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, S.; Wen, Y.; Li, P.; Cheng, S.; Ma, M.; Zhang, L.; Cheng, B.; Qi, X.; Liang, C.; et al. Assessing the genetic relationships between osteoarthritis and human plasma proteins: A large scale genetic correlation scan. Ann. Transl. Med. 2020, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Storm, C.S.; Kia, D.A.; Almramhi, M.M.; Bandres-Ciga, S.; Finan, C.; Hingorani, A.D.; Wood, N.W. Finding genetically-supported drug targets for Parkinson’s disease using Mendelian randomization of the druggable genome. Nat. Commun. 2021, 12, 7342. [Google Scholar] [CrossRef]
- Reay, W.R.; Cairns, M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 2021, 22, 658–671. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, N.A.; Didelez, V.; Burton, P.R.; Tobin, M.D. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008, 5, e177. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J.; et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat. Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Claringbould, A.; Westra, H.J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qin, F.; Li, X.; Du, X.; Li, T. Identification of novel proteins for lacunar stroke by integrating genome-wide association data and human brain proteomes. BMC Med. 2022, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Scott, R.A.; Timpson, N.J.; Davey Smith, G.; Thompson, S.G. Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 2015, 30, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.E.; Rao, S.R.; Schultz, M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.T.; Ou, Y.N.; Wu, B.S.; Yang, Y.X.; Jiang, Y.; Huang, Y.Y.; Liu, Y.; Tan, L.; Dong, Q.; Suckling, J.; et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol. Psychiatry 2022, 27, 2849–2857. [Google Scholar] [CrossRef]
- Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021, 17, e1009440. [Google Scholar] [CrossRef]
- Kamat, M.A.; Blackshaw, J.A.; Young, R.; Surendran, P.; Burgess, S.; Danesh, J.; Butterworth, A.S.; Staley, J.R. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019, 35, 4851–4853. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Nielsen, J.B.; Fritsche, L.G.; Dey, R.; Gabrielsen, M.E.; Wolford, B.N.; LeFaive, J.; VandeHaar, P.; Gagliano, S.A.; Gifford, A.; et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018, 50, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Haberland, V.; Baird, D.; Walker, V.; Haycock, P.C.; Hurle, M.R.; Gutteridge, A.; Erola, P.; Liu, Y.; Luo, S.; et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020, 52, 1122–1131. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Felson, D.T.; Goggins, J.; Niu, J.; Zhang, Y.; Hunter, D.J. The effect of body weight on progression of knee osteoarthritis is dependent on alignment. Arthritis Rheum. 2004, 50, 3904–3909. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, Z.; Yuan, P.; Huang, H.; Wang, J.; Shi, P.; Sun, X. ERK1 loss accelerates the progression of osteoarthritis in aged mice via NRF2/BACH1 signaling. Biochem. Biophys. Res. Commun. 2022, 622, 129–135. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Novak, K.; Haqqi, T.M. ERK1/2-mediated activation of DRP1 regulates mitochondrial dynamics and apoptosis in chondrocytes. Osteoarthr. Cartil. 2022, 30, 315–328. [Google Scholar] [CrossRef]
- Rice, P.L.; Peters, S.L.; Beard, K.S.; Ahnen, D.J. Sulindac independently modulates extracellular signal-regulated kinase 1/2 and cyclic GMP-dependent protein kinase signaling pathways. Mol. Cancer Ther. 2006, 5, 746–754. [Google Scholar] [CrossRef]
- Nishio, T.; Usami, M.; Awaji, M.; Shinohara, S.; Sato, K. Dual effects of acetylsalicylic acid on ERK signaling and Mitf transcription lead to inhibition of melanogenesis. Mol. Cell. Biochem. 2016, 412, 101–110. [Google Scholar] [CrossRef]
- Hu, Y.; Yéléhé-Okouma, M.; Ea, H.K.; Jouzeau, J.Y.; Reboul, P. Galectin-3: A key player in arthritis. Joint Bone Spine 2017, 84, 15–20. [Google Scholar] [CrossRef]
- Ezzat, M.H.; El-Gammasy, T.M.; Shaheen, K.Y.; Osman, A.O. Elevated production of galectin-3 is correlated with juvenile idiopathic arthritis disease activity, severity, and progression. Int. J. Rheum. Dis. 2011, 14, 345–352. [Google Scholar] [CrossRef]
- Janelle-Montcalm, A.; Boileau, C.; Poirier, F.; Pelletier, J.P.; Guévremont, M.; Duval, N.; Martel-Pelletier, J.; Reboul, P. Extracellular localization of galectin-3 has a deleterious role in joint tissues. Arthritis Res. Ther. 2007, 9, R20. [Google Scholar] [CrossRef]
- Filer, A.; Bik, M.; Parsonage, G.N.; Fitton, J.; Trebilcock, E.; Howlett, K.; Cook, M.; Raza, K.; Simmons, D.L.; Thomas, A.M.; et al. Galectin 3 induces a distinctive pattern of cytokine and chemokine production in rheumatoid synovial fibroblasts via selective signaling pathways. Arthritis Rheum. 2009, 60, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Kho, D.H.; Yanagawa, T.; Harazono, Y.; Gao, X.; Hogan, V.; Raz, A. Galectin-3 inhibits osteoblast differentiation through notch signaling. Neoplasia 2014, 16, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Toegel, S.; Bieder, D.; André, S.; Altmann, F.; Walzer, S.M.; Kaltner, H.; Hofstaetter, J.G.; Windhager, R.; Gabius, H.J. Glycophenotyping of osteoarthritic cartilage and chondrocytes by RT-qPCR, mass spectrometry, histochemistry with plant/human lectins and lectin localization with a glycoprotein. Arthritis Res. Ther. 2013, 15, R147. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; van Daalen, K.R.; Crnko, S.; Ten Broeke, T.; Bovenschen, N. Intracellular and Extracellular Roles of Granzyme K. Front. Immunol. 2021, 12, 677707. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Noda, H.; Okaniwa, N.; Adachi, K.; Shinmura, T.; Nakagawa, S.; Ebi, M.; Ogasawara, N.; Funaki, Y.; Zhuo, L.; et al. Serum-Derived Hyaluronan-Associated Protein Is a Novel Biomarker for Inflammatory Bowel Diseases. Digestion 2017, 95, 146–155. [Google Scholar] [CrossRef]
- Chang, Q.H.; Mao, T.; Tao, Y.; Dong, T.; Tang, X.X.; Ge, G.H.; Xu, Z.J. Pan-cancer analysis identifies ITIH1 as a novel prognostic indicator for hepatocellular carcinoma. Aging 2021, 13, 11096–11119. [Google Scholar] [CrossRef]
- Lourido, L.; Balboa-Barreiro, V.; Ruiz-Romero, C.; Rego-Pérez, I.; Camacho-Encina, M.; Paz-González, R.; Calamia, V.; Oreiro, N.; Nilsson, P.; Blanco, F.J. A clinical model including protein biomarkers predicts radiographic knee osteoarthritis: A prospective study using data from the Osteoarthritis Initiative. Osteoarthr. Cartil. 2021, 29, 1147–1154. [Google Scholar] [CrossRef]
- Lourido, L.; Ayoglu, B.; Fernández-Tajes, J.; Oreiro, N.; Henjes, F.; Hellström, C.; Schwenk, J.M.; Ruiz-Romero, C.; Nilsson, P.; Blanco, F.J. Discovery of circulating proteins associated to knee radiographic osteoarthritis. Sci. Rep. 2017, 7, 137. [Google Scholar] [CrossRef]
- Asher, G.; Dym, O.; Tsvetkov, P.; Adler, J.; Shaul, Y. The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry 2006, 45, 6372–6378. [Google Scholar] [CrossRef] [PubMed]
- Ibeas Bih, C.; Chen, T.; Nunn, A.V.; Bazelot, M.; Dallas, M.; Whalley, B.J. Molecular Targets of Cannabidiol in Neurological Disorders. Neurotherapeutics 2015, 12, 699–730. [Google Scholar] [CrossRef] [PubMed]
- Rolf, M.G.; Curwen, J.O.; Veldman-Jones, M.; Eberlein, C.; Wang, J.; Harmer, A.; Hellawell, C.J.; Braddock, M. In vitro pharmacological profiling of R406 identifies molecular targets underlying the clinical effects of fostamatinib. Pharmacol. Res. Perspect. 2015, 3, e00175. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Zhao, Q.; Lawlor, D.A.; Sheehan, N.A.; Thompson, J.; Davey Smith, G. Improving the accuracy of two-sample summary-data Mendelian randomization: Moving beyond the NOME assumption. Int. J. Epidemiol. 2019, 48, 728–742. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, M.; Shao, Z. Proteome-Wide Mendelian Randomization and Colocalization Analysis Identify Therapeutic Targets for Knee and Hip Osteoarthritis. Biomolecules 2024, 14, 355. https://doi.org/10.3390/biom14030355
Zou M, Shao Z. Proteome-Wide Mendelian Randomization and Colocalization Analysis Identify Therapeutic Targets for Knee and Hip Osteoarthritis. Biomolecules. 2024; 14(3):355. https://doi.org/10.3390/biom14030355
Chicago/Turabian StyleZou, Mingrui, and Zhenxing Shao. 2024. "Proteome-Wide Mendelian Randomization and Colocalization Analysis Identify Therapeutic Targets for Knee and Hip Osteoarthritis" Biomolecules 14, no. 3: 355. https://doi.org/10.3390/biom14030355




