The Third-Generation Sequencing Challenge: Novel Insights for the Omic Sciences
Abstract
:1. Introduction
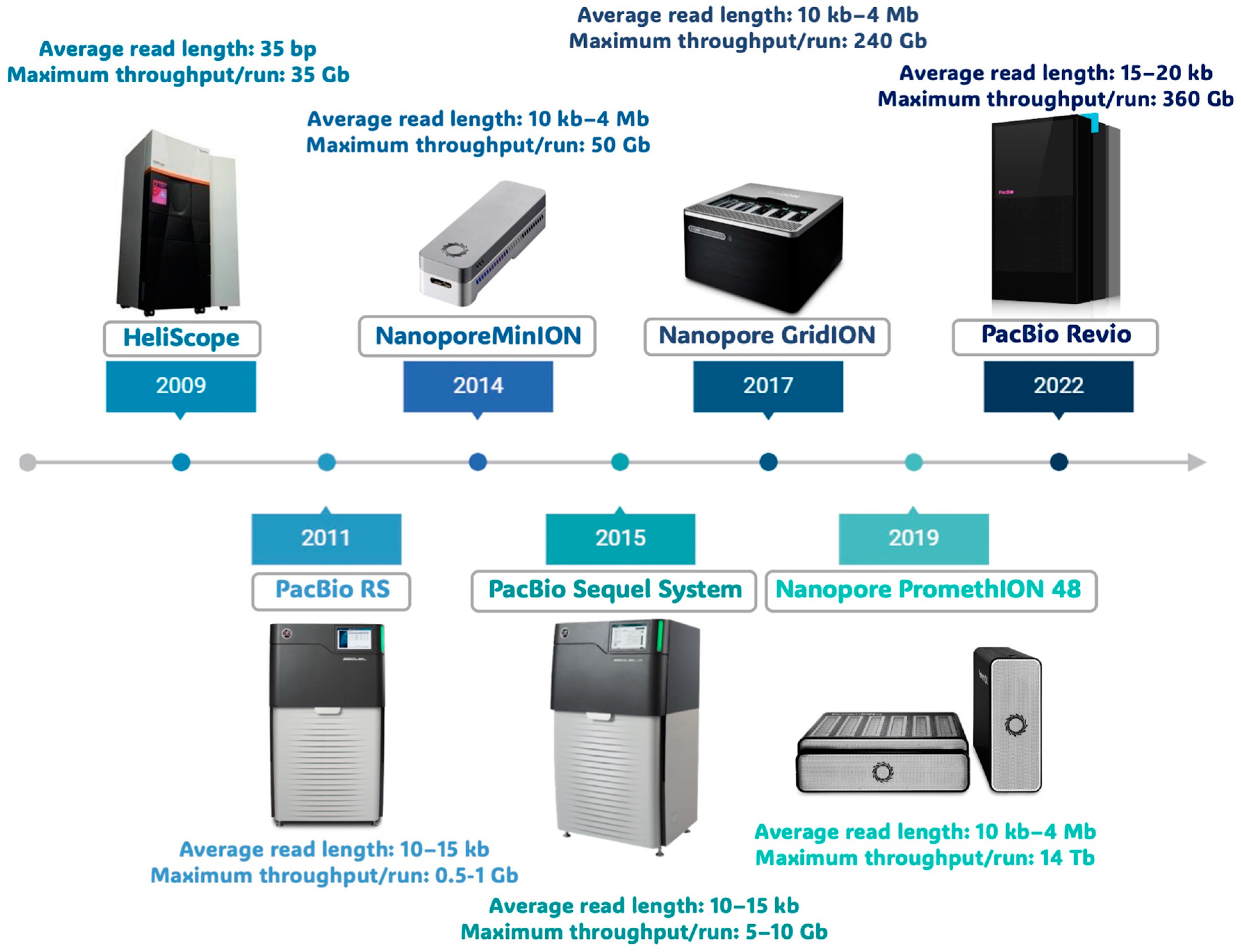
2. Third-Generation Sequencing Methods
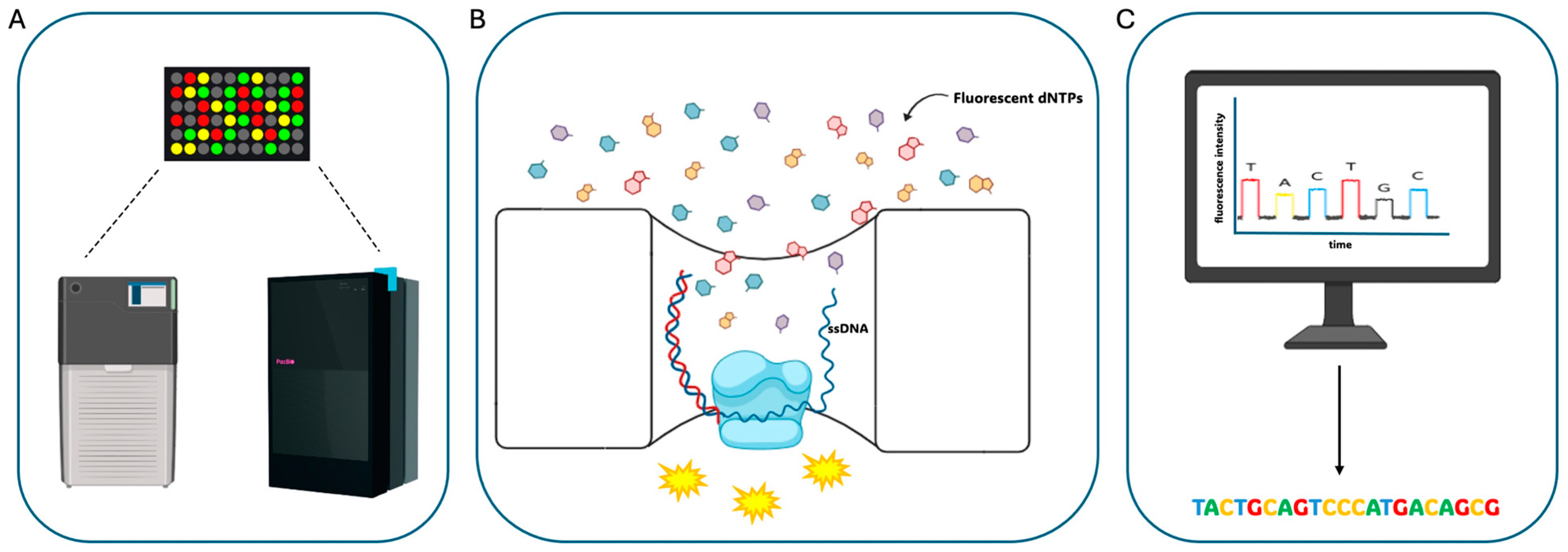
2.1. PacBio Sequencing
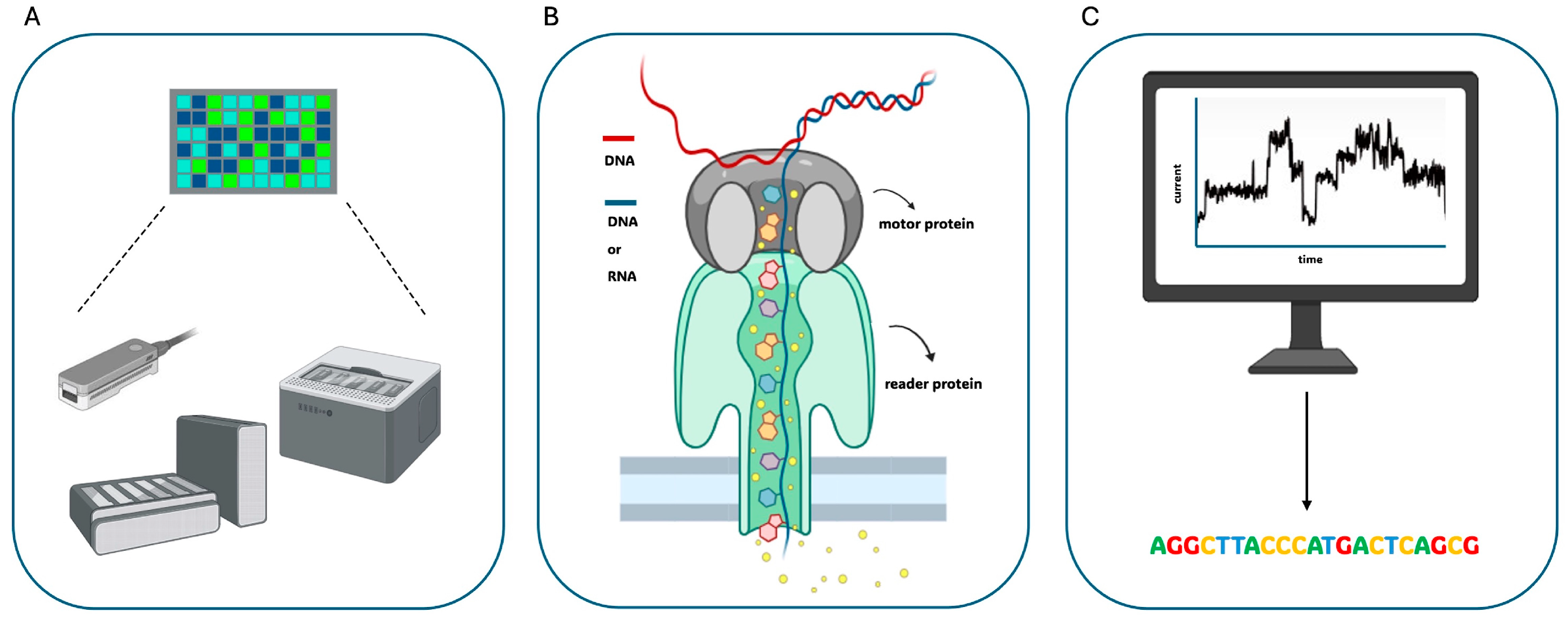
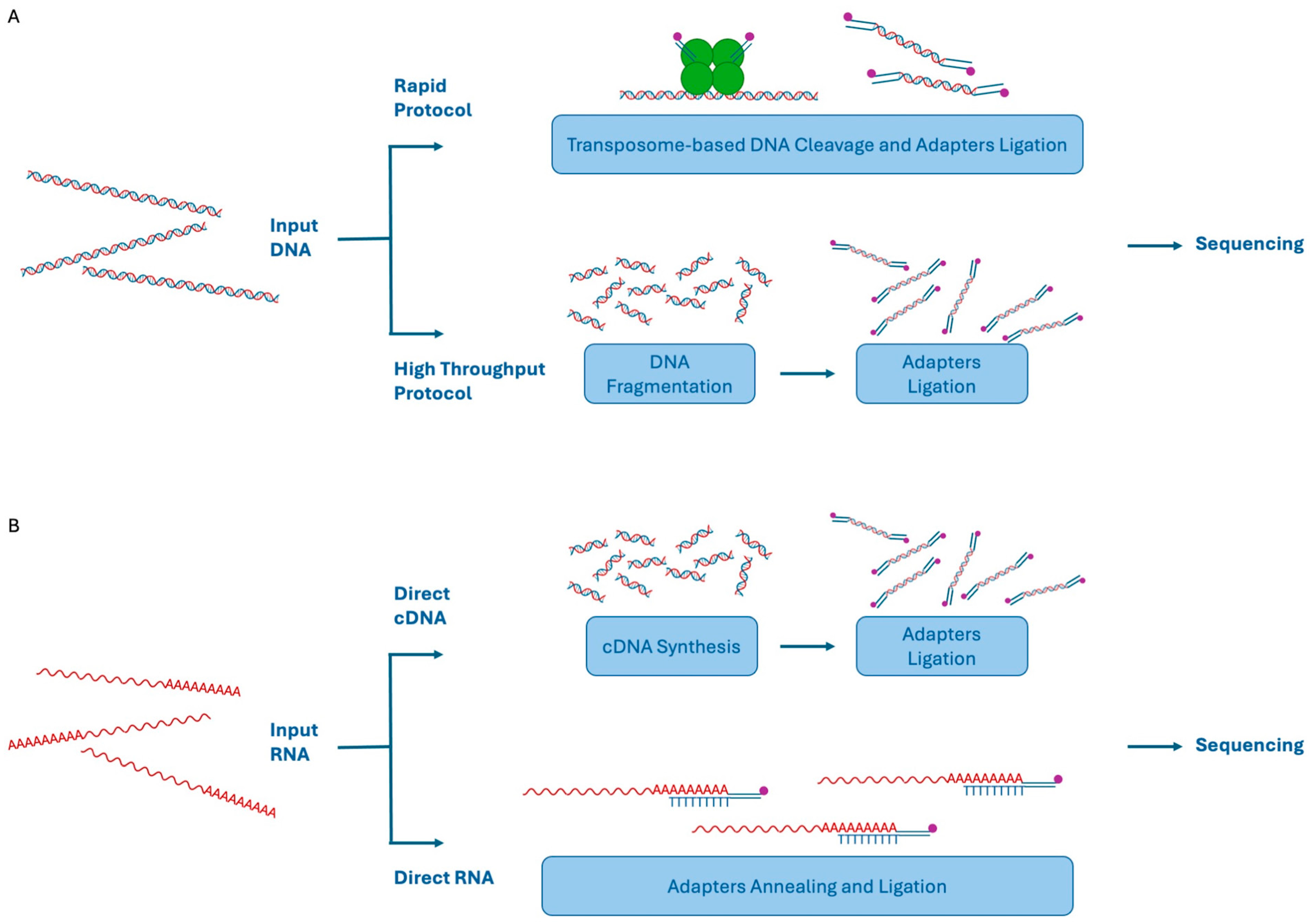
2.2. Oxford Nanopore Technologies
2.3. TGS Data Analysis
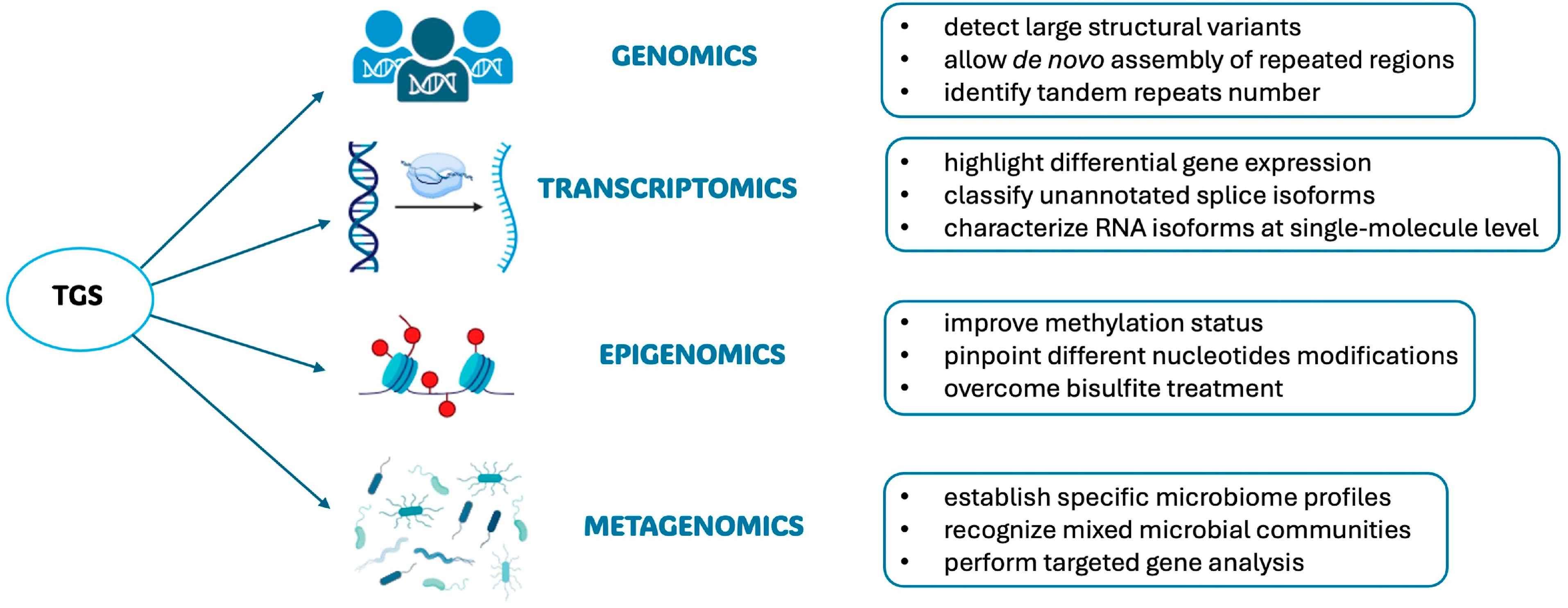
3. Third-Generation Sequencing Applications
3.1. Genome Sequencing
3.2. RNA Sequencing
3.3. Epigenetics
3.4. Metagenomics
3.5. TGS in Single-Cell Multiomics
4. Current Limitations and Future Perspectives of TGS
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Di Resta, C.; D’Argenio, V. Editorial: Whole Genome Sequencing for rare diseases. Front. Med. 2023, 10, 1267930. [Google Scholar] [CrossRef] [PubMed]
- Veneruso, I.; Di Resta, C.; Tomaiuolo, R.; D’Argenio, V. Current Updates on Expanded Carrier Screening: New Insights in the Omics Era. Medicina 2022, 58, 455. [Google Scholar] [CrossRef] [PubMed]
- Precone, V.; Del Monaco, V.; Esposito, M.V.; De Palma, F.D.E.; Ruocco, A.; Salvatore, F.; D’Argenio, V. Cracking the Code of Human Diseases Using Next-Generation Sequencing: Applications, Challenges, and Perspectives. Biomed. Res. Int. 2015, 2015, 161648. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Abdullah, S.T.; Salihi, A.; Sabir, D.K.; Sidiq, K.R.; Rasul, M.F.; Hidayat, H.J.; Ghafouri-Fard, S.; Taheri, M.; Jamali, E. The emerging roles of NGS in clinical oncology and personalized medicine. Pathol. Res. Pract. 2022, 230, 153760. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V. The High-Throughput Analyses Era: Are We Ready for the Data Struggle? High. Throughput 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.F.; Steinmann, K.E. Single molecule sequencing with a HeliScope genetic analysis system. Curr. Protoc. Mol. Biol. 2010, 92, 7.10.1–7.10.14. [Google Scholar] [CrossRef] [PubMed]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Al’Khafaji, A.M.; Smith, J.T.; Garimella, K.V.; Babadi, M.; Popic, V.; Sade-Feldman, M.; Gatzen, M.; Sarkizova, S.; Schwartz, M.A.; Blaum, E.M.; et al. High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat. Biotechnol. 2024, 42, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Shiroma, A.; Shimoji, M.; Tamotsu, H.; Ashimine, N.; Ohki, S.; Shinzato, M.; Minami, M.; Nakanishi, T.; Teruya, K.; et al. Advantages of genome sequencing by long-read sequencer using SMRT technology in medical area. Hum. Cell 2017, 30, 149–161. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Technology and Nanopore Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based Fourth-generation DNA Sequencing Technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Miten, J.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Ermini, L.; Wang, H.; Carty, K.; Cheung, M.S. LongQC: A quality control tool for third generation sequencing long read data. G3 2020, 10, 1193–1196. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C.W. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Yohe, S.; Thyagarajan, B. Review of Clinical Next-Generation Sequencing. Arch. Pathol. Lab. Med. 2017, 141, 1544–1557. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; Fitzhugh, W.; et al. Initial sequencing and analysis of the human genome: International Human Genome Sequencing Consortium. Nature 2001, 409, 860–921, Erratum in Nature 2001, 412, 565–566. [Google Scholar]
- Baker, M. De novo genome assembly: What every biologist should know. Nat. Methods 2012, 9, 333–337. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Symmons, O.; Spitz, F.; Korbel, J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat. Rev. Genet. 2013, 14, 125–138. [Google Scholar] [CrossRef]
- Nolin, S.L.; Glicksman, A.; Ersalesi, N.; Dobkin, C.; Brown, W.T.; Cao, R.; Blatt, E.; Sah, S.; Latham, G.J.; Hadd, A.G. Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med. 2015, 17, 358–364. [Google Scholar] [CrossRef]
- Loomis, E.W.; Eid, J.S.; Peluso, P.; Yin, J.; Hickey, L.; Rank, D.; McCalmon, S.; Hagerman, R.J.; Tassone, F.; Hagerman, P.J. Sequencing the unsequenceable: Expanded CGG-repeat alleles of the fragile X gene. Genome Res. 2013, 23, 121–128. [Google Scholar] [CrossRef]
- McFarland, K.N.; Liu, J.; Landrian, I.; Godiska, R.; Shanker, S.; Yu, F.; Farmerie, W.G.; Ashizawa, T. SMRT Sequencing of Long Tandem Nucleotide Repeats in SCA10 Reveals Unique Insight of Repeat Expansion Structure. PLoS ONE 2015, 10, e0135906. [Google Scholar] [CrossRef] [PubMed]
- Höijer, I.; Tsai, Y.C.; Clark, T.A.; Kotturi, P.; Dahl, N.; Stattin, E.L.; Bondeson, M.L.; Feuk, L.; Gyllensten, U.; Ameur, A. Detailed analysis of HTT repeat elements in human blood using targeted amplification-free long-read sequencing. Hum. Mutat. 2018, 39, 1262–1272. [Google Scholar] [CrossRef]
- Melas, M.; Kautto, E.A.; Franklin, S.J.; Mori, M.; McBride, K.L.; Mosher, T.M.; Pfau, R.B.; Hernandez-Gonzalez, M.E.; McGrath, S.D.; Magrini, V.J.; et al. Long-read whole genome sequencing reveals HOXD13 alterations in synpolydactyly. Hum. Mutat. 2022, 43, 189–199. [Google Scholar] [CrossRef]
- Borràs, D.M.; Vossen, R.H.A.M.; Liem, M.; Buermans, H.P.J.; Dauwerse, H.; van Heusden, D.; Gansevoort, R.T.; den Dunnen, J.T.; Janssen, B.; Peters, D.J.M.; et al. Detecting PKD1 variants in polycystic kidney disease patients by single-molecule long-read sequencing. Hum. Mutat. 2017, 38, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, S.M.; Lawlor, J.M.J.; Handley, L.H.; Ramaker, R.C.; Rogers, B.B.; Partridge, E.C.; Boston, L.B.; Williams, M.; Plott, C.B.; Jenkins, J.; et al. Long-read genome sequencing for the diagnosis of neurodevelopmental disorders. HGG Adv. 2021, 2, 100023. [Google Scholar] [PubMed]
- Pauper, M.; Kucuk, E.; Wenger, A.M.; Chakraborty, S.; Baybayan, P.; Kwint, M.; van der Sanden, B.; Nelen, M.R.; Derks, R.; Brunner, H.G.; et al. Long-read trio sequencing of individuals with unsolved intellectual disability. Eur. J. Hum. Genet. 2021, 29, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Mehinovic, E.; Gray, T.; Campbell, M.; Ekholm, J.; Wenger, A.; Rowell, W.; Grudo, A.; Grimwood, J.; Korlach, J.; Gurnett, C.; et al. Germline mosaicism of a missense variant in KCNC2 in a multiplex family with autism and epilepsy characterized by long-read sequencing. Am. J. Med. Genet. A 2022, 188, 2071–2081. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Wang, C.; Li, L.; Pan, L.; Huang, C.; Zhang, C.; Huang, Y.; Qiu, Y.; Lin, F.; Huang, Y. Third-generation sequencing for genetic disease. Clin. Chim. Acta 2023, 551, 117624. [Google Scholar] [CrossRef]
- Liang, Q.; Gu, W.; Chen, P.; Li, Y.; Liu, Y.; Tian, M.; Zhou, Q.; Qi, H.; Zhang, Y.; He, J.; et al. A More Universal Approach to Comprehensive Analysis of Thalassemia Alleles (CATSA). J. Mol. Diagn. 2021, 23, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Strych, L.; Černá, M.; Hejnalová, M.; Zavoral, T.; Komrsková, P.; Tejcová, J.; Bitar, I.; Sládková, E.; Sýkora, J.; Šubrt, I. Targeted long-read sequencing identified a causal structural variant in X-linked nephrogenic diabetes insipidus. BMC Med. Genom. 2024, 22, 29. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Nakagawa, S.; Takahashi Ueda, M.; Imanishi, T.; Frith, M.C.; Mitsuhashi, H. Nanopore-based single molecule sequencing of the D4Z4 array responsible for facioscapulohumeral muscular dystrophy. Sci. Rep. 2017, 7, 14789. [Google Scholar] [CrossRef]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017, 8, 1326. [Google Scholar] [CrossRef]
- Leija-Salazar, M.; Sedlazeck, F.J.; Toffoli, M.; Mullin, S.; Mokretar, K.; Athanasopoulou, M.; Donald, A.; Sharma, R.; Hughes, D.; Schapira, A.H.V.; et al. Detection of GBA missense mutations and other variants using the Oxford Nanopore MinION. Mol. Genet. Genom. Med. 2019, 7, e564. [Google Scholar] [CrossRef] [PubMed]
- Bruels, C.C.; Little, H.R.; Daugherty, A.L.; Stafki, S.; Estrella, E.A.; McGaughy, E.S.; Truong, D.; Badalamenti, J.P.; Pais, L.; Ganesh, V.S.; et al. Diagnostic capabilities of nanopore long- read sequencing in muscular dystrophy. Ann. Clin. Transl. Neurol. 2022, 9, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Chin, H.L.; O’Neill, K.; Louie, K.; Brown, L.; Schlade-Bartusiak, K.; Eydoux, P.; Rupps, R.; Farahani, A.; Boerkoel, C.F.; Jones, S.J.M. An approach to rapid characterization of DMD copy number variants for prenatal risk assessment. Am. J. Med. Genet. A 2021, 185, 2541–2545. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Deng, J.; Guo, X.; Shan, J.; Luan, X.; Cao, L.; Zhao, J.; Yu, M.; Zhang, W.; Lv, H.; et al. The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain 2021, 144, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shan, J.; Yu, M.; Di, L.; Xie, Z.; Zhang, W.; Lv, H.; Meng, L.; Zheng, Y.; Zhao, Y.; et al. The CGG repeat expansion in RILPL1 is associated with oculopharyngodistal myopathy type 4. Am. J. Hum. Genet. 2022, 109, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, K.; Stacey, A.; Mustafi, D. Targeted long-read sequencing allows for rapid identification of pathogenic disease-causing variants in retinoblastoma. Ophthalmic Genet. 2022, 43, 762–770. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Al-temose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, I.; Gispert, J.D.; Palumbo, E.; Muñoz-Aguirre, M.; Wucher, V.; D’Argenio, V.; Santpere, G.; Navarro, A.; Guigo, R.; Vilor-Tejedor, N. Brain transcriptomic profiling reveals common alterations across neurodegenerative and psychiatric disorders. Comput. Struct. Biotechnol. J. 2022, 20, 4549–4561. [Google Scholar] [CrossRef] [PubMed]
- De Palma, F.D.E.; Del Monaco, V.; Pol, J.G.; Kremer, M.; D’Argenio, V.; Stoll, G.; Montanaro, D.; Uszczyńska-Ratajczak, B.; Klein, C.C.; Vlasova, A.; et al. The abundance of the long intergenic non-coding RNA 01087 differentiates between luminal and triple-negative breast cancers and predicts patient outcome. Pharmacol. Res. 2020, 161, 105249. [Google Scholar] [CrossRef] [PubMed]
- Withanage, M.H.H.; Liang, H.; Zeng, E. RNA-Seq Experiment and Data Analysis. Methods Mol. Biol. 2022, 2418, 405–424. [Google Scholar]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef]
- Cui, J.; Shen, N.; Lu, Z.; Xu, G.; Wang, Y.; Jin, B. Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 2020, 16, 85. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef]
- Oikonomopoulos, S.; Bayega, A.; Fahiminiya, S.; Djambazian, H.; Berube, P.; Ragoussis, J. Methodologies for Transcript Profiling Using Long-Read Technologies. Front. Genet. 2020, 11, 606. [Google Scholar] [CrossRef]
- Sharon, D.; Tilgner, H.; Grubert, F.; Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013, 31, 1009–1014. [Google Scholar] [CrossRef]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Razaghi, R.; Zuzarte, P.C.; Gilpatrick, T.; Payne, A.; Quick, J.; et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 2019, 16, 1297–1305. [Google Scholar] [CrossRef]
- Xie, S.; Wing-Sze Leung, A.; Zheng, Z.; Zhang, D.; Xiao, C.; Luo, R.; Luo, M.; Zhang, S. Applications and potentials of nanopore sequencing in the (epi)genome and (epi)transcriptome era. Innovation 2021, 2, 100153. [Google Scholar] [CrossRef]
- Aw, J.G.A.; Lim, S.W.; Wang, J.X.; Lambert, F.R.P.; Tan, W.T.; Shen, Y.; Zhang, Y.; Kaewsapsak, P.; Li, C.; Ng, S.B.; et al. Determination of isoform-specific RNA structure with nanopore long reads. Nat. Biotechnol. 2021, 39, 336–346. [Google Scholar] [CrossRef]
- Pardo-Palacios, F.J.; Wang, D.; Reese, F.; Diekhans, M.; Carbonell-Sala, S.; Williams, B.; Love-land, J.E.; De María, M.; Adams, M.S.; Balderrama-Gutierrez, G.; et al. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. bioRxiv 2023, 2023.07.25.550582. [Google Scholar]
- Li, J.; Wang, H.; Mao, L.; Yu, H.; Yu, X.; Sun, Z.; Qian, X.; Cheng, S.; Chen, S.; Chen, J.; et al. Rapid genomic characterization of SARS-CoV-2 viruses from clinical specimens using nanopore sequencing. Sci. Rep. 2020, 10, 17492. [Google Scholar] [CrossRef]
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Front. Biosci. 2020, 25, 1058–1109. [Google Scholar]
- Ballestar, E. An Introduction to Epigenetics. Adv. Exp. Med. Biol. 2011, 711, 1–11. [Google Scholar] [PubMed]
- Jin, Z.; Liu, Y. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of tumors in mice by genomic hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Silva, J.M.; Dominguez, G.; Bonilla, F.; Matias-Guiu, X.; Lerma, E.; Bussaglia, E.; Prat, J.; Harkes, I.C.; Repasky, E.A.; et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000, 92, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Gouil, Q.; Keniry, A. Latest techniques to study DNA methylation. Essays Biochem. 2019, 63, 639–648. [Google Scholar] [PubMed]
- Park, Y.; Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 2016, 32, 1446–1453. [Google Scholar] [CrossRef]
- Sandhu, C.; Qureshi, A.; Emili, A. Panomics for Precision Medicine. Trends Mol. Med. 2018, 24, 85–101. [Google Scholar] [CrossRef]
- Li, N.; Ye, M.; Li, Y.; Yan, Z.; Butcher, L.M.; Sun, J.; Han, X.; Chen, Q.; Zhang, X.; Wang, J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 2010, 52, 203–212. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Ku, C.S.; Naidoo, N.; Wu, M.; Soong, R. Studying the epigenome using next generation sequencing. J. Med. Genet. 2011, 48, 721–730. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef]
- Wallace, E.V.; Stoddart, D.; Heron, A.J.; Mikhailova, E.; Maglia, G.; Donohoe, T.J.; Bayley, H. Identification of epigenetic DNA modifications with a protein nanopore. Chem. Commun. 2010, 46, 8195–8197. [Google Scholar] [CrossRef]
- Schreiber, J.; Wescoe, Z.L.; Abu-Shumays, R.; Vivian, J.T.; Baatar, B.; Karplus, K.; Akeson, M. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc. Natl. Acad. Sci. USA 2013, 110, 18910–18915. [Google Scholar] [CrossRef]
- Laszlo, A.H.; Derrington, I.M.; Brinkerhoff, H.; Langford, K.W.; Nova, I.C.; Samson, J.M.; Bartlett, J.J.; Pavlenok, M.; Gundlach, J.H. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA 2013, 110, 18904–18909. [Google Scholar] [CrossRef]
- Wescoe, Z.L.; Schreiber, J.; Akeson, M. Nanopores discriminate among five C5-cytosine variants in DNA. J. Am. Chem. Soc. 2014, 136, 6582–6587. [Google Scholar] [CrossRef]
- Gabrieli, T.; Sharim, H.; Fridman, D.; Arbib, N.; Michaeli, Y.; Ebenstein, Y. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 2018, 46, e87. [Google Scholar] [CrossRef]
- Wongsurawat, T.; Jenjaroenpun, P.; De Loose, A.; Alkam, D.; Ussery, D.W.; Nookaew, I.; Leung, Y.K.; Ho, S.M.; Day, J.D.; Rodriguez, A. A novel Cas9-targeted long-read assay for simultaneous detection of IDH1/2 mutations and clinically relevant MGMT methylation in fresh biopsies of diffuse glioma. Acta Neuropathol. Commun. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, Y.; Qian, P.Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Braekel, J.V.; Fu, Q.; Roosens, N.H.; Keersmaecker, S.C.; Vanneste, K. Targeting the 16s rRNA gene for bacterial identification in complex mixed samples: Comparative evaluation of second (Illumina) and third (Oxford Nanopore Technologies) generation sequencing technologies. Int. J. Mol. Sci. 2019, 21, 298. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Pongpanich, M.; Porntaveetus, T. Unraveling metagenomics through long-read sequencing: A comprehensive review. J. Transl. Med. 2024, 22, 111. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Coupland, P.; Browne, H.P.; Lawley, T.D.; Francis, S.C.; Parkhill, J. Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiol. 2016, 16, 274. [Google Scholar] [CrossRef]
- Earl, J.P.; Adappa, N.D.; Krol, J.; Bhat, A.S.; Balashov, S.; Ehrlich, R.L.; Palmer, J.N.; Workman, A.D.; Blasetti, M.; Sen, B.; et al. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome 2018, 6, 190. [Google Scholar] [CrossRef]
- Whon, T.W.; Chung, W.H.; Lim, M.Y.; Song, E.J.; Kim, P.S.; Hyun, D.W.; Shin, N.R.; Bae, J.W.; Nam, Y.D. The effects of sequencing platforms on phylogenetic resolution in 16 S rRNA gene profiling of human feces. Sci. Data 2018, 5, 180068. [Google Scholar] [CrossRef]
- Hur, M.; Park, S.J. Identification of Microbial Profiles in Heavy-Metal-Contaminated Soil from Full-Length 16S rRNA Reads Sequenced by a PacBio System. Microorganisms 2019, 7, 357. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nishijima, S.; Furuta, Y.; Yoshimura, J.; Suda, W.; Oshima, K.; Hattori, M.; Morishita, S. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 2019, 7, 119. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, H.; Wang, F.; Yang, Y.; Wang, X.; Chen, B.; Stampfli, M.R.; Zhou, H.; Shu, W.; Brightling, C.E.; et al. A Refined View of Airway Microbiome in Chronic Obstructive Pulmonary Disease at Species and Strain-Levels. Front. Microbiol. 2020, 11, 1758. [Google Scholar] [CrossRef] [PubMed]
- Kiguchi, Y.; Nishijima, S.; Kumar, N.; Hattori, M.; Suda, W. Long-read metagenomics of multiple displacement amplified DNA of low-biomass human gut phageomes by SACRA pre-processing chimeric reads. DNA Res. 2021, 28, dsab019. [Google Scholar] [CrossRef]
- Zhang, M.; Tang, H.; Yuan, Y.; Ou, Z.; Chen, Z.; Xu, Y.; Fu, X.; Zhao, Z.; Sun, Y. The Role of Indoor Microbiome and Metabolites in Shaping Children’s Nasal and Oral Microbiota: A Pilot Multi-Omic Analysis. Metabolites 2023, 13, 1040. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Nesme, J.; Santos-Bay, L.; Koziol, A.; Sørensen, S.J.; Alberdi, A.; Aizpurua, O. A comparison of short-read, HiFi long-read, and hybrid strategies for genome-resolved metagenomics. Microbiol. Spectr. 2024, e0359023, online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Debbonaire, A.R.; Sattler, B.; Mur, L.A.J.; Hodson, J.A. Extreme metagenomics using nanopore DNA sequencing: A field report from Svalbard, 78° N. bioRxiv 2016. bioRxiv:073965. [Google Scholar]
- Xiao, M.; Stachler, E.; Bibby, K. Evaluation of oxford nanopore MinIONTM sequencing for 16S rRNA microbiome characterization. bioRxiv 2017. bioRxiv:099960. [Google Scholar]
- Shin, J.; Lee, S.; Go, M.J.; Lee, S.Y.; Kim, S.C.; Lee, C.H.; Cho, B.K. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci. Rep. 2016, 6, 29681. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Portune, K.J.; Sanz, Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION™ portable nanopore sequencer. Gigascience 2016, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Watson, M.; Minot, S.S.; Rivera, M.C.; Franklin, R.B. MinION nanopore sequencing of environmental metagenomes: A synthetic approach. Gigascience 2017, 6, gix007. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Kryukov, K.; Nakagawa, S.; Takeuchi, J.S.; Shiraishi, Y.; Asano, K.; Imanishi, T. A portable system for rapid bacterial composition analysis using a nanopore-based sequencer and laptop computer. Sci. Rep. 2017, 7, 5657. [Google Scholar] [CrossRef]
- Yang, L.; Haidar, G.; Zia, H.; Nettles, R.; Qin, S.; Wang, X.; Shah, F.; Rapport, S.F.; Charalampous, T.; Methé, B.; et al. Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: A feasibility and clinical validity study. Respir. Res. 2019, 20, 265. [Google Scholar] [CrossRef] [PubMed]
- Ibironke, O.; McGuinness, L.R.; Lu, S.E.; Wang, Y.; Hussain, S.; Weisel, C.P.; Kerkhof, L.J. Species-level evaluation of the human respiratory microbiome. Gigascience 2020, 9, giaa038. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.S.; Pearson, J.; Miller, A.; Schmeier, S.; Frizelle, F.A.; Purcell, R.V. MinION Sequencing of colorectal cancer tumour microbiomes-A comparison with amplicon-based and RNA-Sequencing. PLoS ONE 2020, 15, e0233170. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef]
- Chen, Y.W.; Hou, Y.W.; Wang, C.W.; Cheng, S.J.; Kuo, W.T.; Lin, C.P.; Hou, H.H. Oral Lactobacillus zeae exacerbates the pathological manifestation of periodontitis in a mouse model. Mol. Oral. Microbiol. 2024. online ahead of print. [Google Scholar] [CrossRef]
- Brlek, P.; Bulić, L.; Bračić, M.; Projić, P.; Škaro, V.; Shah, N.; Shah, P.; Primorac, D. Implement-ing Whole Genome Sequencing (WGS) in Clinical Practice: Advantages, Challenges, and Future Perspectives. Cells 2024, 13, 504. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, C.; Li, W.; Bai, X.; Zhou, X.; Xie, H.; Wen, L.; Tang, F. SMOOTH-seq: Single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021, 22, 195. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Deng, E.; Wang, J.; Zhou, W.; Ao, J.; Liu, R.; Su, D.; Fan, X. Single-cell third-generation sequencing-based multi-omics uncovers gene expression changes governed by ecDNA and structural variants in cancer cells. Clin. Transl. Med. 2023, 13, e1351. [Google Scholar] [CrossRef] [PubMed]
- Olivucci, G.; Iovino, E.; Innella, G.; Turchetti, D.; Pippucci, T.; Magini, P. Long read sequenc-ing on its way to the routine diagnostics of genetic diseases. Front. Genet. 2024, 15, 1374860. [Google Scholar] [CrossRef]
- Ameur, A.; Kloosterman, W.P.; Hestand, M.S. Single-Molecule Sequencing: Towards Clinical Applications. Trends Biotechnol. 2019, 37, 72–85. [Google Scholar] [CrossRef]

| Third-Generation Sequencing Technologies | Next-Generation Sequencing | ||
|---|---|---|---|
| Features | PacBio | ONT | Illumina |
| Sequencing Chemistry | SMRT | Nanopore-based | Sequencing by synthesis |
| Average reads length | 15–20 kb | 10 kb–4 Mb | 2 × 300 bp 3 |
| Base-calling accuracy | up to 99.95% 1 | 99.9% | 99.9% |
| Maximum throughput/run | 360 Gb 1 | 290 Gb 2 | 8 Tb 4 |
| Cost per Gb * | 65–200 $ | 22–90 $ | 12–27 $ |
| Complex genomic regions (GC-rich, homopolymers) analysis | Yes | Yes | No |
| Direct methylation detection | Yes | Yes | No |
| Pros | Long reads High accuracy Allows direct cDNA analysis Allows direct methylation and other DNA modifications analysis | Very long reads Allows direct RNA analysis Allows direct methylation and other DNA modifications analysis Availability of portable sequencers | High accuracy High sensitivity High multiplexing capacity High versatility in several application fields |
| Cons | High instruments costs Bionformatic requirements | Sequencing cost are still higher than NGS Bionformatic requirements | No long reads Requires PCR amplification Does not allow direct RNA analysis Low accuracy in complex genomic regions analysis Time-consuming workflows |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scarano, C.; Veneruso, I.; De Simone, R.R.; Di Bonito, G.; Secondino, A.; D’Argenio, V. The Third-Generation Sequencing Challenge: Novel Insights for the Omic Sciences. Biomolecules 2024, 14, 568. https://doi.org/10.3390/biom14050568
Scarano C, Veneruso I, De Simone RR, Di Bonito G, Secondino A, D’Argenio V. The Third-Generation Sequencing Challenge: Novel Insights for the Omic Sciences. Biomolecules. 2024; 14(5):568. https://doi.org/10.3390/biom14050568
Chicago/Turabian StyleScarano, Carmela, Iolanda Veneruso, Rosa Redenta De Simone, Gennaro Di Bonito, Angela Secondino, and Valeria D’Argenio. 2024. "The Third-Generation Sequencing Challenge: Novel Insights for the Omic Sciences" Biomolecules 14, no. 5: 568. https://doi.org/10.3390/biom14050568
APA StyleScarano, C., Veneruso, I., De Simone, R. R., Di Bonito, G., Secondino, A., & D’Argenio, V. (2024). The Third-Generation Sequencing Challenge: Novel Insights for the Omic Sciences. Biomolecules, 14(5), 568. https://doi.org/10.3390/biom14050568








