Two Modulators of Skeletal Development: BMPs and Proteoglycans
Abstract
:1. Introduction
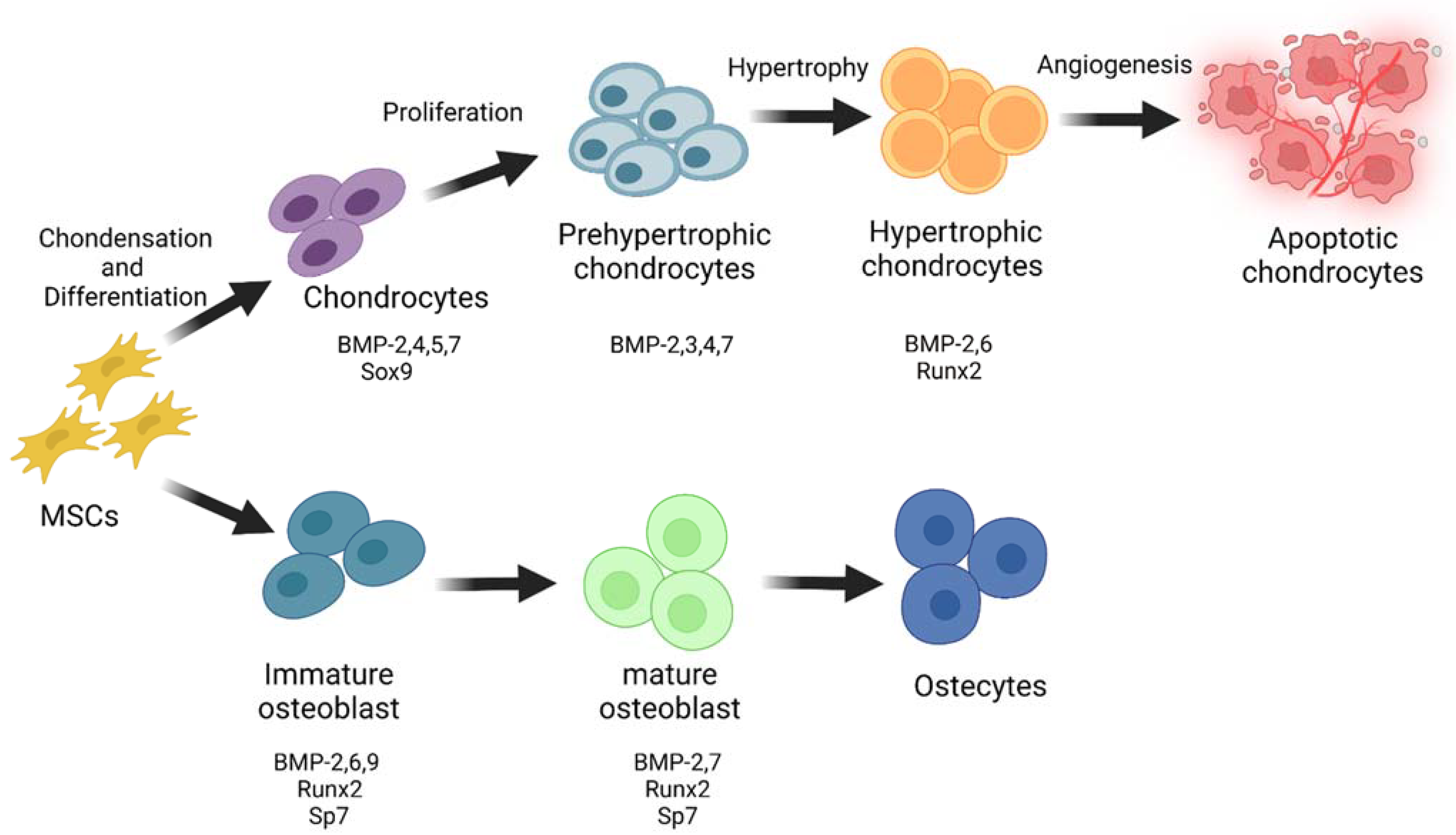
2. Role of BMPs in Skeletal Development
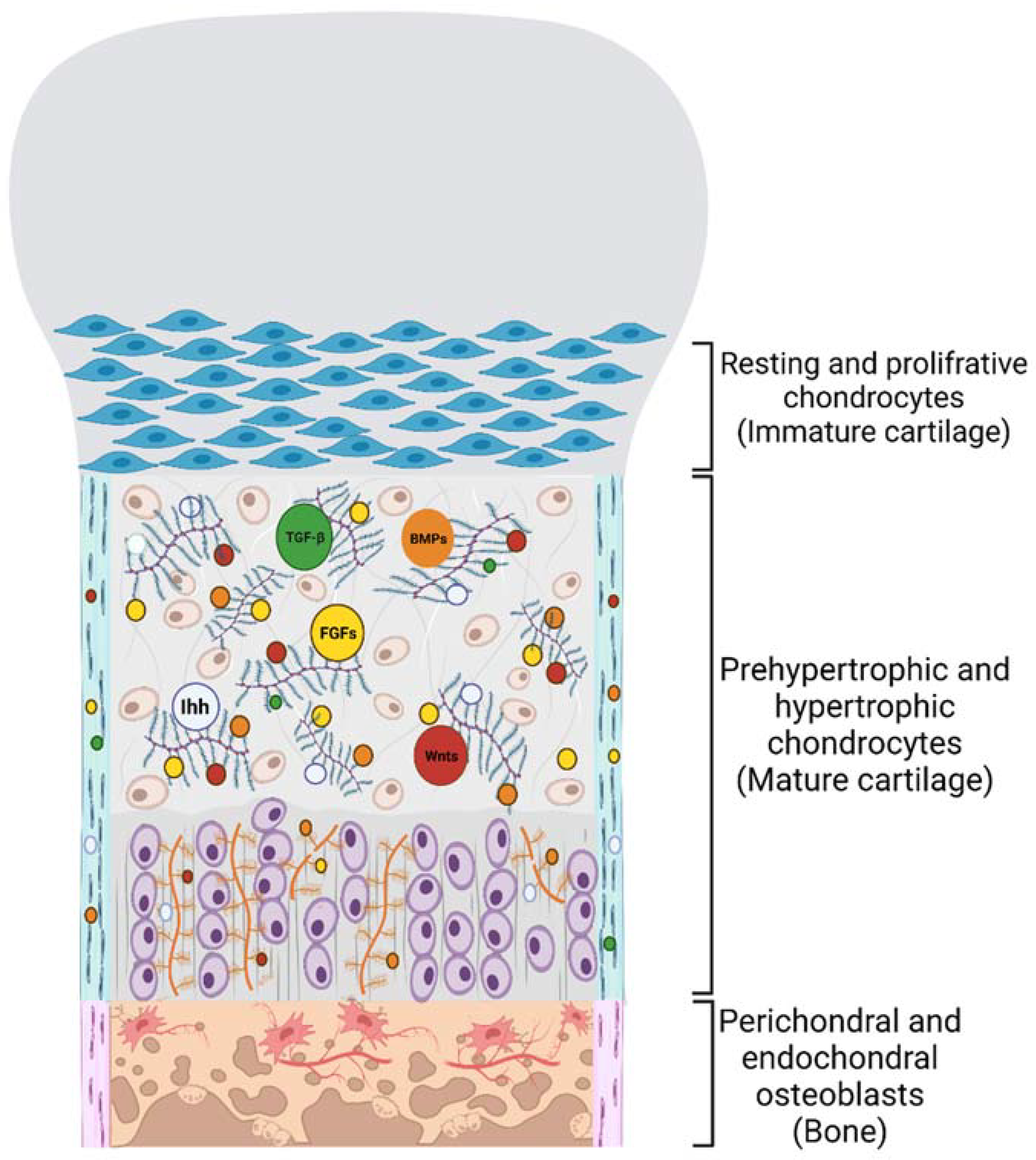
3. PGs’ Function in Cell-ECM Crosstalk and Skeletal Development
4. PGs Modulate BMP Signal Transduction
5. Exogenous Mechanisms to Modulate BMP Signalling Pathway

6. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Urist, M.R. Bone: Formation by autoinduction. Science 1965, 150, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Wozney, J.M. Overview of bone morphogenetic proteins. Spine 2002, 27, S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Newfeld, S.J.; Wisotzkey, R.G.; Kumar, S. Molecular evolution of a developmental pathway: Phylogenetic analyses of transforming growth factor-β family ligands, receptors and Smad signal transducers. Genetics 1999, 152, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Aono, A.; Hazama, M.; Notoya, K.; Taketomi, S.; Yamasaki, H.; Tsukuda, R.; Sasaki, S.; Fujisawa, Y. Potent ectopic bone-inducing activity of bone morphogenetic protein-4/7 heterodimer. Biochem. Biophys. Res. Commun. 1995, 210, 670–677. [Google Scholar] [CrossRef]
- Israel, D.I.; Nove, J.; Kerns, K.M.; Kaufman, R.J.; Rosen, V.; Cox, K.A.; Wozney, J.M. Heterodimeric bone morphogenetic proteins show enhanced activity in vitro and in vivo. Growth Factors 1996, 13, 291–300. [Google Scholar] [CrossRef]
- Kaito, T.; Morimoto, T.; Mori, Y.; Kanayama, S.; Makino, T.; Takenaka, S.; Sakai, Y.; Otsuru, S.; Yoshioka, Y.; Yoshikawa, H. BMP-2/7 heterodimer strongly induces bone regeneration in the absence of increased soft tissue inflammation. Spine J. 2018, 18, 139–146. [Google Scholar] [CrossRef]
- Gomez-Puerto, M.C.; Iyengar, P.V.; García de Vinuesa, A.; ten Dijke, P.; Sanchez-Duffhues, G. Bone morphogenetic protein receptor signal transduction in human disease. J. Pathol. 2019, 247, 9–20. [Google Scholar] [CrossRef]
- Olsen, O.E.; Wader, K.F.; Hella, H.; Mylin, A.K.; Turesson, I.; Nesthus, I.; Waage, A.; Sundan, A.; Holien, T. Activin A inhibits BMP-signaling by binding ACVR2A and ACVR2B. Cell Commun. Signal. 2015, 13, 27. [Google Scholar] [CrossRef] [Green Version]
- Mueller, T.D.; Nickel, J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012, 586, 1846–1859. [Google Scholar] [CrossRef]
- Scharpfenecker, M.; van Dinther, M.; Liu, Z.; van Bezooijen, R.L.; Zhao, Q.; Pukac, L.; Löwik, C.W.; ten Dijke, P. BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J. Cell Sci. 2007, 120, 964–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebisawa, T.; Tada, K.; Kitajima, I.; Tojo, K.; Sampath, T.K.; Kawabata, M.; Miyazono, K.; Imamura, T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. 1999, 112, 3519–3527. [Google Scholar] [CrossRef] [PubMed]
- Ten Dijke, P.; Yamashita, H.; Sampath, T.K.; Reddi, A.H.; Estevez, M.; Riddle, D.L.; Ichijo, H.; Heldin, C.-H.; Miyazono, K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J. Biol. Chem. 1994, 269, 16985–16988. [Google Scholar] [CrossRef]
- De Caestecker, M. The transforming growth factor-β superfamily of receptors. Cytokine Growth Factor Rev. 2004, 15, 1–11. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Massagué, J.; Seoane, J.; Wotton, D. Smad transcription factors. Genes Dev. 2005, 19, 2783–2810. [Google Scholar] [CrossRef] [Green Version]
- Goldring, M.B.; Tsuchimochi, K.; Ijiri, K. The control of chondrogenesis. J. Cell. Biochem. 2006, 97, 33–44. [Google Scholar] [CrossRef]
- Ijiri, K.; Zerbini, L.F.; Peng, H.; Correa, R.G.; Lu, B.; Walsh, N.; Zhao, Y.; Taniguchi, N.; Huang, X.-L.; Otu, H.; et al. A novel role for GADD45β as a mediator of MMP-13 gene expression during chondrocyte terminal differentiation. J. Biol. Chem. 2005, 280, 38544–38555. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, R.; Hata, K.; Ikeda, F.; Matsubara, T.; Yamashita, K.; Ichida, F.; Yoneda, T. The role of Smads in BMP signaling. Front. Biosci. 2003, 8, s275–s284. [Google Scholar] [CrossRef] [Green Version]
- Nohe, A.; Keating, E.; Knaus, P.; Petersen, N.O. Signal transduction of bone morphogenetic protein receptors. Cell. Signal. 2004, 16, 291–299. [Google Scholar] [CrossRef]
- Rosen, V. BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 2009, 20, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Dietz, H.C.; Pyeritz, R.E. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum. Mol. Genet. 1995, 4 (Suppl. 1), 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Gregory, K.E.; Ono, R.N.; Charbonneau, N.L.; Kuo, C.-L.; Keene, D.R.; Bächinger, H.P.; Sakai, L.Y. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J. Biol. Chem. 2005, 280, 27970–27980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakai, L.Y.; Keene, D.R.; Engvall, E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J. Cell Biol. 1986, 103, 2499–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Harris, R.E.; Bayston, L.J.; Ashe, H.L. Type IV collagens regulate BMP signalling in Drosophila. Nature 2008, 455, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Abreu, J.G.; Coffinier, C.; Larraın, J.; Oelgeschläger, M.; De Robertis, E. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 2002, 287, 39–47. [Google Scholar] [CrossRef]
- Andhare, R.; Takahashi, N.; Knudson, W.; Knudson, C. Hyaluronan promotes the chondrocyte response to BMP-7. Osteoarthr. Cartil. 2009, 17, 906–916. [Google Scholar] [CrossRef] [Green Version]
- Peterson, R.S.; Andhare, R.A.; Rousche, K.T.; Knudson, W.; Wang, W.; Grossfield, J.B.; Thomas, R.O.; Hollingsworth, R.E.; Knudson, C.B. CD44 modulates Smad1 activation in the BMP-7 signaling pathway. J. Cell Biol. 2004, 166, 1081–1091. [Google Scholar] [CrossRef]
- Selbi, W.; De La Motte, C.; Hascall, V.; Phillips, A. BMP-7 modulates hyaluronan-mediated proximal tubular cell-monocyte interaction. J. Am. Soc. Nephrol. 2004, 15, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Erlebacher, A.; Filvaroff, E.H.; Gitelman, S.E.; Derynck, R. Toward a molecular understanding of skeletal development. Cell 1995, 80, 371–378. [Google Scholar] [CrossRef] [Green Version]
- Olsen, B.R.; Reginato, A.M.; Wang, W. Bone Development. Annu. Rev. Cell Dev. Biol. 2000, 16, 191–220. [Google Scholar] [CrossRef] [PubMed]
- Soltanoff, C.S.; Chen, W.; Yang, S.; Li, Y.-P. Signaling networks that control the lineage commitment and differentiation of bone cells. Crit. Rev. Eukaryot. Gene Expr. 2009, 19, 1–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caplan, A. Endochondral bone formation: The lineage cascade. Mech. Bone Dev. Growth 1994, 1–46. [Google Scholar]
- Cormack, D.H.; Ham, A.W. Ham’s Histology; Lippincott: New York, NY, USA, 1987. [Google Scholar]
- Eames, B.F.; De La Fuente, L.; Helms, J.A. Molecular ontogeny of the skeleton. Birth Defects Res. Part C Embryo Today Rev. 2003, 69, 93–101. [Google Scholar] [CrossRef]
- Eames, B.F.; Yan, Y.-L.; Swartz, M.E.; Levic, D.S.; Knapik, E.W.; Postlethwait, J.H.; Kimmel, C.B. Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 2011, 7, e1002246. [Google Scholar] [CrossRef] [Green Version]
- Kronenberg, H.M. Developmental regulation of the growth plate. Nature 2003, 423, 332–336. [Google Scholar] [CrossRef]
- Hendriks, M.; Ramasamy, S.K. Blood vessels and vascular niches in bone development and physiological remodeling. Front. Cell Dev. Biol. 2020, 8, 1481. [Google Scholar] [CrossRef]
- Gerber, H.-P.; Vu, T.H.; Ryan, A.M.; Kowalski, J.; Werb, Z.; Ferrara, N. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 1999, 5, 623–628. [Google Scholar] [CrossRef]
- Cheng, H.; Jiang, W.; Phillips, F.M.; Haydon, R.C.; Peng, Y.; Zhou, L.; Luu, H.H.; An, N.; Breyer, B.; Vanichakarn, P.; et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J. Bone Jt. Surg. 2003, 85, 1544–1552. [Google Scholar] [CrossRef]
- Satoh, A.; Suzuki, M.; Amano, T.; Tamura, K.; Ide, H. Joint development in Xenopus laevis and induction of segmentations in regenerating froglet limb (spike). Dev. Dyn. 2005, 233, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Dudley, A.T.; Robertson, E.J. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev. Dyn. 1997, 208, 349–362. [Google Scholar] [CrossRef]
- Kingsley, D.M. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994, 10, 16–21. [Google Scholar] [CrossRef]
- Lyons, K.M.; Hogan, B.L.; Robertson, E.J. Colocalization of BMP 7 and BMP 2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. Mech. Dev. 1995, 50, 71–83. [Google Scholar] [CrossRef]
- McMahon, J.A.; Takada, S.; Zimmerman, L.B.; Fan, C.-M.; Harland, R.M.; McMahon, A.P. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998, 12, 1438–1452. [Google Scholar] [CrossRef] [Green Version]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.; Hall, B.K. Chondrogenic cell differentiation from membrane bone periostea. Anat. Embryol. 1997, 196, 349–362. [Google Scholar] [CrossRef]
- Nakashima, K.; de Crombrugghe, B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003, 19, 458–466. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; De Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.C.; Hoang, B.; Thomas, J.T.; Vukicevic, S.; Luyten, F.P.; Ryba, N.; Kozak, C.A.; Reddi, A.H.; Moos, M. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J. Biol. Chem. 1994, 269, 28227–28234. [Google Scholar] [CrossRef]
- Jones, C.M.; Lyons, K.M.; Hogan, B. Involvement of Bone Morphogenetic Protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development 1991, 111, 531–542. [Google Scholar] [CrossRef]
- Lyons, K.M.; Pelton, R.; Hogan, B. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for bone morphogenetic protein-2A (BMP-2A). Development 1990, 109, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Macias, D.; Ganan, Y.; Sampath, T.; Piedra, M.; Ros, M.; Hurle, J. Role of BMP-2 and OP-1 (BMP-7) in programmed cell death and skeletogenesis during chick limb development. Development 1997, 124, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.; Schneider, S.; Rosowski, M.; Lauster, R.; Vortkamp, A. Expression of Fgf and Tgfβ signaling related genes during embryonic endochondral ossification. Gene Expr. Patterns 2005, 6, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Wozney, J.M.; Rosen, V.; Celeste, A.J.; Mitsock, L.M.; Whitters, M.J.; Kriz, R.W.; Hewick, R.M.; Wang, E.A. Novel regulators of bone formation: Molecular clones and activities. Science 1988, 242, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Settle, S.H., Jr.; Rountree, R.B.; Sinha, A.; Thacker, A.; Higgins, K.; Kingsley, D.M. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 2003, 254, 116–130. [Google Scholar] [CrossRef] [Green Version]
- Storm, E.E.; Kingsley, D.M. GDF5 coordinates bone and joint formation during digit development. Dev. Biol. 1999, 209, 11–27. [Google Scholar] [CrossRef] [Green Version]
- Wolfman, N.M.; Hattersley, G.; Cox, K.; Celeste, A.J.; Nelson, R.; Yamaji, N.; Dube, J.L.; DiBlasio-Smith, E.; Nove, J.; Song, J.J.; et al. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-beta gene family. J. Clin. Investig. 1997, 100, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Minina, E.; Wenzel, H.M.; Kreschel, C.; Karp, S.; Gaffield, W.; McMahon, A.P.; Vortkamp, A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development 2001, 128, 4523–4534. [Google Scholar] [CrossRef]
- Minina, E.; Kreschel, C.; Naski, M.C.; Ornitz, D.M.; Vortkamp, A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev. Cell 2002, 3, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Ohba, S. Hedgehog signaling in endochondral ossification. J. Dev. Biol. 2016, 4, 20. [Google Scholar] [CrossRef]
- Long, F.; Chung, U.-I.; Ohba, S.; McMahon, J.; Kronenberg, H.M.; McMahon, A.P. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 2004, 131, 1309–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St-Jacques, B.; Hammerschmidt, M.; McMahon, A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999, 13, 2072–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, K.; Hata, A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. J. Biol. Chem. 2004, 279, 18544–18549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathi, S.; Rutenberg, J.B.; Johnson, R.L.; Vortkamp, A. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Dev. Biol. 1999, 209, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Kawai, S.; Sugiura, T. Characterization of human bone morphogenetic protein (BMP)-4 and -7 gene promoters: Activation of BMP promoters by Gli, a sonic hedgehog mediator. Bone 2001, 29, 54–61. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994, 127, 1755–1766. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, A.; Katagiri, T.; Ikeda, T.; Wozney, J.M.; Rosen, V.; Wang, E.A.; Kahn, A.J.; Suda, T.; Yoshiki, S. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 1991, 113, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, A.; Komori, T.; Suda, T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev. 2000, 21, 393–411. [Google Scholar] [CrossRef]
- Chen, G.; Deng, C.; Li, Y.-P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272. [Google Scholar] [CrossRef] [Green Version]
- Dorman, L.J.; Tucci, M.; Benghuzzi, H. In vitro effects of bmp-2, bmp-7, and bmp-13 on proliferation and differentation of mouse mesenchymal stem cells. Biomed. Sci. Instrum. 2012, 48, 81–87. [Google Scholar]
- Hata, K.; Nishimura, R.; Ikeda, F.; Yamashita, K.; Matsubara, T.; Nokubi, T.; Yoneda, T. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor γ during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell 2003, 14, 545–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Q.; Song, W.-X.; Luo, Q.; Tang, N.; Luo, J.; Luo, X.; Chen, J.; Bi, Y.; He, B.-C.; Park, J.K.; et al. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009, 18, 545–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Ren, P.-G.; Ma, T.; Smith, R.L.; Goodman, S.B. Modulating osteogenesis of mesenchymal stem cells by modifying growth factor availability. Cytokine 2010, 51, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-S.; Kim, H.-J.; Li, Q.-L.; Chi, X.-Z.; Ueta, C.; Komori, T.; Wozney, J.M.; Kim, E.-G.; Choi, J.-Y.; Ryoo, H.-M.; et al. Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 2000, 20, 8783–8792. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.H.; Javed, A.; Kim, H.J.; Shin, H.I.; Gutierrez, S.; Choi, J.Y.; Rosen, V.; Stein, J.L.; Van Wijnen, A.J.; Stein, G.S.; et al. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem. 1999, 73, 114–125. [Google Scholar] [CrossRef]
- Lee, M.-H.; Kim, Y.-J.; Kim, H.-J.; Park, H.-D.; Kang, A.-R.; Kyung, H.-M.; Sung, J.-H.; Wozney, J.M.; Ryoo, H.-M. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-β1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 2003, 278, 34387–34394. [Google Scholar] [CrossRef] [Green Version]
- Luo, T.; Matsuo-Takasaki, M.; Lim, J.; Sargent, T.D. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int. J. Dev. Biol. 2004, 45, 681–684. [Google Scholar]
- Miyama, K.; Yamada, G.; Yamamoto, T.S.; Takagi, C.; Miyado, K.; Sakai, M.; Ueno, N.; Shibuya, H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev. Biol. 1999, 208, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Noël, D.; Gazit, D.; Bouquet, C.; Apparailly, F.; Bony, C.; Plence, P.; Millet, V.; Turgeman, G.; Perricaudet, M.; Sany, J.; et al. Short-term BMP-2 expression is sufficient for in vivo osteochondral differentiation of mesenchymal stem cells. Stem Cells 2004, 22, 74–85. [Google Scholar] [CrossRef]
- Gu, K.; Zhang, L.; Jin, T.; Rutherford, R.B. Identification of potential modifiers of Runx2/Cbfa1 activity in C2C12 cells in response to bone morphogenetic protein-7. Cells Tissues Organs 2004, 176, 28–40. [Google Scholar] [CrossRef]
- Shen, B.; Wei, A.; Whittaker, S.; Williams, L.A.; Tao, H.; Ma, D.D.; Diwan, A.D. The role of BMP-7 in chondrogenic and osteogenic differentiation of human bone marrow multipotent mesenchymal stromal cells in vitro. J. Cell. Biochem. 2010, 109, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, T.E.; Fosang, A.J. Proteoglycans: Many forms and many functions. FASEB J. 1992, 6, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Murdoch, A.D. Proteoglycans of the extracellular environment: Clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996, 10, 598–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wight, T.N.; Toole, B.P.; Hascall, V.C. Hyaluronan and the aggregating proteoglycans. In The Extracellular Matrix: An Overview; Mecham, R.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 147–195. [Google Scholar]
- Cortes, M.; Baria, A.T.; Schwartz, N.B. Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development 2009, 136, 1697–1706. [Google Scholar] [CrossRef] [Green Version]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Hardingham, T.E.; MuIR, H. Binding of oligosaccharides of hyaluronic acid to proteoglycans. Biochem. J. 1973, 135, 905–908. [Google Scholar] [CrossRef] [Green Version]
- Kimata, K.; Oike, Y.; Tani, K.; Shinomura, T.; Yamagata, M.; Uritani, M.; Suzuki, S. A large chondroitin sulfate proteoglycan (PG-M) synthesized before chondrogenesis in the limb bud of chick embryo. J. Biol. Chem. 1986, 261, 13517–13525. [Google Scholar] [CrossRef]
- Schwartz, N.; Hennig, A.; Krueger, R., Jr.; Krzystolik, M.; Li, H.; Mangoura, D. Developmental expression of S103L cross-reacting proteoglycans in embryonic chick. Prog. Clin. Biol. Res. 1993, 383, 505–514. [Google Scholar]
- Kiani, C.; Liwen, C.; Wu, Y.J.; Albert, J.Y.; Burton, B.Y. Structure and function of aggrecan. Cell Res. 2002, 12, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Domowicz, M.S.; Cortes, M.; Henry, J.G.; Schwartz, N.B. Aggrecan modulation of growth plate morphogenesis. Dev. Biol. 2009, 329, 242–257. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Li, S.; Shi, Z.; Yue, Y.; Sun, J.; Chen, J.; Fu, Q.; Hughes, C.E.; Caterson, B. Articular cartilage metabolism in patients with Kashin–Beck disease: An endemic osteoarthropathy in China. Osteoarthr. Cartil. 2008, 16, 680–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Lei, R.; Tiainen, M.; Wu, S.; Zhang, Q.; Pei, F.; Guo, X. Disordered glycometabolism involved in pathogenesis of Kashin–Beck disease, an endemic osteoarthritis in China. Exp. Cell Res. 2014, 326, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Gleghorn, L.; Ramesar, R.; Beighton, P.; Wallis, G. A mutation in the variable repeat region of the aggrecan gene (AGC1) causes a form of spondyloepiphyseal dysplasia associated with severe, premature osteoarthritis. Am. J. Hum. Genet. 2005, 77, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Tompson, S.W.; Merriman, B.; Funari, V.A.; Fresquet, M.; Lachman, R.S.; Rimoin, D.L.; Nelson, S.F.; Briggs, M.D.; Cohn, D.H.; Krakow, D. A recessive skeletal dysplasia, SEMD aggrecan type, results from a missense mutation affecting the C-type lectin domain of aggrecan. Am. J. Hum. Genet. 2009, 84, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krueger, R.C.; Kurima, K.; Schwartz, N.B. Completion of the mouse aggrecan gene structure and identification of the defect in the cmd-Bc mouse as a near complete deletion of the murine aggrecan gene. Mamm. Genome 1999, 10, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Kimata, K.; Line, S.; Strong, D.; Gao, L.-Y.; Kozak, C.A.; Yamada, Y. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat. Genet. 1994, 7, 154–157. [Google Scholar] [CrossRef]
- Watanabe, H.; Nakata, K.; Kimata, K.; Nakanishi, I.; Yamada, Y. Dwarfism and age-associated spinal degeneration of heterozygote cmd mice defective in aggrecan. Proc. Natl. Acad. Sci. USA 1997, 94, 6943–6947. [Google Scholar] [CrossRef] [Green Version]
- Yoo, T.; Cho, H.; Yamada, Y. Hearing impairment in mice with the cmd/cmd (cartilage matrix deficiency) mutant gene. Ann. N. Y. Acad. Sci. 1991, 630, 265–267. [Google Scholar] [CrossRef]
- Primorac, D.; Stover, M.; Clark, S.; Rowe, D. Molecular basis of nanomelia, a heritable chondrodystrophy of chicken. Matrix Biol. 1994, 14, 297–305. [Google Scholar] [CrossRef]
- Stirpe, N.S.; Argraves, W.S.; Goetinck, P.F. Chondrocytes from the cartilage proteoglycan-deficient mutant, nanomelia, synthesize greatly reduced levels of the proteoglycan core protein transcript. Dev. Biol. 1987, 124, 77–81. [Google Scholar] [CrossRef]
- Vertel, B.M.; Grier, B.L.; Li, H.; Schwartz, N.B. The chondrodystrophy, nanomelia: Biosynthesis and processing of the defective aggrecan precursor. Biochem. J. 1994, 301, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vertel, B.M.; Walters, L.M.; Grier, B.; Maine, N.; Goetinck, P.F. Nanomelic chondrocytes synthesize, but fail to translocate, a truncated aggrecan precursor. J. Cell Sci. 1993, 104, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Benito-Arenas, R.; Doncel-Pérez, E.; Fernández-Gutiérrez, M.; Garrido, L.; García-Junceda, E.; Revuelta, J.; Bastida, A.; Fernández-Mayoralas, A. A holistic approach to unravelling chondroitin sulfation: Correlations between surface charge, structure and binding to growth factors. Carbohydr. Polym. 2018, 202, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, K.; Wakao, M.; Suda, Y. Syntheses of chondroitin sulfate tetrasaccharide structures containing 4, 6-disulfate patterns and analysis of their interaction with glycosaminoglycan-binding protein. Bioorganic Med. Chem. Lett. 2015, 25, 1552–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadanaka, S.; Ishida, M.; Ikegami, M.; Kitagawa, H. Chondroitin 4-O-sulfotransferase-1 modulates Wnt-3a signaling through control of E disaccharide expression of chondroitin sulfate. J. Biol. Chem. 2008, 283, 27333–27343. [Google Scholar] [CrossRef] [Green Version]
- Klüppel, M.; Wight, T.N.; Chan, C.; Hinek, A.; Wrana, J.L. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development 2005, 132, 3989–4003. [Google Scholar] [CrossRef] [Green Version]
- Fenwick, S.; Gregg, P.; Kumar, S.; Smith, J.; Rooney, P. Intrinsic control of vascularization in developing cartilage rudiments. Int. J. Exp. Pathol. 1997, 78, 187–196. [Google Scholar] [CrossRef]
- Carlevaro, M.F.; Cermelli, S.; Cancedda, R.; Cancedda, F.D. Vascular endothelial growth factor (VEGF) in cartilage neovascularization and chondrocyte differentiation: Auto-paracrine role during endochondral bone formation. J. Cell Sci. 2000, 113, 59–69. [Google Scholar] [CrossRef]
- Grodzinsky, A.J. Electromechanical and physicochemical properties of connective tissue. Crit. Rev. Biomed. Eng. 1983, 9, 133–199. [Google Scholar]
- Mongiat, M.; Taylor, K.; Otto, J.; Aho, S.; Uitto, J.; Whitelock, J.M.; Iozzo, R.V. The protein core of the proteoglycan perlecan binds specifically to fibroblast growth factor-7. J. Biol. Chem. 2000, 275, 7095–7100. [Google Scholar] [CrossRef] [Green Version]
- Muddasani, P.; Norman, J.C.; Ellman, M.; Van Wijnen, A.J.; Im, H.-J. Basic fibroblast growth factor activates the MAPK and NFκB pathways that converge on Elk-1 to control production of matrix metalloproteinase-13 by human adult articular chondrocytes. J. Biol. Chem. 2007, 282, 31409–31421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nummenmaa, E.; Hämäläinen, M.; Moilanen, T.; Vuolteenaho, K.; Moilanen, E. Effects of FGF-2 and FGF receptor antagonists on MMP enzymes, aggrecan, and type II collagen in primary human OA chondrocytes. Scand. J. Rheumatol. 2015, 44, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.; McLean, C.; Full, L.; Peston, D.; Saklatvala, J. FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthr. Cartil. 2007, 15, 752–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansouri, R.; Jouan, Y.; Hay, E.; Blin-Wakkach, C.; Frain, M.; Ostertag, A.; Le Henaff, C.; Marty, C.; Geoffroy, V.; Marie, P.J.; et al. Correction: Osteoblastic heparan sulfate glycosaminoglycans control bone remodeling by regulating Wnt signaling and the crosstalk between bone surface and marrow cells. Cell Death Dis. 2018, 9, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koike, T.; Izumikawa, T.; Tamura, J.-I.; Kitagawa, H. Chondroitin sulfate-E fine-tunes osteoblast differentiation via ERK1/2, Smad3 and Smad1/5/8 signaling by binding to N-cadherin and cadherin-11. Biochem. Biophys. Res. Commun. 2012, 420, 523–529. [Google Scholar] [CrossRef]
- Choi, Y.; Chung, H.; Jung, H.; Couchman, J.R.; Oh, E.-S. Syndecans as cell surface receptors: Unique structure equates with functional diversity. Matrix Biol. 2011, 30, 93–99. [Google Scholar] [CrossRef]
- Xian, X.; Gopal, S.; Couchman, J.R. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 2010, 339, 31–46. [Google Scholar] [CrossRef]
- David, G.; Bai, X.M.; Van der Schueren, B.; Marynen, P.; Cassiman, J.-J.; Van den Berghe, H. Spatial and temporal changes in the expression of fibroglycan (syndecan-2) during mouse embryonic development. Development 1993, 119, 841–854. [Google Scholar] [CrossRef]
- Gutierrez, J.; Osses, N.; Brandan, E. Changes in secreted and cell associated proteoglycan synthesis during conversion of myoblasts to osteoblasts in response to bone morphogenetic protein-2: Role of decorin in cell response to BMP-2. J. Cell. Physiol. 2006, 206, 58–67. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Haupt, L.M.; Ling, L.; Dombrowski, C.; Mun, F.K.; Nathan, S.S.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Cool, S.M. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J. Cell. Biochem. 2009, 107, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Dieudonné, F.X.; Marion, A.; Marie, P.J.; Modrowski, D. Targeted inhibition of T-cell factor activity promotes syndecan-2 expression and sensitization to doxorubicin in osteosarcoma cells and bone tumors in mice. J. Bone Miner. Res. 2012, 27, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Marion, A.; Dieudonné, F.X.; Patiño-Garcia, A.; Lecanda, F.; Marie, P.J.; Modrowski, D. Calpain-6 is an endothelin-1 signaling dependent protective factor in chemoresistant osteosarcoma. Int. J. Cancer 2012, 130, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Orosco, A.; Fromigué, O.; Haÿ, E.; Marie, P.J.; Modrowski, D. Dual involvement of protein kinase C δ in apoptosis induced by syndecan-2 in osteoblasts. J. Cell. Biochem. 2006, 98, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Li, Y.; Seghatoleslami, M.R.; Dealy, C.N.; Kosher, R.A. Heparan sulfate proteoglycans including syndecan-3 modulate BMP activity during limb cartilage differentiation. Matrix Biol. 2006, 25, 27–39. [Google Scholar] [CrossRef]
- Iozzo, R.V. The family of the small leucine-rich proteoglycans: Key regulators of matrix assembly and cellular growth. Crit. Rev. Biochem. Mol. Biol. 1997, 32, 141–174. [Google Scholar] [CrossRef]
- Berendsen, A.D.; Fisher, L.W.; Kilts, T.M.; Owens, R.T.; Robey, P.G.; Gutkind, J.S.; Young, M.F. Modulation of canonical Wnt signaling by the extracellular matrix component biglycan. Proc. Natl. Acad. Sci. USA 2011, 108, 17022–17027. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-D.; Fisher, L.W.; Robey, P.G.; Young, M.F. The small leucine-rich proteoglycan biglycan modulates BMP-4-induced osteoblast differentiation. FASEB J. 2004, 18, 948–958. [Google Scholar] [CrossRef]
- Young, M.F.; Bi, Y.; Ameye, L.; Chen, X.-D. Biglycan knockout mice: New models for musculoskeletal diseases. Glycoconj. J. 2002, 19, 257–262. [Google Scholar] [CrossRef]
- Bi, Y.; Stuelten, C.H.; Kilts, T.; Wadhwa, S.; Iozzo, R.V.; Robey, P.G.; Chen, X.-D.; Young, M.F. Extracellular matrix proteoglycans control the fate of bone marrow stromal cells. J. Biol. Chem. 2005, 280, 30481–30489. [Google Scholar] [CrossRef] [Green Version]
- Eames, B.F.; Singer, A.; Smith, G.A.; Wood, Z.A.; Yan, Y.-L.; He, X.; Polizzi, S.J.; Catchen, J.M.; Rodriguez-Mari, A.; Linbo, T.; et al. UDP xylose synthase 1 is required for morphogenesis and histogenesis of the craniofacial skeleton. Dev. Biol. 2010, 341, 400–415. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.R.; Atwood, T.S.; Eames, B.F.; Eberhart, J.K.; Yan, Y.-L.; Postlethwait, J.H.; Johnson, E.A. RAD marker microarrays enable rapid mapping of zebrafish mutations. Genome Biol. 2007, 8, R105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, N. Regulation of chondroitin sulfate synthesis. Effect of beta-xylosides on synthesis of chondroitin sulfate proteoglycan, chondroitin sulfate chains, and core protein. J. Biol. Chem. 1977, 252, 6316–6321. [Google Scholar] [CrossRef]
- Koike, T.; Izumikawa, T.; Tamura, J.-I.; Kitagawa, H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan–protein linkage region. Biochem. J. 2009, 421, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Tagliabracci, V.S.; Engel, J.L.; Wiley, S.E.; Xiao, J.; Gonzalez, D.J.; Appaiah, H.N.; Koller, A.; Nizet, V.; White, K.E.; Dixon, J.E. Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. USA 2014, 111, 5520–5525. [Google Scholar] [CrossRef] [Green Version]
- Iozzo, R.V. Series Introduction: Heparan sulfate proteoglycans: Intricate molecules with intriguing functions. J. Clin. Investig. 2001, 108, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, R.; Hoffmann, E.; Sebald, W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur. J. Biochem. 1996, 237, 295–302. [Google Scholar] [CrossRef]
- Olivares, G.H.; Carrasco, H.; Aroca, F.; Carvallo, L.; Segovia, F.; Larraín, J. Syndecan-1 regulates BMP signaling and dorso-ventral patterning of the ectoderm during early Xenopus development. Dev. Biol. 2009, 329, 338–349. [Google Scholar] [CrossRef] [Green Version]
- Paine-Saunders, S.; Viviano, B.L.; Zupicich, J.; Skarnes, W.C.; Saunders, S. glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev. Biol. 2000, 225, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Grisaru, S.; Cano-Gauci, D.; Tee, J.; Filmus, J.; Rosenblum, N.D. Glypican-3 modulates BMP-and FGF-mediated effects during renal branching morphogenesis. Dev. Biol. 2001, 231, 31–46. [Google Scholar] [CrossRef] [Green Version]
- Kirkbride, K.C.; Townsend, T.A.; Bruinsma, M.W.; Barnett, J.V.; Blobe, G.C.; Kawai, Y.; Morinaga, H.; Kondo, H.; Miyoshi, N.; Nakamura, Y.; et al. Bone morphogenetic proteins signal through the transforming growth factor-β type III receptor. J. Biol. Chem. 2008, 283, 7628–7637. [Google Scholar] [CrossRef] [Green Version]
- DeCarlo, A.A.; Belousova, M.; Ellis, A.L.; Petersen, D.; Grenett, H.; Hardigan, P.; O’Grady, R.; Lord, M.; Whitelock, J.M. Perlecan domain 1 recombinant proteoglycan augments BMP-2 activity and osteogenesis. BMC Biotechnol. 2012, 12, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, R.; Kirn-Safran, C.; Farach-Carson, M.; Carson, D. Perlecan: An important component of the cartilage pericellular matrix. J. Musculoskelet. Neuronal Interact. 2002, 2, 511. [Google Scholar] [PubMed]
- Mochida, Y.; Parisuthiman, D.; Yamauchi, M. Biglycan is a positive modulator of BMP-2 induced osteoblast differentiation. In Tissue Engineering; Springer: Boston, MA, USA, 2006; pp. 101–113. [Google Scholar]
- Kawashima, K.; Ogawa, H.; Komura, S.; Ishihara, T.; Yamaguchi, Y.; Akiyama, H.; Matsumoto, K. Heparan sulfate deficiency leads to hypertrophic chondrocytes by increasing bone morphogenetic protein signaling. Osteoarthr. Cartil. 2020, 28, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Inubushi, T.; Nozawa, S.; Matsumoto, K.; Irie, F.; Yamaguchi, Y. Aberrant perichondrial BMP signaling mediates multiple osteochondroma genesis in mice. JCI Insight 2017, 2, e90049. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Billings, P.C.; O’Connell, M.P.; Kaplan, F.S.; Shore, E.M.; Glaser, D.L. Heparan sulfate proteoglycans (HSPGs) modulate BMP2 osteogenic bioactivity in C2C12 cells. J. Biol. Chem. 2007, 282, 1080–1086. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, Y.; Matsumoto, K.; Irie, F.; Fukushi, J.-I.; Stallcup, W.B.; Yamaguchi, Y. Conditional ablation of the heparan sulfate-synthesizing enzyme Ext1 leads to dysregulation of bone morphogenic protein signaling and severe skeletal defects. J. Biol. Chem. 2010, 285, 19227–19234. [Google Scholar] [CrossRef] [Green Version]
- Fujise, M.; Takeo, S.; Kamimura, K.; Matsuo, T.; Aigaki, T.; Izumi, S.; Nakato, H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development 2003, 130, 1515–1522. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, T.; Kamimura, K.; Firkus, C.; Takeo, S.; Shimmi, O.; Nakato, H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev. Biol. 2008, 313, 408–419. [Google Scholar] [CrossRef] [Green Version]
- Ohkawara, B.; Iemura, S.-I.; ten Dijke, P.; Ueno, N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr. Biol. 2002, 12, 205–209. [Google Scholar] [CrossRef] [Green Version]
- Irie, A.; Habuchi, H.; Kimata, K.; Sanai, Y. Heparan sulfate is required for bone morphogenetic protein-7 signaling. Biochem. Biophys. Res. Commun. 2003, 308, 858–865. [Google Scholar] [CrossRef]
- Jasuja, R.; Allen, B.L.; Pappano, W.N.; Rapraeger, A.C.; Greenspan, D.S. Cell-surface heparan sulfate proteoglycans potentiate chordin antagonism of bone morphogenetic protein signaling and are necessary for cellular uptake of chordin. J. Biol. Chem. 2004, 279, 51289–51297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paine-Saunders, S.; Viviano, B.L.; Economides, A.N.; Saunders, S. Heparan sulfate proteoglycans retain Noggin at the cell surface: A potential mechanism for shaping bone morphogenetic protein gradients. J. Biol. Chem. 2002, 277, 2089–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pegge, J.; Tatsinkam, A.J.; Rider, C.C.; Bell, E. Heparan sulfate proteoglycans regulate BMP signalling during neural crest induction. Dev. Biol. 2020, 460, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Do, A.-T.; Kusche-Gullberg, M.; Lindahl, U.; Lu, K.; Emerson, C.P. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 2006, 281, 4969–4976. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.D.; Moore, W.M.; Guiral, E.C.; Holme, A.D.; Turnbull, J.E.; Pownall, M.E. Extracellular regulation of developmental cell signaling by XtSulf1. Dev. Biol. 2008, 320, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Lai, J.-P.; Chien, J.; Strome, S.E.; Staub, J.; Montoya, D.P.; Greene, E.L.; Smith, D.I.; Roberts, L.R.; Shridhar, V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene 2004, 23, 1439–1447. [Google Scholar] [CrossRef] [Green Version]
- Meyers, J.R.; Planamento, J.; Ebrom, P.; Krulewitz, N.; Wade, E.; Pownall, M.E. Sulf1 modulates BMP signaling and is required for somite morphogenesis and development of the horizontal myoseptum. Dev. Biol. 2013, 378, 107–121. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Lee, J.Y.; Park, J.H.; Park, J.B.; Suh, J.S.; Choi, Y.S.; Lee, S.J.; Chung, C.-P.; Park, Y.J. The identification of a heparin binding domain peptide from bone morphogenetic protein-4 and its role on osteogenesis. Biomaterials 2010, 31, 7226–7238. [Google Scholar] [CrossRef]
- Billings, P.C.; Yang, E.; Mundy, C.; Pacifici, M. Domains with highest heparan sulfate–binding affinity reside at opposite ends in BMP2/4 versus BMP5/6/7: Implications for function. J. Biol. Chem. 2018, 293, 14371–14383. [Google Scholar] [CrossRef] [Green Version]
- Cappato, S.; Tonachini, L.; Giacopelli, F.; Tirone, M.; Galietta, L.J.; Sormani, M.; Giovenzana, A.; Spinelli, A.E.; Canciani, B.; Brunelli, S. High-throughput screening for modulators of ACVR1 transcription: Discovery of potential therapeutics for fibrodysplasia ossificans progressiva. Dis. Models Mech. 2016, 9, 685–696. [Google Scholar] [CrossRef] [Green Version]
- Jinnin, M.; Ihn, H.; Tamaki, K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-β1-induced extracellular matrix expression. Mol. Pharmacol. 2006, 69, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.; Cappato, S.; Giacopelli, F.; Ravazzolo, R.; Bocciardi, R. The role of the 3′ UTR region in the regulation of the ACVR1/Alk-2 gene expression. PLoS ONE 2012, 7, e50958. [Google Scholar] [CrossRef] [PubMed]
- Olsen, O.E.; Sankar, M.; Elsaadi, S.; Hella, H.; Buene, G.; Darvekar, S.R.; Misund, K.; Katagiri, T.; Knaus, P.; Holien, T. BMPR2 inhibits activin and BMP signaling via wild-type ALK2. J. Cell Sci. 2018, 131, jcs213512. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Cai, J.; de Gorter, D.J.; Sanchez-Duffhues, G.; Kemaladewi, D.U.; Hoogaars, W.M.; Aartsma-Rus, A.; ’t Hoen, P.A.; ten Dijke, P. Antisense-oligonucleotide mediated exon skipping in activin-receptor-like kinase 2: Inhibiting the receptor that is overactive in fibrodysplasia ossificans progressiva. PLoS ONE 2013, 8, e69096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammerschmidt, M.; Serbedzija, G.N.; McMahon, A.P. Genetic analysis of dorsoventral pattern formation in the zebrafish: Requirement of a BMP-like ventralizing activity and its dorsal repressor. Genes Dev. 1996, 10, 2452–2461. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, B.; Seibel, W. Compound C/dorsomorphin: Its use and misuse as an AMPK inhibitor. In AMPK; Humana Press: New York, NY, USA, 2018; pp. 195–202. [Google Scholar]
- Paul, B.Y.; Hong, C.C.; Sachidanandan, C.; Babitt, J.L.; Deng, D.Y.; Hoyng, S.A.; Lin, H.Y.; Bloch, K.D.; Peterson, R.T. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008, 4, 33–41. [Google Scholar]
- Wrighton, K.H.; Lin, X.; Paul, B.Y.; Feng, X.-H. Transforming growth factor β can stimulate Smad1 phosphorylation independently of bone morphogenic protein receptors. J. Biol. Chem. 2009, 284, 9755–9763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, J.; Ho, J.N.; Lewis, J.A.; Karim, K.A.; Daniels, R.N.; Gentry, P.R.; Hopkins, C.R.; Lindsley, C.W.; Hong, C.C. In vivo structure–activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem. Biol. 2010, 5, 245–253. [Google Scholar] [CrossRef] [Green Version]
- Cuny, G.D.; Paul, B.Y.; Laha, J.K.; Xing, X.; Liu, J.-F.; Lai, C.S.; Deng, D.Y.; Sachidanandan, C.; Bloch, K.D.; Peterson, R.T. Structure–activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorganic Med. Chem. Lett. 2008, 18, 4388–4392. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-C.; Cheng, C.-J.; Bilen, M.A.; Lu, J.-F.; Satcher, R.L.; Yu-Lee, L.-Y.; Gallick, G.E.; Maity, S.N.; Lin, S.-H. BMP4 promotes prostate tumor growth in bone through osteogenesis. Cancer Res. 2011, 71, 5194–5203. [Google Scholar] [CrossRef] [Green Version]
- Zilberberg, L.; ten Dijke, P.; Sakai, L.Y.; Rifkin, D.B. A rapid and sensitive bioassay to measure bone morphogenetic protein activity. BMC Cell Biol. 2007, 8, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, B.Y.; Deng, D.Y.; Lai, C.S.; Hong, C.C.; Cuny, G.D.; Bouxsein, M.L.; Hong, D.W.; McManus, P.M.; Katagiri, T.; Sachidanandan, C.; et al. BMP type I receptor inhibition reduces heterotopic ossification. Nat. Med. 2008, 14, 1363–1369. [Google Scholar]
- Kaplan, F.S.; Xu, M.; Seemann, P.; Connor, J.M.; Glaser, D.L.; Carroll, L.; Delai, P.; Fastnacht-Urban, E.; Forman, S.J.; Gillessen-Kaesbach, G.; et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum. Mutat. 2009, 30, 379–390. [Google Scholar] [CrossRef] [Green Version]
- Pacifici, M.; Shore, E.M. Common mutations in ALK2/ACVR1, a multi-faceted receptor, have roles in distinct pediatric musculoskeletal and neural orphan disorders. Cytokine Growth Factor Rev. 2016, 27, 93–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pignolo, R.J.; Shore, E.M.; Kaplan, F.S. Fibrodysplasia ossificans progressiva: Clinical and genetic aspects. Orphanet J. Rare Dis. 2011, 6, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, J.; Traynor, R.; Sapkota, G.P. The specificities of small molecule inhibitors of the TGFß and BMP pathways. Cell. Signal. 2011, 23, 1831–1842. [Google Scholar] [CrossRef] [Green Version]
- Owens, P.; Pickup, M.W.; Novitskiy, S.V.; Giltnane, J.M.; Gorska, A.E.; Hopkins, C.R.; Hong, C.C.; Moses, H.L. Inhibition of BMP signalling suppresses metastasis in mammary cancer. Oncogene 2015, 34, 2437–2449. [Google Scholar] [CrossRef] [Green Version]
- Augeri, D.J.; Langenfeld, E.; Castle, M.; Gilleran, J.A.; Langenfeld, J. Inhibition of BMP and of TGFβ receptors downregulates expression of XIAP and TAK1 leading to lung cancer cell death. Mol. Cancer 2016, 15, 27. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.H.; Augeri, D.J.; NeMoyer, R.; Malhotra, J.; Langenfeld, E.; Chesson, C.B.; Dobias, N.S.; Lee, M.J.; Tarabichi, S.; Jhawar, S.R.; et al. Novel bone morphogenetic protein receptor inhibitor JL5 suppresses tumor cell survival signaling and induces regression of human lung cancer. Oncogene 2018, 37, 3672–3685. [Google Scholar] [CrossRef]
- Boergermann, J.; Kopf, J.; Yu, P.; Knaus, P. Dorsomorphin and LDN-193189 inhibit BMP-mediated Smad, p38 and Akt signalling in C2C12 cells. Int. J. Biochem. Cell Biol. 2010, 42, 1802–1807. [Google Scholar] [CrossRef]
- Langenfeld, E.; Hong, C.C.; Lanke, G.; Langenfeld, J. Bone morphogenetic protein type I receptor antagonists decrease growth and induce cell death of lung cancer cell lines. PLoS ONE 2013, 8, e61256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Wang, X.-L.; Zettervall, S.L.; Cai, Y.; Guzman, R.J. DMH1, a Highly Selective Small Molecule BMP Inhibitor, Suppresses Arterial Medial Calcification. J. Vasc. Surg. 2017, 66, 586. [Google Scholar] [CrossRef] [PubMed]

| Compound | Chemical Name | Structure |
|---|---|---|
| Dorsomorphin | 6-[4-[2-(1-Piperidinyl)ethoxy]phenyl]-3-(4-pyridinyl)-pyrazolo[1,5-a]pyrimidine |  |
| LDN-193189 | 4-[6-[4-(1-Piperazinyl)phenyl]pyrazolo[1,5-a]pyrimidin-3-yl]-quinoline dihydrochloride | 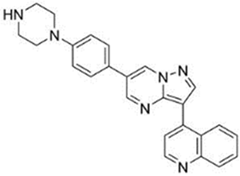 |
| DMH1 | 4-[6-(4-Propan-2-yloxyphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline | 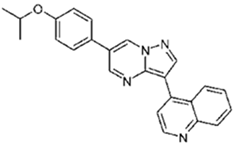 |
| DMH2 | 4-(2-(4-(3-(quinolin-4-yl)pyrazolo[1,5-a]pyrimidin-6-yl)phenoxy)ethyl)morpholine |  |
| JL5 | 4-(3-(4-(3-(quinolin-4-yl)pyrazolo[1,5-a]pyrimidin-6-yl)phenyl)propyl)morpholine |  |
| DMH3 | N,N-dimethyl-3-(4-(3-(quinolin-4-yl)pyrazolo[1,5-a]pyrimidin-6-yl)phenoxy)propan-1-amine |  |
| DMH4 | 4-(2-(4-(3-phenylpyrazolo[1,5-a]pyrimidin-6-yl)phenoxy)ethyl)morpholine | 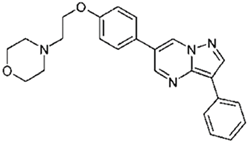 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koosha, E.; Eames, B.F. Two Modulators of Skeletal Development: BMPs and Proteoglycans. J. Dev. Biol. 2022, 10, 15. https://doi.org/10.3390/jdb10020015
Koosha E, Eames BF. Two Modulators of Skeletal Development: BMPs and Proteoglycans. Journal of Developmental Biology. 2022; 10(2):15. https://doi.org/10.3390/jdb10020015
Chicago/Turabian StyleKoosha, Elham, and B. Frank Eames. 2022. "Two Modulators of Skeletal Development: BMPs and Proteoglycans" Journal of Developmental Biology 10, no. 2: 15. https://doi.org/10.3390/jdb10020015





