Functional and Comparative Genomics of Hoxa2 Gene cis-Regulatory Elements: Evidence for Evolutionary Modification of Ancestral Core Element Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tol2 Plasmid Construction
2.2. Medaka Genomic DNA Extraction
2.3. Amplification of Medaka hoxa2a and ψhoxa2b UER(K20-RE5)s
2.4. PCR-Mediated Deletion Mutagenesis of the Medaka hoxa2a and ψhoxa2b UER(K20-RE5)s
2.5. Microinjection of Medaka Embryos
2.6. Generation and Visualization of Transient and Stable-Line Transgenic Medaka Embryos
2.7. Whole-Mount in Situ Hybridization
2.8. Comparative Genomic Sequence Analysis
3. Results
3.1. Validation of the Tol2 Transposon System for Medaka Embryos
3.2. Functional Genomic Analysis of the Medaka Hoxa2a UER(K20-RE5)
3.3. Functional Genomic Analysis of the Medaka ψHoxa2b UER(K20-RE5)
4. Discussion
4.1. The Use of Medaka in Reporter Gene Expression Analyses
4.2. Medaka Hoxa2a-Directed Gene Expression in the Hindbrain
4.3. Hoxa2a-Directed Gene Expression in the Cranial Neural Crest Cells
4.4. Functional Nature of the Medaka ψHoxa2b r3/5 Enhancer Region
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| A-P | anterior-posterior |
| bp | base pair |
| CNCC | cranial neural crest cell |
| CNE | conserved noncoding element |
| CRE | cis-regulatory element |
| eGFP | enhanced green fluorescent protein |
| ERM | embryo rearing medium |
| E. coli | Escherichia coli |
| GFP | green fluorescent protein |
| NC | neural crest |
| PA | pharyngeal arch |
| PG | paralog group |
| r | rhombomere |
| RE | rhombomeric element |
| UER | upstream enhancer region |
| UER(K20-RE5) | upstream enhancer region spanning 5′ from Krox20 binding element to the 3′ RE5 element |
References
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Ferrier, D.E.; Minguillon, C.; Holland, P.W.; Garcia-Fernandez, J. The amphioxus Hox cluster: Deuterostome posterior flexibility and Hox14. Evolut. Dev. 2000, 2, 284–293. [Google Scholar] [CrossRef]
- Holland, P.W.; Garcia-Fernandez, J. Hox genes and chordate evolution. Dev. Biol. 1996, 173, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Powers, T.P.; Amemiya, C.T. Evidence for a Hox14 paralog group in vertebrates. Curr. Biol. 2004, 14, R183–R184. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Force, A.; Yan, Y.L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.L.; et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Suzuki, T.; Yan, Y.L.; Pomeroy, J.; Singer, A.; Amemiya, C.; Postlethwait, J.H. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoegg, S.; Boore, J.L.; Kuehl, J.V.; Meyer, A. Comparative phylogenomic analyses of teleost fish Hox gene clusters: Lessons from the cichlid fish Astatotilapia burtoni. BMC Genom. 2007, 8, 317. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, H.; Ferguson, M.; Danzmann, R. Evolution of Hox Clusters in Salmonidae: A Comparative Analysis Between Atlantic Salmon (Salmo salar) and Rainbow Trout (Oncorhynchus mykiss). J. Mol. Evol. 2005, 61, 636–649. [Google Scholar] [CrossRef] [PubMed]
- Mungpakdee, S.; Seo, H.; Angotzi, A.R.; Dong, X.; Akalin, A.; Chourrout, D. Differential evolution of the 13 Atlantic salmon hox clusters. Mol. Biol. Evol. 2008, 25, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Prince, V. The Hox paradox: More complex(es) than imagined. Dev. Biol. 2002, 249, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Stellwag, E.J. Hox gene duplication in fish. Semin. Cell Dev. Biol. 1999, 10, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.; Scemama, J.L.; Stellwag, E.J. Japanese medaka Hox paralog group 2: Insights into the evolution of Hox PG2 gene composition and expression in the Osteichthyes. J. Exp. Zool. B Mol. Dev. Evol. 2008, 310, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.; Stellwag, E.J. Spatio-temporal patterns of Hox paralog group 3–6 gene expression during Japanese medaka (Oryzias latipes) embryonic development. Gene Expr. Patterns 2010, 10, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Le Pabic, P.; Stellwag, E.J.; Brothers, S.N.; Scemama, J.L. Comparative analysis of Hox paralog group 2 gene expression during Nile tilapia (Oreochromis niloticus) embryonic development. Dev. Genes Evol. 2007, 217, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Lyon, R.S.; Davis, A.; Scemama, J.L. Spatio-temporal expression patterns of anterior Hox genes during Nile tilapia (Oreochromis niloticus) embryonic development. Gene Expr. Patterns 2013, 13, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Hurley, I.A.; Scemama, J.; Prince, V.E. Consequences of Hoxb1 duplication in teleost fish. Evol. Dev. 2007, 9, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Le Pabic, P.; Stellwag, E.J.; Scemama, J. Embryonic development and skeletogenesis of the pharyngeal jaw apparatus in the cichlid Nile tilapia (Oreochromis niloticus). Anat. Rec. 2009, 292, 1780–1800. [Google Scholar] [CrossRef] [PubMed]
- Mungpakdee, S.; Seo, H.; Chourrout, D. Spatio-temporal expression patterns of anterior Hox genes in Atlantic salmon (Salmo salar). Gene Expr. Patterns 2008, 8, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Scemama, J.L.; Vernon, J.L.; Stellwag, E. Differential expression of Hoxa2a and Hoxa2b genes during striped bass embryonic development. Gene Expr. Patterns 2006, 6, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, N.; Dewaele, R.; Janvier, P.; Krumlauf, R.; Duboule, D. Duplications of hox gene clusters and the emergence of vertebrates. Dev. Biol. 2013, 378, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Cambronero, F.; Wiedemann, L.M.; Krumlauf, R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes). Proc. Natl. Acad. Sci. USA 2006, 103, 5419–5424. [Google Scholar] [CrossRef] [PubMed]
- Gendron-Maguire, M.; Mallo, M.; Zhang, M.; Gridley, T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell 1993, 75, 1317–1331. [Google Scholar] [CrossRef]
- Rijli, F.M.; Mark, M.; Lakkaraju, S.; Dierich, A.; Dolle, P.; Chambon, P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 1993, 75, 1333–1349. [Google Scholar] [CrossRef]
- Amin, S.; Donaldson, I.J.; Zannino, D.A.; Hensman, J.; Rattray, M.; Losa, M.; Spitz, F.; Ladam, F.; Sagestrom, C.; Bobola, N. Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 2015, 32, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Frasch, M.; Chen, X.; Lufkin, T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development 1995, 121, 957–974. [Google Scholar] [PubMed]
- Lampe, X.; Samad, O.A.; Guiguen, A.; Matis, C.; Remacle, S.; Picard, J.J.; Rijli, F.M.; Rezsohazy, R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008, 36, 3214–3225. [Google Scholar] [CrossRef] [PubMed]
- Maconochie, M.K.; Krishnamurthy, R.; Nonchev, S.; Meier, P.; Manzanares, M.; Mitchell, P.J.; Krumlauf, R. Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family. Development 1999, 126, 1483–1494. [Google Scholar] [PubMed]
- Maconochie, M.K.; Nonchev, S.; Manzanares, M.; Marshall, H.; Krumlauf, R. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev. Biol. 2001, 233, 468–481. [Google Scholar] [CrossRef] [PubMed]
- McEllin, J.A.; Alexander, T.B.; Tümpel, S.; Wiedemann, L.M.; Krumlauf, R. Analyses of fugu hoxa2 genes provide evidence for subfunctionalization of neural crest cell and rhombomere cis-regulatory modules during vertebrate evolution. Dev. Biol. 2016, 409, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Nonchev, S.; Maconochie, M.; Vesque, C.; Aparicio, S.; Ariza-McNaughton, L.; Manzanares, M.; Maruthainar, K.; Kuroiwa, A.; Brenner, S.; Charnay, P.; et al. The conserved role of Krox-20 in directing Hox gene expression during vertebrate hindbrain segmentation. Proc. Natl. Acad. Sci. USA 1996, 93, 9339–9345. [Google Scholar] [CrossRef] [PubMed]
- Nonchev, S.; Vesque, C.; Maconochie, M.; Seitanidou, T.; Ariza-McNaughton, L.; Frain, M.; Marshall, H.; Sham, M.H.; Krumlauf, R.; Charnay, P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 1996, 122, 543–554. [Google Scholar] [PubMed]
- Tümpel, S.; Maconochie, M.; Wiedemann, L.M.; Krumlauf, R. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev. Biol. 2002, 246, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Cambronero, F.; Ferretti, E.; Blasi, F.; Wiedemann, L.M.; Krumlauf, R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev. Biol. 2007, 302, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Cambronero, F.; Sims, C.; Krumlauf, R.; Wiedemann, L.M. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc. Natl. Acad. Sci. USA 2008, 105, 20077–20082. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.A.; Bronner, M.E.; Krumlauf, R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 2014, 514, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Sham, M.H.; Vesque, C.; Nonchev, S.; Marshall, H.; Frain, M.; Gupta, R.D.; Whiting, J.; Wilkinson, D.; Charnay, P.; Krumlauf, R. The zinc finger gene Krox20 regulates HoxB2 (Hox2.8) during hindbrain segmentation. Cell 1993, 72, 183–196. [Google Scholar] [CrossRef]
- Ferretti, E.; Marshall, H.; Popperl, H.; Maconochie, M.; Krumlauf, R.; Blasi, F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 2000, 127, 155–166. [Google Scholar] [PubMed]
- Vesque, C.; Maconochie, M.; Nonchev, S.; Ariza-McNaughton, L.; Kuroiwa, A.; Charnay, P.; Krumlauf, R. Hoxb-2 transcriptional activation in rhombomeres 3 and 5 requires an evolutionarily conserved cis-acting element in addition to the Krox-20 binding site. EMBO J. 1996, 15, 5383–5396. [Google Scholar] [PubMed]
- Barrow, J.R.; Capecchi, M.R. Targeted disruption of the Hoxb-2 locus in mice interferes with expression of Hoxb-1 and Hoxb-4. Development 1996, 122, 3817–3828. [Google Scholar] [PubMed]
- Barrow, J.R.; Stadler, H.S.; Capecchi, M.R. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development 2000, 127, 933–944. [Google Scholar] [PubMed]
- Davenne, M.; Maconochie, M.K.; Neun, R.; Pattyn, A.; Chambon, P.; Krumlauf, R.; Rijli, F.M. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron 1999, 22, 677–691. [Google Scholar] [CrossRef]
- Gavalas, A.; Davenne, M.; Lumsden, A.; Chambon, P.; Rijli, F.M. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development 1997, 124, 3693–3702. [Google Scholar] [PubMed]
- Gavalas, A.; Ruhrberg, C.; Livet, J.; Henderson, C.E.; Krumlauf, R. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development 2003, 130, 5663–5679. [Google Scholar] [CrossRef] [PubMed]
- Oury, F.; Murakami, Y.; Renaud, J.S.; Pasqualetti, M.; Charnay, P.; Ren, S.Y.; Rijli, F.M. Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science 2006, 313, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Baltzinger, M.; Ori, M.; Pasqualetti, M.; Nardi, I.; Rijli, F.M. Hoxa2 knockdown in Xenopus results in hyoid to mandibular homeosis. Dev. Dyn. 2005, 234, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Grammatopoulos, G.A.; Bell, E.; Toole, L.; Lumsden, A.; Tucker, A.S. Homeotic transformation of branchial arch identity after Hoxa2 overexpression. Development 2000, 127, 2355–5365. [Google Scholar]
- Kitazawa, T.; Fujisawa, K.; Narboux-Nême, N.; Arima, Y.; Kawamura, Y.; Inoue, T.; Wada, Y.; Kohro, T.; Aburatani, H.; Kodama, T.; et al. Distinct effects of Hoxa2 overexpression in cranial neural crest populations reveal that the mammalian hyomandibular-ceratohyal boundary maps within the styloid process. Dev. Biol. 2015, 402, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Minoux, M.; Antonarakis, G.S.; Kmita, M.; Duboule, D.; Rijli, F.M. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development 2009, 136, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Pasqualetti, M.; Ori, M.; Nardi, I.; Rijli, F.M. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development 2000, 127, 5367–5378. [Google Scholar] [PubMed]
- Hurley, I.A.; Mueller, R.L.; Dunn, K.A.; Schmidt, K.J.; Friedman, M.; Ho, R.K.; Prince, V.E.; Yang, Z.; Thomas, M.G.; Coates, M.I. A new time-scale for ray-finned fish evolution. Proc. R. Soc. B 2007, 274, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.; Amores, A.; Cresko, W.; Singer, A.; Yan, Y.L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004, 20, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Santini, F.; Harmon, L.J.; Carnevale, G.; Alfaro, M.E. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol. Biol. 2009, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; van de Peer, Y.; Meyer, A. Revisiting recent challenges to the ancient fish-specific genome duplication hypothesis. Curr. Biol. 2001, 11, R1005–R1008. [Google Scholar] [CrossRef]
- Moens, C.B.; Selleri, L. Hox cofactors in vertebrate development. Dev. Biol. 2006, 291, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.; Prince, V.E. Zebrafish Hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev. Biol. 2002, 247, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Prince, V.; Lumsden, A. Hoxa-2 expression in normal and transposed rhombomeres: Independent regulation in the neural tube and neural crest. Development 1994, 120, 911–923. [Google Scholar] [PubMed]
- Steinke, D.; Salzburger, W.; Meyer, A. Novel relationships among ten fish model species based on a phylogenomic analysis using ESTs. J. Mol. Evol. 2006, 62, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Wiedemann, L.M.; Krumlauf, R. Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 2009, 88, 103–137. [Google Scholar] [PubMed]
- Davidson, E.H.; Erwin, D.H. Gene regulatory networks and the evolution of animal body plans. Science 2006, 311, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Barrallo-Gimeno, A.; Holzschuh, J.; Driever, W.; Knapik, E.W. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development 2004, 131, 1463–1477. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.G.; Swartz, M.E.; Eberhart, J.K.; Kimmel, C.B. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development 2006, 133, 2661–2669. [Google Scholar] [CrossRef] [PubMed]
- Deflorian, G.; Tiso, N.; Ferretti, E.; Meyer, D.; Blasi, F.; Bortolussi, M.; Argenton, F. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development 2004, 131, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Le Pabic, P.; Scemama, J.; Stellwag, E.J. Role of Hox PG2 genes in Nile tilapia pharyngeal arch specification: Implications for gnathostome pharyngeal arch evolution. Evol. Dev. 2010, 12, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Melvin, V.S.; Feng, W.; Hernandez-Lagunas, L.; Artinger, K.B.; Williams, T. A morpholino-based screen to identify novel genes involved in craniofacial morphogenesis. Dev. Dyn. 2013, 242, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Selleri, L.; Depew, M.J.; Jacobs, Y.; Chanda, S.K.; Tsang, K.Y.; Cheah, K.S.; Rubenstein, J.L.; O’Gorman, S.; Cleary, M.L. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 2001, 128, 3543–3557. [Google Scholar] [PubMed]
- Ferretti, E.; Cambronero, F.; Tümpel, S.; Longobardi, E.; Wiedemann, L.M.; Blasi, F.; Krumlauf, R. The Hoxb1 enhancer and control of rhombomere 4 expression: Complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 2005, 25, 8541–8552. [Google Scholar] [CrossRef] [PubMed]
- Pöpperl, H.; Bienz, M.; Studer, M.; Chan, S.; Aparicio, S.; Brenner, S.; Mann, R.; Krumlauf, R. Segmental expression of Hoxb1 is controlled by a highly conserved autoregulatory loop dependent on exd/Pbx. Cell 1995, 81, 1031–1042. [Google Scholar] [CrossRef]
- Urasaki, A.; Morvan, G.; Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequences and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 2006, 174, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K. Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. 2007, 8, S7. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, G.; Takamatsu, N.; Takahashi, M.; Sumitomo, M.; Sanaka, E.; Yamada, K.; Nishii, K.; Matsuda, M.; Asakawa, S.; Ishiguro, H.; et al. Organization and structure of hox gene loci in medaka genome and comparison with those of pufferfish and zebrafish genomes. Gene 2006, 370, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004, 12, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Oxendine, S.L.; Cowden, J.; Hinton, D.E.; Padilla, S. Adapting the medaka embryo assay to a high-throughput approach for developmental toxicity testing. Neuro Toxicol. 2006, 27, 840–845. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Hofsten, J.V.; Olsson, P.E. Generating transparent zebrafish: A refined method to improve detection of gene expression during embryonic development. Mar. Biotechnol. 2001, 3, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Sandelin, A.; Alkema, W.; Engstrom, P.; Wasserman, W.W.; Lenhard, B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004, 32, D91–D94. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Kaufmann, M.; Morgenstern, B. DIALIGN-TX: Greedy and Progressive Approaches for Segment-Based Multiple Sequence Alignment. Algorithms Mol Biol. 2008, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, C.T.; Powers, T.P.; Prohaska, S.J.; Grimwood, J.; Schmutz, J.; Dickson, M.; Miyake, T.; Schoenborn, M.A.; Myers, R.M.; Ruddle, F.H.; et al. Complete Hox cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc. Natl. Acad. Sci. USA 2010, 107, 3622–3627. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Dewar, K.; Wagner, G.P.; Takahashi, K.; Ruddle, F.; Ledje, C.; Bartsch, P.; Scemama, J.; Stellwag, E.J.; Fried, C.; et al. Bichir HoxA cluster sequence reveals surpising trends in ray-finned fish genomic evolution. Genome Res. 2004, 14, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Church, D.M.; Goodstadt, L.; Hillier, L.W.; Zody, M.C.; Goldstein, S.; She, X.; Bult, C.J.; Agarwala, R.; Cherry, J.L.; DiCuccio, M.; et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009, 7, e1000112. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.B.; Amemiya, C.; Bailey, W.; Kawasaki, K.; Mezey, J.; Miller, W.; Minoshima, S.; Shimizu, N.; Wagner, G.; Ruddle, F. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl. Acad. Sci. USA 2000, 97, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.P.; Koh, E.G.L.; Tay, A.; Brenner, S.; Venkatesh, B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc. Natl. Acad. Sci. USA 2006, 103, 6994–6999. [Google Scholar] [CrossRef] [PubMed]
- Malaga-Trillo, E.; Meyer, A. Genome duplications and accelerated evolution of Hox genes and cluster architecture in teleost fishes. Am. Zool. 2001, 41, 676–686. [Google Scholar]
- Oulion, S.; Debiais-Thibaud, M.; D’Aubenton-Carafa, Y.; Thermes, C.; da Silva, C.; Bernard-Samain, S.; Gavory, F.; Wincker, P.; Mazan, S.; Casane, D. Evolution of Hox gene clusters in gnathostomes: Insights from a survey of a shark (Scyliorhinus canicula) transcriptome. Mol. Biol. Evol. 2010, 27, 2829–2838. [Google Scholar] [CrossRef] [PubMed]
- Raincrow, J.D. Hox Cluster Intergenic Sequence Evolution. Ph.D. Thesis, Rutgers University, NJ, USA, 2010. [Google Scholar]
- Goode, D.K.; Callaway, H.A.; Cerda, G.A.; Lewis, K.E.; Elgar, G. Minor change, major divergence: Divergent functions of highly conserved cis-regulatory elements subsequent to whole genome duplication events. Development 2011, 138, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Santagati, F.; Minoux, M.; Ren, S.Y.; Rijli, F.M. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development 2005, 132, 4927–4936. [Google Scholar] [CrossRef] [PubMed]
- Scemama, J.L.; Hunter, M.; McCallum, J.; Prince, V.; Stellwag, E. Evolutionary divergence of vertebrate Hoxb2 expression patterns and transcriptional regulatory loci. J. Exp. Zool. B Mol. Evol. Dev. 2002, 294, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Caroll, S.B. Evo-Devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Capellini, T.D.; di Giacomo, G.; Salsi, V.; Brendolan, A.; Ferretti, E.; Srivastava, D.; Zappavigna, V.; Selleri, L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 2006, 133, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Kmita, M.; Tarchini, B.; Zakany, J.; Logan, M.; Tabin, C.J.; Duboule, D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 2005, 435, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Mercader, N.; Leonardo, E.; Azpiazu, N.; Serrano, A.; Morata, G.; Martinez, C.; Torres, M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 1999, 402, 425–429. [Google Scholar] [PubMed]
- Tamura, K.; Yokouchi, Y.; Kuroiwa, A.; Ide, H. Retinoic acid changes the proximodistal developmental competence and affinity of distal cells in the developing chick limb bud. Dev. Biol. 1997, 188, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Yonei-Tamura, S.; Yano, T.; Yokoyama, H.; Ide, H. The autopod: Its formation during limb development. Dev. Growth Differ. 2008, 50, S177–S187. [Google Scholar] [CrossRef] [PubMed]
- Zeller, R. The temporal dynamics of vertebrate limb development, teratogenesis and evolution. Curr. Opin. Genet. Dev. 2010, 20, 384–390. [Google Scholar] [CrossRef] [PubMed]








| Primer | Sequence 5′ to 3′ | 5′ Start Site |
|---|---|---|
| Medaka hoxa2a Genomic Primers | ||
| A2a For | TTATTCCCACAACCCTTTCATTTCG | −2691 |
| A2a Rev | CACACTCAGCCACAATCTCTTCTTC | 1846 |
| Medaka ψhoxa2b Genomic Primers | ||
| A2b For | ACACAGCAGGGGTCAACAATAGGTC | −3093 |
| A2b Rev | ATAGGCAGAGCACGAAAACAAAATG | 3193 |
| Medaka hoxa2a UER(K20-RE5) Forward Primers | ||
| AF1 | GATCGATATCGAACAGGCTGAAATCCACTGAATGC | −1778 |
| AF2 | GATCGATATCGCTTCTAATCTGAGAAGCCAGTGTTTC | −1468 |
| AF3 | GATCGATATCATGTGTTGCGAGGGCACCGAGCTGTC | −1392 |
| AF4 | GATCGATATCGAGTAAGATTGATCGCGCACAGGCTTC | −1354 |
| Medaka hoxa2a UER(K20-RE5) Reverse Primers | ||
| AR1 | GATCGAATTCGTTTGCTGTGGAACAGAGGAAAGAAG | −1247 |
| AR2 | GATCGAATTCTTATATACCAAACAAAGAGTCCTGG | −1303 |
| AR3 | GATCGAATTCTTACTCGCCAAAAGGTCTGACAGCTC | −1348 |
| Medaka ψhoxa2b UER(K20-RE5) Forward Primers | ||
| BF1 | GATCGATATCATGTGCCAACACCCACTCACCCCAG | −1068 |
| BF2 | GATCGATATCCTTCGCTCCGCACCGAGGGCATCCTC | −868 |
| BF3 | GATCGATATCATGTTCTCTAAGGGCAAAGAGCTGTC | −803 |
| BF4 | GATCGATATCTGGAAAGATTGATCACACAGAATACC | −765 |
| Medaka ψhoxa2b UER(K20-RE5) Reverse Primers | ||
| BR1 | GATCGAATTCAAAAAGCTGCAGGAAAAGGAGGGGATC | −671 |
| BR2 | GATCGAATTCCCGGGCTCTGAACAAAAGATTCCTG | −715 |
| BR3 | GATCGAATTCTTTCCAGCCAAGAGCTCTGACAGCTC | −759 |
| Construct | Primer Pairs | Amplicon Length | Construct Schematic | Hindbrain Expression (F0) | CNCC Expression (F0)) | F1s |
|---|---|---|---|---|---|---|
| Medaka hoxa2a UER(K20-RE5) construct design and transgenic analysis | ||||||
| 1 | AF1/AR1 | 531 bp |  | 42/48 (87.5%) | 42/48 (87.5%) | 3 |
| 2 | AF2/AR1 | 221 bp |  | 64/84 (76%) | 56/84 (67%) | 3 |
| 3 | AF3/AR1 | 145 bp | 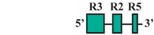 | 39/49 (80%) | 41/49 (84%) | 0 |
| 4 | AF4/AR1 | 107 bp |  | 7/47 (15%) * | 23/49 (49%) * | 0 |
| 5 | AF1/AR2 | 475 bp | 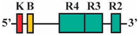 | 42/50 (84%) | 42/50 (84%) | 4 |
| 6 | AF1/AR3 | 430 bp | 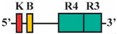 | 0/52 (0%) | 0/52 (0%) | 0 |
| 7 | AF3/AR2 | 89 bp | 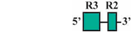 | 52/62 (84%) | 52/62 (84%) | 4 |
| Medaka ψhoxa2b UER(K20-RE5) construct design and transgenic analysis | ||||||
| 8 | BF1/BR1 | 397 bp |  | 46/51 (90%) | 46/51 (90%) | 2 |
| 9 | BF2/BR1 | 197 bp |  | 47/52 (90%) | 47/52 (90%) | 4 |
| 10 | BF3/BR1 | 132 bp | 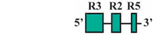 | 33/38 (87%) | 33/38 (87%) | 3 |
| 11 | BF4/BR1 | 94 bp |  | 27/52 (52%) * | 27/52 (52%)* | 0 |
| 12 | BF1/BR2 | 353 bp | 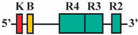 | 33/41 (80%) | 33/41 (80%) | 3 |
| 13 | BF1/BR3 | 309 bp | 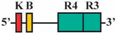 | 9/64 (14%) * | 15/64 (23%) * | 0 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davis, A.; Reubens, M.C.; Stellwag, E.J. Functional and Comparative Genomics of Hoxa2 Gene cis-Regulatory Elements: Evidence for Evolutionary Modification of Ancestral Core Element Activity. J. Dev. Biol. 2016, 4, 15. https://doi.org/10.3390/jdb4020015
Davis A, Reubens MC, Stellwag EJ. Functional and Comparative Genomics of Hoxa2 Gene cis-Regulatory Elements: Evidence for Evolutionary Modification of Ancestral Core Element Activity. Journal of Developmental Biology. 2016; 4(2):15. https://doi.org/10.3390/jdb4020015
Chicago/Turabian StyleDavis, Adam, Michael C. Reubens, and Edmund J. Stellwag. 2016. "Functional and Comparative Genomics of Hoxa2 Gene cis-Regulatory Elements: Evidence for Evolutionary Modification of Ancestral Core Element Activity" Journal of Developmental Biology 4, no. 2: 15. https://doi.org/10.3390/jdb4020015
APA StyleDavis, A., Reubens, M. C., & Stellwag, E. J. (2016). Functional and Comparative Genomics of Hoxa2 Gene cis-Regulatory Elements: Evidence for Evolutionary Modification of Ancestral Core Element Activity. Journal of Developmental Biology, 4(2), 15. https://doi.org/10.3390/jdb4020015





