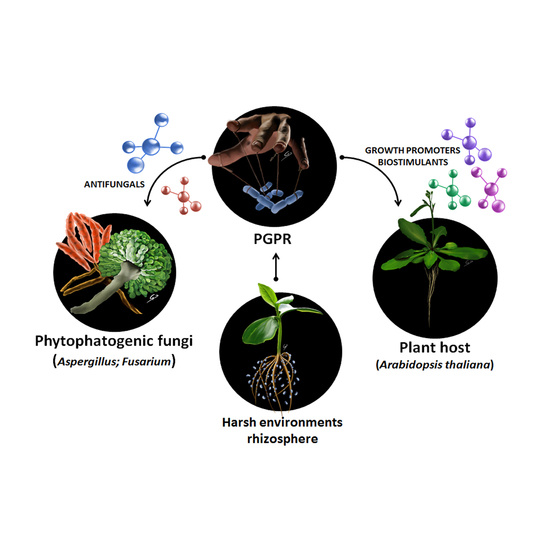
Phyto-Beneficial Traits of Rhizosphere Bacteria: In Vitro Exploration of Plant Growth Promoting and Phytopathogen Biocontrol Ability of Selected Strains Isolated from Harsh Environments
Abstract
:1. Introduction
2. Results
2.1. Evaluation of the Bacterial Strains Properties
2.2. Effect of Bacterial Inoculation on A. thaliana Morphological Features
2.3. Antifungal Activity against Selected Phytopathogenic Fungi
2.4. Identification of Potentially Beneficial Molecules for Plant in Bacterial Broths
3. Discussion
4. Materials and Methods
4.1. Microorganisms Used in This Study and Growth Conditions
4.2. Assessment of PGP Traits of Bacterial Strains
4.3. Seed Bacterial Inoculation and Plants Growth Parameters
4.4. Direct Antifungal Activity Assay
4.5. Identification of Potential Plant Growth Beneficial Molecules by Bacterial Strains
4.5.1. Detection of Functional Groups
4.5.2. UPLC Determination
4.5.3. LC–ESI–MS/MS Determination
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Ann. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, D.; Backer, R.; Subramanian, S.; Smith, D. Phytomicrobiome coordination signals hold potential for climate change-resilient agriculture. Front. Plant Sci. 2020, 11, 634. [Google Scholar] [CrossRef]
- Bertola, M.; Ferrarini, A.; Visioli, G. Improvement of Soil Microbial Biodiversity through Sustainable Agricultural Practices and Its Evaluation by -Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety. Microorganisms 2021, 9, 1400. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Grube, M.; Koberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, B.R.; Gamalero, E. Recent Developments in the Study of Plant Microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef]
- Pirttilä, A.M.; Mohammad Parast Tabas, H.; Baruah, N.; Koskimäki, J.J. Biofertilizers and Biocontrol Agents for Agriculture: How to Identify and Develop New Potent Microbial Strains and Traits. Microorganisms 2021, 9, 817. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Kloepper, J.W. Plant–microbes interactions in enhanced fertilizer-use efficiency. Appl. Microbiol. Biotechnol. 2009, 85, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, X.; Takishita, Y.; Guisheng, Z.; Smith, D.L. Plant Associated Rhizobacteria for Biocontrol and Plant Growth Enhancement. Front. Plant Sci. 2021, 12, 420. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Jošić, D.; Ćirić, A.; Soković, M.; Stanojković-Sebić, A.; Pivić, R.; Lepšanović, Z.; Glamočlija, J. Antifungal activities of indigenous plant growth promoting Pseudomonas spp. from alfalfa and clover rhizosphere. Front. Life Sci. 2015, 8, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Singh, K.P.; Singh, M.; Bhareti, P.; Singh, U.P. Antifungal activity of some strains of plant growth-promoting rhizobacteria. J. Pharmacogn. Phytochem. 2017, 6, 577–582. [Google Scholar]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Hameed, S.; Shahid, M.; Iqbal, M.; Lazarovits, G.; Imran, A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol. Res. 2020, 232, 126389. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, M.; Pagnani, G.; Bernardi, M.; Mattedi, A.; Spera, D.M.; Gallo, M.D. Cell-Free Supernatants of Plant Growth-Promoting Bacteria: A Review of Their Use as Biostimulant and Microbial Biocontrol Agents in Sustainable Agriculture. Sustainability 2020, 12, 9917. [Google Scholar] [CrossRef]
- Manwar, A.V.; Khandelwal, S.R.; Chaudhari, B.L.; Meyer, J.M.; Chincholkar, S.B. Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl. Biochem. Biotechnol. 2004, 118, 243–251. [Google Scholar] [CrossRef]
- Sulochana, M.B.; Jayachandra, S.Y.; Kumar, S.K.; Dayanand, A. Antifungal attributes of siderophore produced by the Pseudomonas aeruginosa JAS-25. J. Basic. Microbiol. 2014, 54, 418–424. [Google Scholar] [CrossRef]
- Maindad, D.V.; Kasture, V.M.; Chaudhari, H.; Dhavale, D.D.; Chopade, B.A.; Sachdev, D.P. Characterization and Fungal Inhibition Activity of Siderophore from Wheat Rhizosphere Associated Acinetobacter calcoaceticus Strain HIRFA32. Indian J. Microbiol. 2014, 54, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J. Bacteriol. 2017, 200, e00345-17. [Google Scholar] [CrossRef] [Green Version]
- Bakker, P.A.H.M.; Ran, L.; Mercado-Blanco, J. Rhizobacterial salicylate production provokes headaches! Plant Soil 2014, 382, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.X.; Kong, X.W.; Li, S.Y.; Chen, X.J.; Chen, X.J. Antibiotics of Pseudomonas protegens FD6 are essential for biocontrol activity. Australas. Plant Pathol. 2020, 49, 307–317. [Google Scholar] [CrossRef]
- Mandal, S.; Mallick, N.; Mitra, A. Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. Biochem. 2009, 47, 642–649. [Google Scholar] [CrossRef]
- Da Rocha Neto, A.C.; Maraschin, M.; Di Piero, R.M. Antifungal activity of salicylic acid against Penicillium expansum and its possible mechanisms of action. Internat. J. Food Microbiol. 2015, 215, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Panahirad, S.; Zaare-Nahandi, F.; Safaralizadeh, R.; Alizadeh-Salteh, S. Postharvest Control of Rhizopus stolonifer. J. Food Saf. 2012, 32, 502–507. [Google Scholar] [CrossRef]
- Antenozio, M.L.; Giannelli, G.; Marabottini, R.; Brunetti, P.; Allevato, E.; Marzi, D.; Capobianco, G.; Bonifazi, G.; Serranti, S.; Visioli, G.; et al. Phytoextraction efficiency of Pteris vittata grown on a naturally As-rich soil and characterization of As-restistant rhizopshere bacteria. Sci. Rep. 2021, 11, 6794. [Google Scholar] [CrossRef]
- Visioli, G.; D’Egidio, S.; Vamerali, T.; Mattarozzi, M.; Sanangelantoni, A.M. Culturable endophytic bacteria enhance Ni translocation in the hyperaccumulator Noccaea caerulescens. Chemosphere 2014, 117, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Bertola, M.; Mattarozzi, M.; Careri, M.; Sanangelantoni, A.M.; Visioli, G. PGPB colonizing three-year biochar amended soil: Towards biochar-mediated biofertilization. J. Soil Sci. Plant Nutr. 2019, 19, 841–850. [Google Scholar] [CrossRef]
- Rizzo, P.; Malerba, M.; Bucci, A.; Sanangelantoni, A.M.; Remelli, S.; Celico, F. Potential Enhancement of the In-Situ Bioremediation of Contaminated Sites through the Isolation and Screening of Bacterial Strains in Natural Hydrocarbon Springs. Water 2020, 12, 2090. [Google Scholar] [CrossRef]
- Bach, H.J.; Munch, J. Identification of bacterial sources of soil peptidases. Biol. Fertil. Soils 2000, 31, 219–224. [Google Scholar] [CrossRef]
- Sessitsch, A.; Pfaffenbichler, N.; Mitter, B. Microbiome Applications from Lab to Field: Facing Complexity. Trends Plant Sci. 2019, 24, 194–198. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Cesa-Luna, C.; Baez, A.; Aguayo-Acosta, A.; Llano-Villarreal, R.C.; Juárez-González, V.R.; Gaytán, P.; del Rocío Bustillos-Cristales, M.; Rivera-Urbalejo, A.; Muñoz-Rojas, J.; Quintero-Hernández, V. Growth inhibition of pathogenic microorganisms by Pseudomonas protegens EMM-1 and partial characterization of inhibitory substances. PLoS ONE 2020, 15, e0240545. [Google Scholar] [CrossRef]
- Mannaa, M.; Oh, J.; Kim, K. Microbe-mediated control of Aspergillus flavus in stored rice grains with a focus on aflatoxin inhibition and biodegradation. Ann. Appl. Biol. 2017, 171, 376–392. [Google Scholar] [CrossRef]
- Andreolli, M.; Zapparoli, G.; Angelini, E.; Lucchetta, G.; Lampis, S.; Vallini, G. Pseudomonas protegens MP12: A plant growth-promoting endophytic bacterium with broad-spectrum antifungal activity against grapevine phytopathogens. Microbiol. Res. 2019, 219, 123–131. [Google Scholar] [CrossRef]
- Chu, T.N.; Bui, L.V.; Hoang, M.T.T. Pseudomonas PS01 Isolated from Maize Rhizosphere Alters Root System Architecture and Promotes Plant Growth. Microorganisms 2020, 8, 471. [Google Scholar] [CrossRef] [Green Version]
- Safdarian, M.; Askari, H.; Shariati, J.V.; Nematzadeh, G. Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 2019, 9, 1792. [Google Scholar] [CrossRef]
- Tchuisseu Tchakounté, G.V.; Berger, B.; Patz, S.; Fankem, H.; Ruppel, S. Community structure and plant growth-promoting potential of cultivable bacteria isolated from Cameroon soil. Microbiol. Res. 2018, 214, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Maurya, B.R.; Raghuwanshi, R. Isolation and characterization of PGPR and their effect on growth, yield and nutrient content in wheat (Triticum aestivum L.). Biocatal. Agric. Biotechnol. 2014, 3, 121–128. [Google Scholar] [CrossRef]
- Ruiz, J.A.; Bernar, E.M.; Jung, K. Production of siderophores increases resistance to fusaric acid in Pseudomonas protegens Pf-5. PLoS ONE 2015, 10, e0117040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C.W.; He, Y.F.; Tang, C.X.; Wu, P.; Zheng, S.J. Mechanisms of microbially enhanced Fe acquisition in red clover (Trifolium pratense L.). Plant Cell Environ. 2006, 29, 888–897. [Google Scholar] [CrossRef] [Green Version]
- Vansuyt, G.; Robin, A.; Briat, J.; Catherine Curie, C.; Lemanceau, P. Iron Acquisition from Fe-Pyoverdine by Arabidopsis thaliana. Mol. Plant Microbe Interact. 2007, 20, 441–447. [Google Scholar] [CrossRef] [Green Version]
- Masalha, J.; Kosegarten, H.; Elmaci, Ö.; Mengel, K. The central role of microbial activity for iron acquisition in maize and sunflower. Biol. Fertil. Soils 2000, 30, 433–439. [Google Scholar] [CrossRef]
- Yu, S.; Teng, C.; Liang, J.; Song, T.; Dong, L.; Bai, X.; Jin, Y.; Qu, J. Characterization of siderophore produced by Pseudomonas syringae BAF.1 and its inhibitory effects on spore germination and mycelium morphology of Fusarium oxysporum. J. Microbiol. 2017, 55, 877–884. [Google Scholar] [CrossRef]
- Codd, R.; Richardson-Sanchez, T.; Telfer, T.J.; Gotsbacher, M.P. Advances in the Chemical Biology of Desferrioxamine B. ACS Chem. Biol. 2018, 13, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, D.; Remelli, M. Deferoxamine B: A Natural, Excellent and Versatile Metal Chelator. Molecules 2021, 26, 3255. [Google Scholar] [CrossRef]
- Nagoba, B.; Vedpathak, D. Medical Applications of Siderophores. Eur. J. Gen. Med. 2011, 8, 229–235. [Google Scholar] [CrossRef]
- Khodr, H.H.; Hider, R.C.; Duhme-Klair, A.K. The iron-binding properties of aminochelin, the mono(catecholamide) siderophore of Azotobacter vinelandii. J. Biol. Inorg. Chem. 2002, 7, 891–896. [Google Scholar] [CrossRef]
- Mossialos, D.; Meyer, J.M.; Budzikiewicz, H.; Wolff, U.; Koedam, N.; Baysse, C.; Anjaiah, V.; Cornelis, P. Quinolobactin, a new siderophore of Pseudomonas fluorescens ATCC 17400, the production of which is repressed by the cognate pyoverdine. Appl. Environ. Microbiol. 2000, 66, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, T.; Nelson, R.; Jesi, S.J. Screening of rhizobacteria for plant growth promoting traits and antifungal activity against charcoal rot pathogen Macrophomina phaseolina. Int. J. Pharm. Bio Sci. 2013, 4, 177–186. [Google Scholar]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Sun Moon, H.; Nam, K. Enhanced uptake and translocation of arsenic in cretan brake fern (Pteris cretica L.) through siderophorearsenic complex formation with an aid of rhizospheric bacterial activity. J. Hazard. Mat. 2014, 280, 536–543. [Google Scholar] [CrossRef]
- Payne, S.M. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994, 235, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Snow, G.A. Mycobactin, a growth factor for Mycobacterium johnei: II. Degradation and identification of fragments. J. Chem. Soc. 1954, 11, 2588–2596. [Google Scholar] [CrossRef]
- Arnow, L.E. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine tyrosine mixtures. J. Biol. Chem. 1937, 118, 531–537. [Google Scholar] [CrossRef]
- Ferreira, C.M.H.; Vilas-Boas, Â.; Sousa, C.A.; Soares, H.M.V.; Soares, E.V. Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express 2019, 9, 78. [Google Scholar] [CrossRef]
- Dimkpa, C.; Svatos, A.; Merten, D.; Büchel, G.; Kothe, E. Hydroxamate siderophores produced by Streptomyces acidiscabies E13 bind nickel and promote growth in cowpea (Vigna unguiculata L.) under nickel stress. Can. J. Microbiol. 2008, 54, 163–172. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 13 May 2011).



| Strains | Homology | Siderophore Production (PSU) (a) | IAA Production (mg L−1) | ACC Deaminase Activity (b) | Phosphate Solubilization Ability (c) | Protease Activity (d) | Biofilm Formation (Abs Units) | References | |
|---|---|---|---|---|---|---|---|---|---|
| SMS | Succinic | ||||||||
| Pvr_5 | Paenarthrobacter ureafaciens (98.16%) | 88.64 ± 0.74 | 91.5 ± 1.05 | 62.48 ± 6.3 | + | - | + | 0.037 ± 0.010 | [25] |
| Prv_9 | Beijerinckia fluminensis (100%) | 91.90 ± 0.11 | 70.7 ± 2.60 | 82.08 ± 1.7 | + | - | - | 1.048 ± 0.141 | [25] |
| Bioch_2 | Arthrobacter defluii (98%) | 91.29 ± 0.56 | 85.91 ± 4.70 | 44.02 ± 2.3 | + | - | + | 0.1 ± 0.007 | [26] |
| Bioch_7 | Arthrobacter nicotinovorans (99%) | 92.33 ± 0.70 | 89.02 ± 1.12 | 58.65 ± 4.2 | + | - | + | 0.216 ± 0.032 | [26] |
| NCr-1 | Arthrobacter sp. (99%) | 93.04 ± 0.08 | 58.78 ± 2.78 | 25.6 ± 1.3 | + | - | + | 0.059 ± 0.003 | [27] |
| PHA_1 | Pseudomonas protegens (98%) | 90.38 ± 0.09 | 76.89 ± 4.94 | n.d. | n.d. | + | - | 0.134 ± 0.007 | [28] |
| Isolates | Functional Group | SMS Medium | Succinic Medium |
|---|---|---|---|
| Pvr_5 | Carboxylate | Salicylic Acid (UPLC–MS) | n.d. |
| Hydroxamate | Desferrioxamine B (UPLC–MS) | n.d. | |
| Pvr_9 | Carboxylate | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) |
| Catecholate | n.d. | Aminochelin (LC–ESI–MS/MS) | |
| PHA_1 | Carboxylate | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) |
| NCr-1 | Carboxylate | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) | Salicylic Acid (UPLC–MS; LC–ESI–MS/MS) |
| Hydroxamate | n.d. | Asperchrome B (UPLC–MS) | |
| Bioch_2 | Carboxylate | Quinolobactin (UPLC–MS) | n.d. |
| Bioch_7 | Carboxylate | Salicylic Acid (UPLC–MS) | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannelli, G.; Bisceglie, F.; Pelosi, G.; Bonati, B.; Cardarelli, M.; Antenozio, M.L.; Degola, F.; Visioli, G. Phyto-Beneficial Traits of Rhizosphere Bacteria: In Vitro Exploration of Plant Growth Promoting and Phytopathogen Biocontrol Ability of Selected Strains Isolated from Harsh Environments. Plants 2022, 11, 230. https://doi.org/10.3390/plants11020230
Giannelli G, Bisceglie F, Pelosi G, Bonati B, Cardarelli M, Antenozio ML, Degola F, Visioli G. Phyto-Beneficial Traits of Rhizosphere Bacteria: In Vitro Exploration of Plant Growth Promoting and Phytopathogen Biocontrol Ability of Selected Strains Isolated from Harsh Environments. Plants. 2022; 11(2):230. https://doi.org/10.3390/plants11020230
Chicago/Turabian StyleGiannelli, Gianluigi, Franco Bisceglie, Giorgio Pelosi, Beatrice Bonati, Maura Cardarelli, Maria Luisa Antenozio, Francesca Degola, and Giovanna Visioli. 2022. "Phyto-Beneficial Traits of Rhizosphere Bacteria: In Vitro Exploration of Plant Growth Promoting and Phytopathogen Biocontrol Ability of Selected Strains Isolated from Harsh Environments" Plants 11, no. 2: 230. https://doi.org/10.3390/plants11020230