Biocontrol Effect of Clonostachys rosea on Fusarium graminearum Infection and Mycotoxin Detoxification in Oat (Avena sativa)
Abstract
:1. Introduction
2. Results
2.1. C. rosea Reduces F. graminearum Biomass and Mycotoxin Content in Mature Oat Kernels
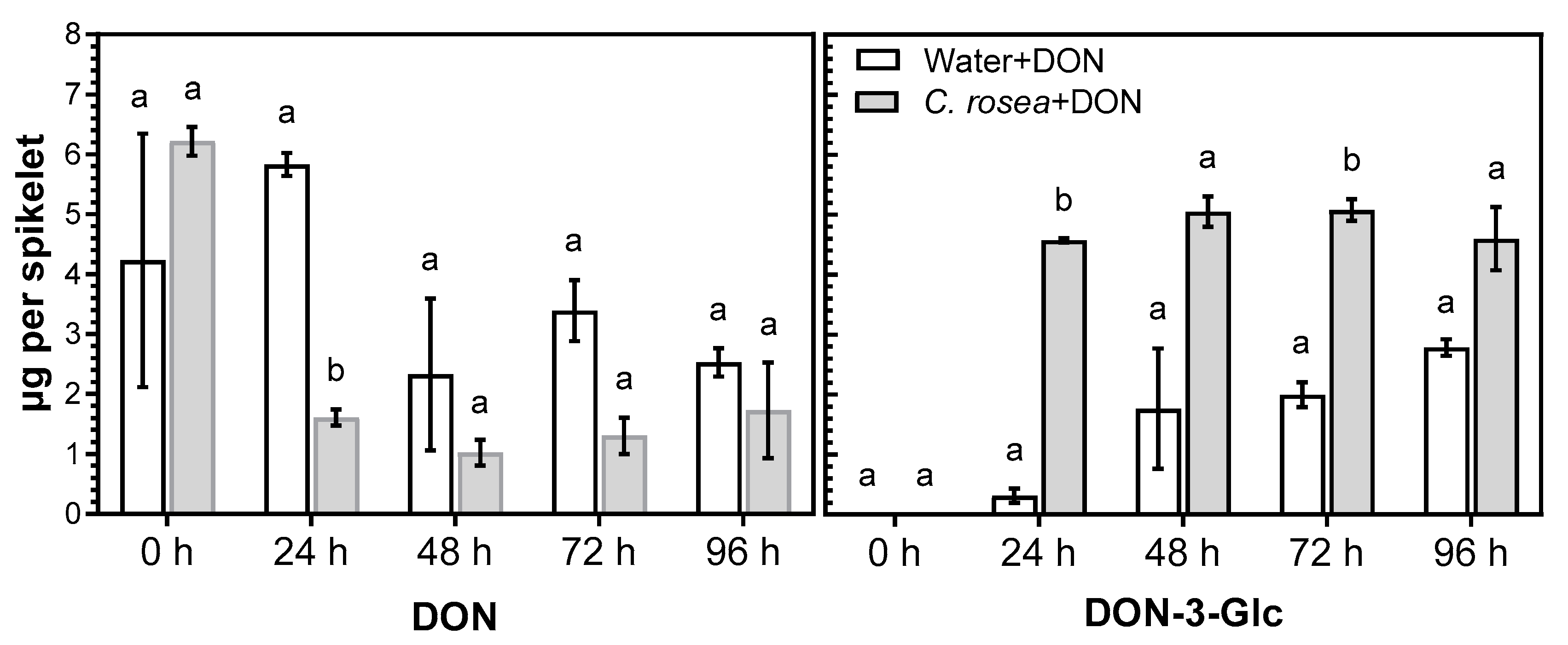
2.2. C. rosea Enhances Conversion of DON into DON-3-Glc in Oat Spikelets
2.3. C. rosea Enhances Expression of Oat UDP-glucosyltransferases in DON-Treated Oat Spikelets
2.4. C. rosea Induces Expression of Genes of Oat PR-Proteins and WRKY23 Transcription Factor
3. Discussion
3.1. C. rosea Reduces F. graminearum Biomass and Mycotoxin Content in Mature Oat Kernels
3.2. Treatment of C. rosea-Inoculated Spikelets with DON Increased Conversion of DON into DON-3-Glc and Enhanced Expression of Two Glucosyltransferase Genes
3.3. C. rosea Inoculation Induces Expression of PR-Genes and a WRKY Transcription Factor
4. Conclusions
5. Materials and Methods
5.1. Plant and Fungal Material
5.2. Application of C. rosea, F. graminearum and DON to Flowering Spikelets
5.3. Quantification of F. graminearum DNA
5.4. Analysis of DON-Induced UGT Expression in Oat
5.5. Analysis of the Expression of Oat PR-Proteins Genes and WRKY-Transcription Factors
5.6. UPLC-MS/MS Analysis of DON, 3-ADON and DON-3-Glc in Mature Oat Kernels
5.7. UHPLC-MS Analysis of DON and its Conjugates in Green Oat Spikelets
5.8. Statistics Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, A.; Cowan, S.; Edwards, S.; Griffiths, I.; Howarth, C.; Langdon, T.; White, E. Crops that feed the world 9. Oats- a cereal crop for human and livestock feed with industrial applications. Food Secur. 2013, 5, 13–33. [Google Scholar] [CrossRef]
- Strychar, R. World Oat Production, Trade, and Usage. In Oats: Chemistry and Technology, 2nd ed.; Webster, F.H., Wood, P.J., Eds.; AACC International: St. Paul, MN, USA, 2016. [Google Scholar] [CrossRef]
- Ames, N.; Rhymer, C.; Storsley, J. Food oat quality throughout the value chain. In Oats Nutrition and Technology; Wiley: New York, NY, USA, 2013; pp. 33–70. [Google Scholar] [CrossRef]
- Gorash, A.; Armonien, R.; Fetch, J.M.; Liatukas, Ž.; Danyt, V.; Gorash, C.A. Aspects in oat breeding: Nutrition quality, nakedness and disease resistance, challenges and perspectives. Ann. Appl. Biol. 2017, 171, 281–302. [Google Scholar] [CrossRef]
- Paudel, D.; Dhungana, B.; Caffe, M.; Krishnan, P. A review of health-beneficial properties of oats. Foods 2021, 10, 2591. [Google Scholar] [CrossRef] [PubMed]
- Hietaniemi, V.; Rämö, S.; Yli-Mattila, T.; Jestoi, M.; Peltonen, S.; Kartio, M.; Sieviläinen, E.; Koivisto, T.; Parikka, P. Updated survey of Fusarium species and toxins in Finnish cereal grains. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk. Assess. 2016, 33, 831–848. [Google Scholar] [CrossRef]
- Islam, M.N.; Tabassum, M.; Banik, M.; Daayf, F.; Dilantha Fernando, W.G.; Harris, L.J.; Sura, S.; Wang, X. Naturally occurring Fusarium species and mycotoxins in oat grains from Manitoba, Canada. Toxins 2021, 13, 670. [Google Scholar] [CrossRef] [PubMed]
- Khodaei, D.; Javanmardi, F.; Khaneghah, A.M. The global overview of the occurrence of mycotoxins in cereals: A three-year survey. Curr. Opin. Food Sci. 2021, 39, 36–42. [Google Scholar] [CrossRef]
- Luo, S.; Du, H.; Kebede, H.; Liu, Y.; Xing, F. Contamination status of major mycotoxins in agricultural product and food stuff in Europe. Food Control 2021, 127, 108120. [Google Scholar] [CrossRef]
- Fredlund, E.; Lindblad, M.; Gidlund, A.; Sulyok, M.; Börjesson, T.; Krska, R.; Olsen, M.; Lindblad, M. Deoxynivalenol and other selected Fusarium toxins in Swedish oats—Occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 2013, 167, 284–291. [Google Scholar] [CrossRef]
- Hofgaard, I.S.; Aamot, H.U.; Torp, T.; Jestoi, M.; Lattanzio, V.M.T.; Klemsdal, S.S.; Waalwijk, C.; van der Lee, T.; Brodal, G. Associations between Fusarium species and mycotoxins in oats and spring wheat from farmers fields in Norway over a six-year period. World Mycotoxin J. 2016, 9, 365–378. [Google Scholar] [CrossRef]
- Pinheiro, M.; Iwase, C.H.T.; Bertozzi, B.G.; Caramês, E.T.S.; Carnielli-Queiroz, L.; Langaro, N.C.; Furlong, E.B.; Correa, B.; Rocha, L.O. Survey of freshly harvested oat grains from southern Brazil reveals high incidence of type b trichothecenes and associated Fusarium species. Toxins 2021, 13, 855. [Google Scholar] [CrossRef]
- Cundliffe, E.; Cannon, M.; Davies, J. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. USA 1974, 71, 30–34. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Zhu, Y.; Abraham, N.; Li, X.Z.; Kimber, M.; Zhou, T. The ribosome-binding mode of trichothecene mycotoxins rationalizes their structure—Activity relationships. Int. J. Mol. Sci. 2021, 22, 1604. [Google Scholar] [CrossRef]
- Zinedine, A.; Soriano, J.M.; Moltó, J.C.; Mañes, J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: An oestrogenic mycotoxin. Food Chem. Toxicol. 2007, 45, 1–18. [Google Scholar] [CrossRef]
- European Commission (EC). Commission Regulation No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union 2006, L364, 5–24. [Google Scholar]
- EFSA CONTAM Panel (EFSA Panel on Contaminants in the Food Chain). Scientific opinion on the appropriateness to set a group health-based guidance value for zearalenone and its modified forms. EFSA J. 2016, 14, e04425. [Google Scholar] [CrossRef]
- Edwards, S.G.; Godley, N.P. Reduction of Fusarium head blight and deoxynivalenol in wheat with early fungicide applications of prothioconazole. Food Addit. Contam. Part A 2010, 27, 629–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freije, A.N.; Wise, K.A. Impact of Fusarium graminearum inoculum availability and fungicide application timing on Fusarium head blight in wheat. Crop Prot. 2015, 77, 139–147. [Google Scholar] [CrossRef]
- Takemoto, J.Y.; Wegulo, S.N.; Yuen, G.Y.; Stevens, J.A.; Jochum, C.C.; Chang, C.W.T.; Kawasaki, Y.; Miller, G.W. Suppression of wheat Fusarium head blight by novel amphiphilic aminoglycoside fungicide K20. Fungal Biol. 2018, 122, 465–470. [Google Scholar] [CrossRef]
- Misonoo, G. Ecological and physiological studies on the blooming of oat flowers. J. Fac. Agric. Hokkaido Imp. Univ. 1936, 37, 211–337. [Google Scholar]
- Collinge, D.B.; Jensen, D.F.; Rabiey, M.; Sarrocco, S.; Shaw, M.W.; Shaw, R.H. Biological control of plant diseases—What has been achieved and what is the direction? Plant Pathol. 2022, 71, 1024–1047. [Google Scholar] [CrossRef]
- Xue, A.G.; Voldeng, H.D.; Savard, M.E.; Fedak, G.; Tian, X.; Hsiang, T. Biological control of Fusarium head blight of wheat with Clonostachys rosea strain ACM941. Can. J. Plant Pathol. 2009, 31, 169–179. [Google Scholar] [CrossRef]
- Xue, A.G.; Chen, Y.; Voldeng, H.D.; Fedak, G.; Savard, M.E.; Längle, T.; Zhang, J.; Harman, G.E. Concentration and cultivar effects on efficacy of CLO-1 biofungicide in controlling Fusarium head blight of wheat. Biol. Control 2014, 73, 2–7. [Google Scholar] [CrossRef]
- Gimeno, A.; Leimgruber, M.; Kägi, A.; Jenny, E.; Vogelgsang, S. UV protection and shelf life of the biological control agent Clonostachys rosea against Fusarium graminearum. Biol. Control 2021, 158, 104600. [Google Scholar] [CrossRef]
- Bai, G.H.; Desjardins, A.E.; Plattner, R.D. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection but does not cause disease spread in wheat spikes. Mycopathologia 2002, 153, 91–98. [Google Scholar] [CrossRef]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection pattern in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Michlmayr, H.; Malachova, A.; Koutnik, A.; Maloku, I.; Berthiller, F.; Adam, G.; Krska, R.; et al. Biotransformation of the mycotoxin deoxynivalenol in Fusarium resistant and susceptible near isogenic wheat lines. PLoS ONE 2015, 10, e0119656. [Google Scholar] [CrossRef]
- Berthiller, F.; Dall’asta, C.; Corradini, R.; Marchelli, R.; Sulyok, M.; Krska, R.; Adam, G.; Schuhmacher, R. Occurrence of deoxynivalenol and its 3-β-D-glucoside in wheat and maize. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk. Assess. 2009, 26, 507–511. [Google Scholar] [CrossRef] [Green Version]
- Mandala, G.; Tundo, S.; Francesconi, S.; Gevi, F.; Zolla, L.; Ceoloni, C.; D’Ovidio, R. Deoxynivalenol detoxification in transgenic wheat confers resistance to Fusarium head blight and crown rot diseases. Mol. Plant-Microbe Interact. 2019, 32, 583–592. [Google Scholar] [CrossRef]
- Li, X.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Shin, S.; Huang, Y.; Dong, Y.; Wiesenberger, G.; McCormick, S.; Lemmens, M.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium head blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, M.; Humpf, H.U.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagl, V.; Schwartz, H.; Krska, R.; Moll, W.D.; Knasmüller, S.; Ritzmann, M.; Adam, G.; Berthiller, F. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in rats. Toxicol. Lett. 2012, 213, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Uhlig, S.; Stanic, A.; Hofgaard, I.S.; Kluger, B.; Schuhmacher, R.; Miles, C.O. Glutathione-conjugates of deoxynivalenol in naturally contaminated grain are primarily linked via the epoxide group. Toxins 2016, 8, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khairullina, A.; Renhuldt, N.T.; Wiesenberger, G.; Bentzer, J.; Collinge, D.B.; Adam, G.; Bülow, L. Identification and functional characterisation of two oat UDP-glucosyltransferases involved in deoxynivalenol detoxification. Toxins 2022, 14, 446. [Google Scholar] [CrossRef]
- Fontana, D.C.; de Paula, S.; Torres, A.G.; de Souza, V.H.M.; Pascholati, S.F.; Schmidt, D.; Neto, D.D. Endophytic fungi: Biological control and induced resistance to phytopathogens and abiotic stresses. Pathogens 2021, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Roberti, R.; Veronesi, A.R.; Cesari, A.; Cascone, A.; di Berardino, I.; Bertini, L.; Caruso, C. Induction of PR proteins and resistance by the biocontrol agent Clonostachys rosea in wheat plants infected with Fusarium culmorum. Plant Sci. 2008, 175, 339–347. [Google Scholar] [CrossRef]
- Kamou, N.N.; Cazorla, F.; Kandylas, G.; Lagopodi, A.L. Induction of defense-related genes in tomato plants after treatments with the biocontrol agents Pseudomonas chlororaphis ToZa7 and Clonostachys rosea IK726. Arch. Microbiol. 2020, 202, 257–267. [Google Scholar] [CrossRef]
- Pritsch, C.; Muehlbauer, G.J.; Bushnell, W.R.; Somers, D.A.; Vance, C.P. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant-Microbe Interact. 2000, 13, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Jensen, J.D.; Svensson, B.; Jørgensen, H.J.L.; Collinge, D.B.; Finnie, C.F. Analysis of early events in the interaction between Fusarium graminearum and the susceptible barley (Hordeum vulgare) cultivar Scarlett. Proteomics 2010, 10, 3748–3755. [Google Scholar] [CrossRef]
- Trümper, C.; Paffenholz, K.; Smit, I.; Kössler, P.; Karlovsky, P.; Braun, H.; Pawelzik, E. Identification of regulated proteins in naked barley grains (Hordeum vulgare nudum) after Fusarium graminearum infection at different grain ripening stages. J Proteomics 2016, 133, 86–92. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boddu, J.; Cho, S.; Kruger, W.M.; Muehlbauer, G.J. Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol. Plant-Microbe Interact. 2006, 19, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Erayman, M.; Turktas, M.; Akdogan, G.; Gurkok, T.; Inal, B.; Ishakoglu, E.; Ilhan, E.; Unver, T. Transcriptome analysis of wheat inoculated with Fusarium graminearum. Front. Plant Sci. 2015, 6, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kage, U.; Yogendra, K.N.; Kushalappa, A.C. TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 2017, 7, 13–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karre, S.; Kumar, A.; Yogendra, K.; Kage, U.; Kushalappa, A.; Charron, J.B. HvWRKY23 regulates flavonoid glycoside and hydroxycinnamic acid amide biosynthetic genes in barley to combat Fusarium head blight. Plant Mol. Biol. 2019, 100, 591–605. [Google Scholar] [CrossRef]
- Mouekouba, L.D.O.; Zhang, L.; Guan, X.; Chen, X.; Chen, H.; Zhang, J.; Zhang, J.; Li, J.; Yang, Y.; Wang, A. Analysis of Clonostachys rosea-induced resistance to tomato gray mold disease in tomato leaves. PLoS ONE 2014, 9, e10269. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Chen, X.; Chai, X.; Xue, D.; Zheng, W.; Shi, Y.; Wang, A. The involvement of jasmonic acid, ethylene, and salicylic acid in the signaling pathway of Clonostachys rosea-induced resistance to gray mold disease in tomato. Phytopathology 2019, 109, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Lv, R.; Cheng, M.; Mo, F.; Zhang, N.; Qi, H.; Liu, J.; Chen, X.; Liu, Y.; Ghanizadeh, H.; et al. Insights into the molecular basis of biocontrol of Botrytis cinerea by Clonostachys rosea in tomato. Sci. Hortic. 2022, 291, 110547. [Google Scholar] [CrossRef]
- Jensen, B.; Knudsen, I.M.B.; Jensen, D.F. Biological seed treatment of cereals with fresh and long-term stored formulations of Clonostachys rosea: Biocontrol efficacy against Fusarium culmorum. Eur. J. Plant Pathol. 2000, 106, 233–242. [Google Scholar] [CrossRef]
- Karlsson, M.; Durling, M.B.; Choi, J.; Kosawang, C.; Lackner, G.; Tzelepis, G.D.; Nygren, K.; Dubey, M.K.; Kamou, N.; Levasseur, A.; et al. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol. Evol. 2015, 7, 465–480. [Google Scholar] [CrossRef]
- Marinelli, E. Evaluation of Potential Biological Control Agents against Fusarium graminearum and Deoxynivalenol Production under In Vitro and Greenhouse Conditions. Master’s Thesis, University of Copenhagen, Copenhagen, Denmark, 2021; p. 94. [Google Scholar]
- Jensen, F.D.; Dubey, M.; Jensen, B.; Karlsson, M. Clonostachys rosea to control plant diseases. In Microbial Bioprotectants for Plant Disease Management, 1st ed.; Köhl, J., Ravensberg, W., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2021; pp. 429–472. [Google Scholar] [CrossRef]
- Abdallah, M.F.; de Boevre, M.; Landschoot, S.; de Saeger, S.; Haesaert, G.; Audenaert, K. Fungal endophytes control Fusarium graminearum and reduce trichothecenes and zearalenone in maize. Toxins 2018, 10, 493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, A.; Kägi, A.; Drakopoulos, D.; Bänziger, I.; Lehmann, E.; Forrer, H.R.; Keller, B.; Vogelgsang, S. From laboratory to the field: Biological control of Fusarium graminearum on infected maize crop residues. J. Appl. Microbiol. 2020, 129, 680–694. [Google Scholar] [CrossRef] [Green Version]
- Góral, T.; Wiśniewska, H.; Ochodzki, P.; Nielsen, L.K.; Walentyn-Góral, D.; Stępień, Ł. Relationship between Fusarium head blight, kernel damage, concentration of Fusarium biomass, and Fusarium toxins in grain of winter wheat inoculated with Fusarium culmorum. Toxins 2019, 11, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakker, M.G.; Brown, D.W.; Kelly, A.C.; Kim, H.S.; Kurtzman, C.P.; Mccormick, S.P.; O’Donnell, K.L.; Proctor, R.H.; Vaughan, M.M.; Ward, T.J. Fusarium mycotoxins: A trans-disciplinary overview. Can. J. Plant Pathol. 2018, 40, 161–171. [Google Scholar] [CrossRef]
- Schmeitzl, C.; Varga, E.; Warth, B.; Kugler, K.G.; Malachová, A.; Michlmayr, H.; Wiesenberger, G.; Mayer, K.F.X.; Mewes, H.W.; Krska, R.; et al. Identification and characterization of carboxylesterases from Brachypodium distachyon deacetylating trichothecene mycotoxins. Toxins 2015, 8, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demissie, Z.A.; Witte, T.; Robinson, K.A.; Sproule, A.; Foote, S.J.; Johnston, A.; Harris, L.J.; Overy, D.P.; Loewen, M.C. Transcriptomic and exometabolomic profiling reveals antagonistic and defensive modes of Clonostachys rosea action against Fusarium graminearum. Mol. Plant-Microbe Interact. 2020, 33, 842–858. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.G. Edwards, S. Zearalenone risk in European wheat. World Mycotoxin J. 2011, 4, 433–438. [Google Scholar] [CrossRef]
- Kharbikar, L.L.; Dickin, E.T.; Edwards, S.G.; Kharbikar, L.L.; Dickin, E.T.; Impact, S.G.E. Food Additives & contaminants: Part A impact of post-anthesis rainfall, fungicide and harvesting time on the concentration of deoxynivalenol and zearalenone in wheat. Food Addit. Contam. Part A 2015, 32, 2075–2085. [Google Scholar] [CrossRef]
- Nygren, K.; Dubey, M.; Zapparata, A.; Iqbal, M.; Tzelepis, G.D.; Brandström, M.; Dan, D.; Jensen, F. The mycoparasitic fungus Clonostachys rosea responds with both common and specific gene expression during interspecific interactions with fungal prey. Evol. Appl. 2018, 11, 931–949. [Google Scholar] [CrossRef] [Green Version]
- Yuan, G.; He, X.; Li, H.; Xiang, K.; Liu, L.; Zou, C.; Lin, H.; Wu, J.; Zhang, Z.; Pan, G. Transcriptomic responses in resistant and susceptible maize infected with Fusarium graminearum. Crop J. 2019, 8, 153–163. [Google Scholar] [CrossRef]
- Shin, S.; Mackintosh, C.A.; Lewis, J.; Heinen, S.J.; Radmer, L. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 2008, 59, 2371–2378. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.; Chilosi, G.; Caporale, C.; Leonardi, L.; Bertini, L.; Magro, P.; Buonocore, V. Induction of pathogenesis-related proteins in germinating wheat seeds infected with Fusarium culmorum. Plant Sci. 1999, 140, 87–97. [Google Scholar] [CrossRef]
- Geddes, J.; Eudes, F.; Laroche, A.; Selinger, L.B. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics 2008, 8, 545–554. [Google Scholar] [CrossRef]
- Latz, M.A.C.; Jensen, B.; Collinge, D.B.; Jørgensen, H.J.L. Endophytic fungi as biocontrol agents: Elucidating mechanisms in disease suppression. Plant Ecol. Divers 2018, 11, 555–567. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N.; et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef] [Green Version]
- Quarta, A.; Mita, G.; Haidukowski, M.; Logrieco, A.; Mulè, G.; Visconti, A. Multiplex PCR assay for the identification of nivalenol, 3- and 15-acetyl-deoxynivalenol chemotypes in Fusarium. FEMS Microbiol. Lett. 2006, 259, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Bai, G.H.; Shaner, G. Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 1996, 8, 975–979. [Google Scholar] [CrossRef]
- Waalwijk, C.; van der Heide, R.; de Vries, I.; van der Lee, T.; Schoen, C.; Costrel-de Corainville, G.; Häuser-Hahn, I.; Kastelein, P.; Köhl, J.; Lonnet, P.; et al. Quantitative detection of Fusarium species in wheat using Taqman. Eur. J. Plant Pathol. 2004, 110, 481–494. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef] [PubMed]
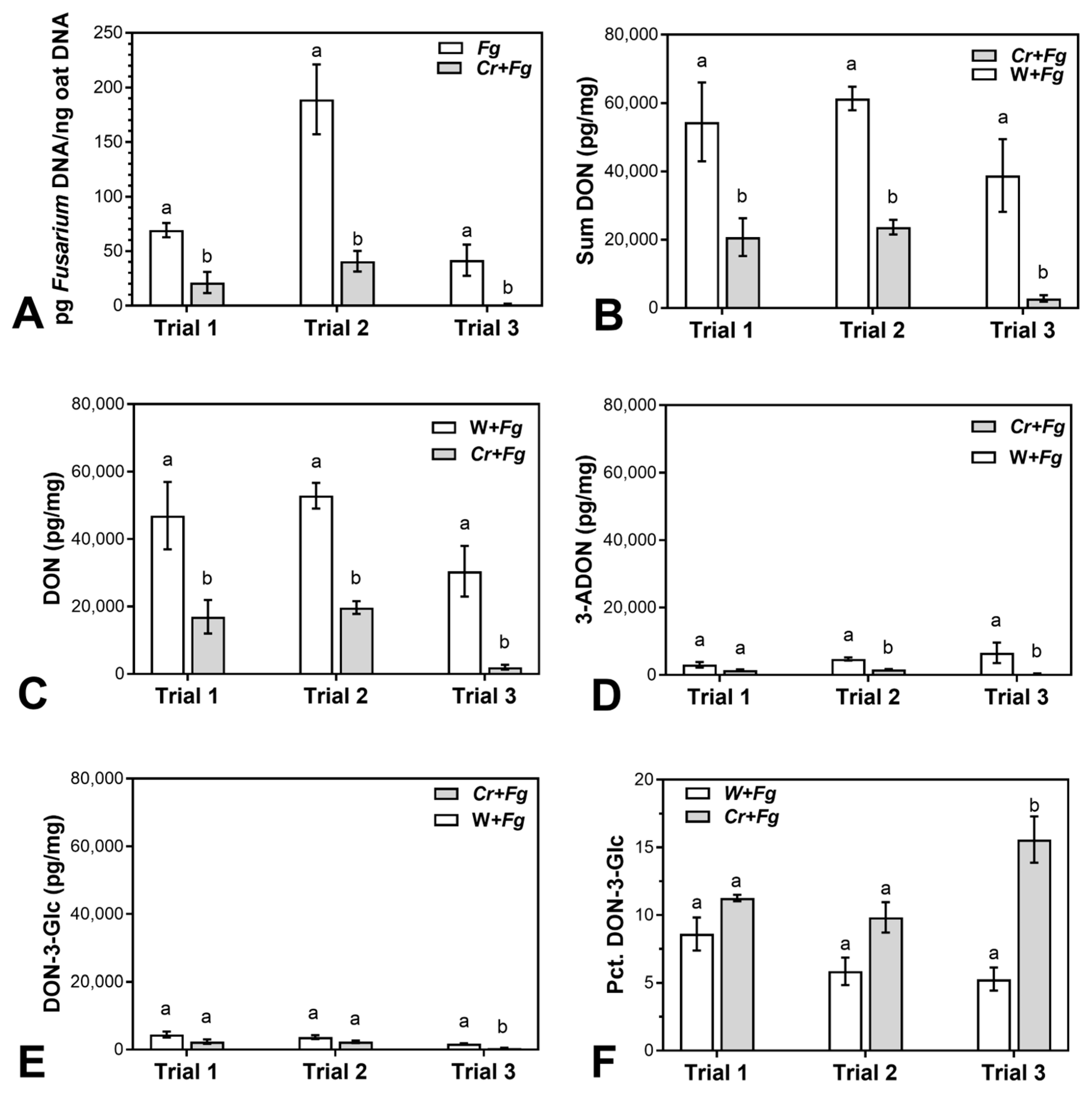


| Trial | F. graminearum DNA | Sum of DONs | DON | 3-ADON | DON-3-Glc | DON-3-Glc/Sum of DONs 1 |
|---|---|---|---|---|---|---|
| 1 | 69.3 | 61.9 | 64.0 | 64.0 | 40.5 | 30.9 |
| 2 | 78.4 | 61.4 | 62.7 | 62.7 | 37.3 | 68.0 |
| 3 | 97.1 | 92.8 | 93.3 | 93.3 | 77.7 | 195.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khairullina, A.; Micic, N.; Jørgensen, H.J.L.; Bjarnholt, N.; Bülow, L.; Collinge, D.B.; Jensen, B. Biocontrol Effect of Clonostachys rosea on Fusarium graminearum Infection and Mycotoxin Detoxification in Oat (Avena sativa). Plants 2023, 12, 500. https://doi.org/10.3390/plants12030500
Khairullina A, Micic N, Jørgensen HJL, Bjarnholt N, Bülow L, Collinge DB, Jensen B. Biocontrol Effect of Clonostachys rosea on Fusarium graminearum Infection and Mycotoxin Detoxification in Oat (Avena sativa). Plants. 2023; 12(3):500. https://doi.org/10.3390/plants12030500
Chicago/Turabian StyleKhairullina, Alfia, Nikola Micic, Hans J. Lyngs Jørgensen, Nanna Bjarnholt, Leif Bülow, David B. Collinge, and Birgit Jensen. 2023. "Biocontrol Effect of Clonostachys rosea on Fusarium graminearum Infection and Mycotoxin Detoxification in Oat (Avena sativa)" Plants 12, no. 3: 500. https://doi.org/10.3390/plants12030500







