NMR Metabolite Profiling for the Characterization of Vessalico Garlic Ecotype and Bioactivity against Xanthomonas campestris pv. campestris
Abstract
:1. Introduction
2. Results
2.1. 1H-NMR Compound Identification
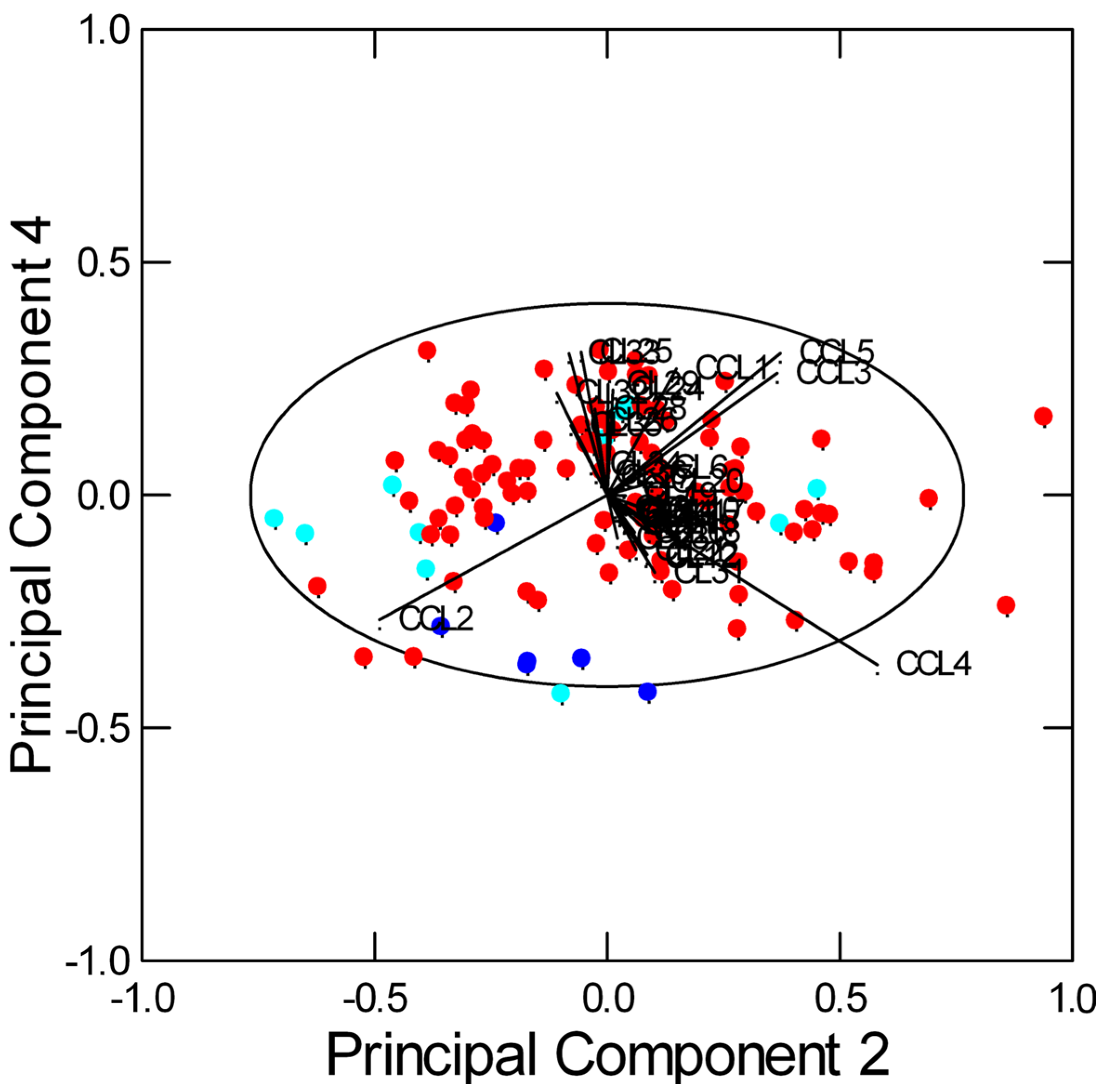
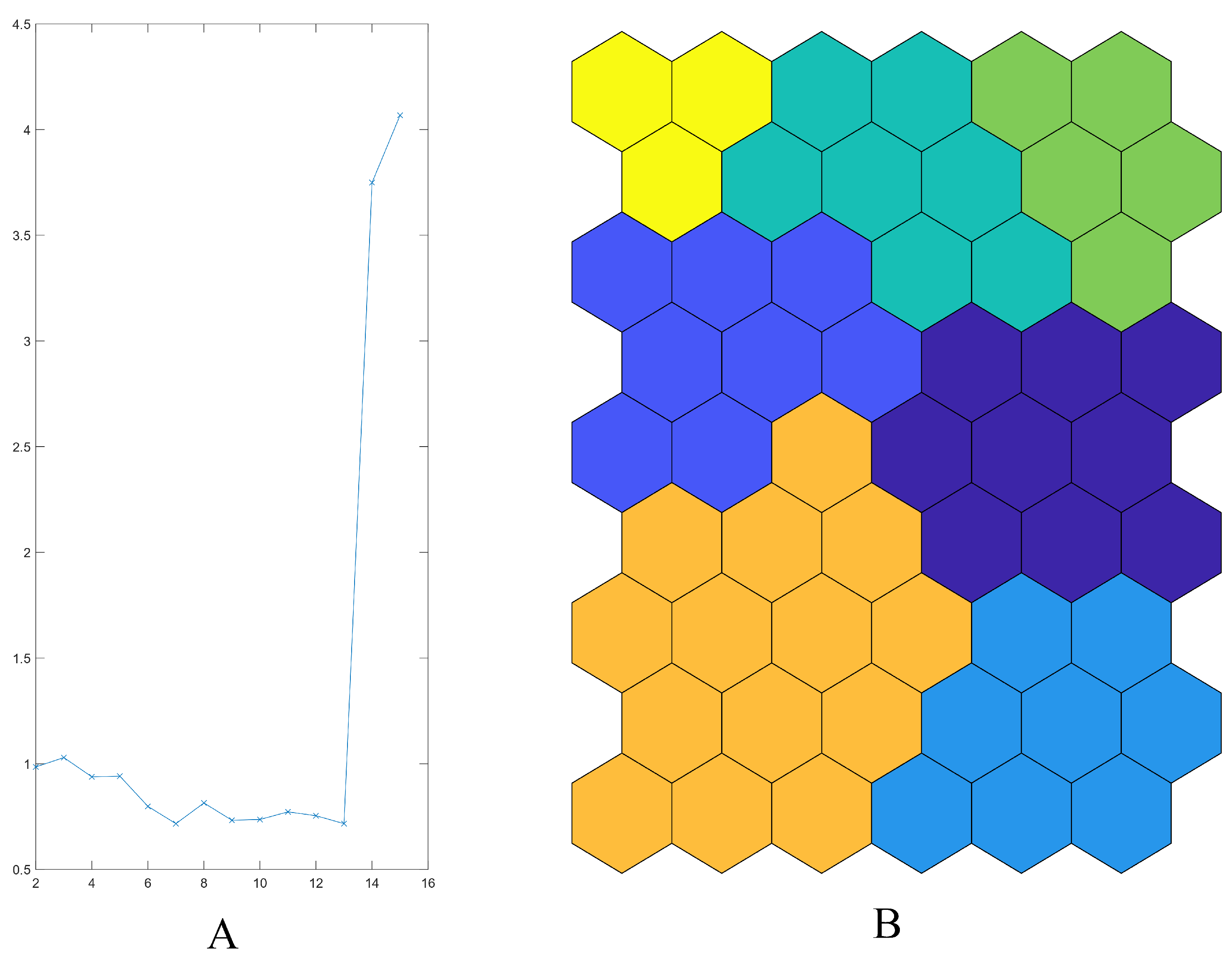
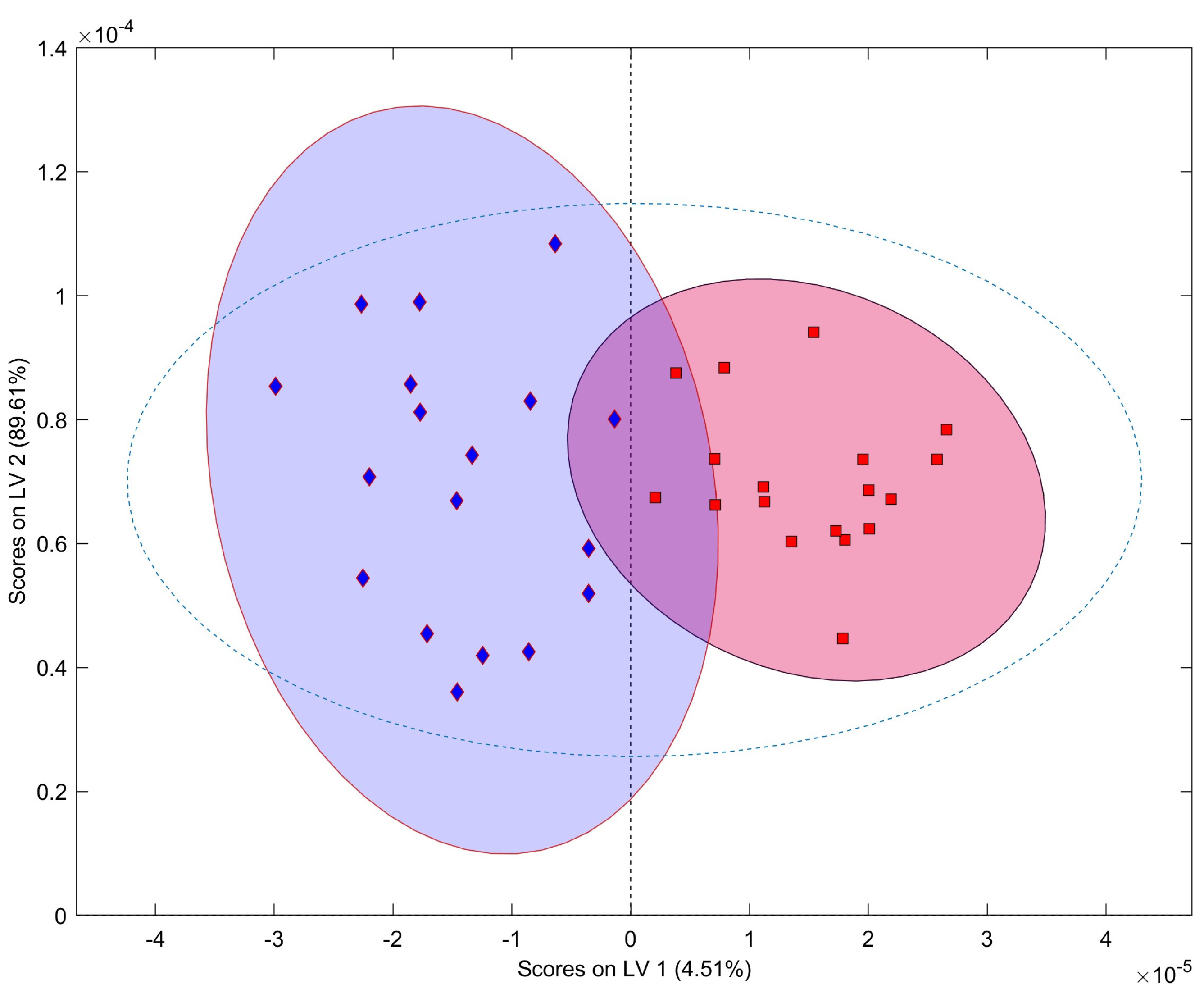
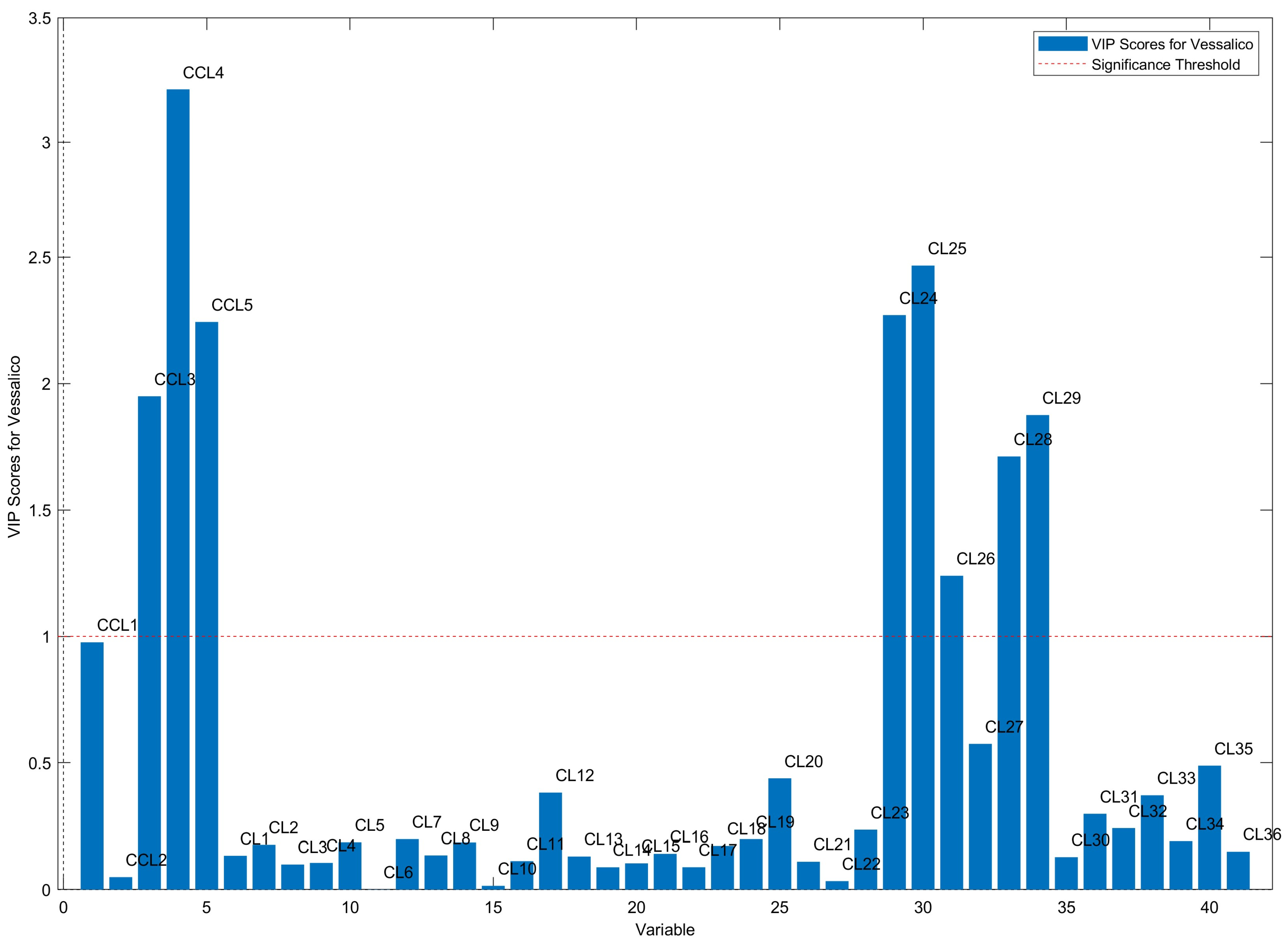
2.2. Multivariate Data Analysis
2.3. S-Allyl-L-Cysteine Quantification
2.4. Antibacterial Activity
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
4.3. Sample Collection and Preparation
4.4. NMR Spectroscopy and Processing
4.5. NMR Data Analysis
4.6. Multivariate Data Analysis
4.7. S-Allyl-L-Cysteine Quantification
4.8. Antibacterial Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Casals, J.; Rivera, A.; Campo, S.; Aymerich, E.; Isern, H.; Fenero, D.; Garriga, A.; Palou, A.; Monfort, A.; Howad, W.; et al. Phenotypic diversity and distinctiveness of the Belltall garlic landrace. Front. Plant Sci. 2023, 13, 1004069. [Google Scholar] [CrossRef] [PubMed]
- Egea, L.A.; Mérida-García, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology “genotyping-by-sequencing” platform (DArTseq). Front. Genet. 2017, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Volk, G.M.; Henk, A.D.; Richards, C.M. Genetic diversity among U.S. garlic clones as detected using AFLP methods. J. Am. Soc. Hort. Sci. 2004, 129, 559–569. [Google Scholar] [CrossRef]
- Montaño, A.; Beato, V.M.; Mansilla, F.; Orgaz, F. Effect of genetic characteristics and environmental factors on organosulfur compounds in garlic (Allium sativum L.) grown in Andalusia, Spain. J. Agric. Food. Chem. 2011, 59, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Volk, G.M. Phenotypic characteristics of ten garlic cultivars grown at different North American locations. Hort. Sci. 2009, 44, 1238–1247. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Shen, D.; Oiu, Y.; Song, J. Diversity evaluation of morphological traits and allicin content in garlic (Allium sativum L.) from China. Euphytica 2014, 198, 243–254. [Google Scholar] [CrossRef]
- Leite, V.S.A.; Reis, M.R.; Pinto, F.G. Untargeted metabolomics reveals metabolic changes linked to bulb purpling in garlic (Allium sativum L.). ACS Food Sci. Technol. 2021, 1, 242–248. [Google Scholar] [CrossRef]
- Lanzotti, V. Bioactive polar natural compounds from garlic and onions. Phytochem. Rev. 2012, 11, 179–196. [Google Scholar] [CrossRef]
- Lawson, L.D. Bioactive organosulfur compounds of garlic and garlic products. In Human Medicinal Agents from Plants; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1993; Volume 534, pp. 306–330. [Google Scholar]
- Block, E. The organosulfur chemistry of the genus Allium. Implications for the organic chemistry of sulfur. Angew. Chem. 1992, 31, 1135–1178. [Google Scholar] [CrossRef]
- Lawson, L.D. Garlic: A review of its medicinal effects and indicated active compounds. In Phytomedicines of Europe; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1998; Volume 691, pp. 176–209. [Google Scholar]
- Abdelrahman, M.; Hirata, S.; Mukae, T.; Yamada, T.; Sawada, Y.; El-Syaed, M.; Yamada, Y.; Sato, M.; Hirai, M.Y.; Shigyo, M. Comprehensive metabolite profiling in genetic resources of garlic (Allium sativum L.) collected from different geographical regions. Molecules 2021, 26, 1415. [Google Scholar] [CrossRef]
- Molino, R.; Rellin, K.F.B.; Nellas, R.B.; Junio, H.A. Small in size, big on taste: Metabolomics analysis of flavor compounds from Philippine garlic. PLoS ONE 2021, 16, e0247289. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Weng, R.; Sheng, X.; Wang, X.; Zhang, W.; Qian, Y.; Qiu, J. Profiling of organosulfur compounds and amino acids in garlic from different regions of China. Food Chem. 2020, 305, 125499. [Google Scholar] [CrossRef] [PubMed]
- Hirata, S.; Abdelrahman, M.; Yamauchi, N.; Shigyo, M. Characteristics of chemical components in genetic resources of garlic Allium sativum collected from all over the world. Genet. Resour. Crop Evol. 2016, 63, 35–45. [Google Scholar] [CrossRef]
- Jabbes, N.; Arnault, I.; Auger, J.; Al Mohandes Dridi, B.; Hannachi, C. Agro-morphological markers and organo-sulphur compounds to assess diversity in Tunisian garlic landraces. Sci. Hortic. 2012, 148, 47–54. [Google Scholar] [CrossRef]
- González, R.E.; Soto, V.C.; Sance, M.M.; Camargo, A.B.; Galmarini, C.R. Variability of solids, organosulfur compounds, pungency and health-enhancing traits in garlic (Allium sativum L.) cultivars belonging to different ecophysiological groups. J. Agric. Food. Chem. 2009, 57, 10282–10288. [Google Scholar] [CrossRef] [PubMed]
- Kamenetsky, R.; London Shafir, I.; Khassanov, F.; Kik, C.; van Heusden, A.W.; Vrielink-van Ginkel, M.; Burger-Meijer, K.; Auger, J.; Arnault, I.; Rabinowitch, H.D. Diversity in fertility potential and organo-sulphur compounds among garlics from Central Asia. Biodivers. Conserv. 2005, 14, 281–295. [Google Scholar] [CrossRef]
- Sommano, S.; Saratan, N.; Suksathan, R.; Pusadee, T. Chemical composition and comparison of genetic variation of commonly available Thai garlic used as food supplement. J. Appl. Bot. Food Qual. 2016, 89, 235–242. [Google Scholar] [CrossRef]
- Szychowski, K.A.; Rybczyńska-Tkaczyk, K.; Gaweł-Bęben, K.; Świeca, M.; Karaś, M.; Jakubczyk, A.; Matysiak, M.; Binduga, U.E.; Gmiński, J. Characterization of active compounds of different garlic (Allium sativum L.) cultivars. Pol. J. Food Nutr. Sci. 2018, 68, 73–81. [Google Scholar] [CrossRef]
- MIPAAF. Aggiornamento Dell’elenco Nazionale dei Prodotti Agroalimentari Tradizionali ai Sensi Dell’articolo 12, Comma 1, della Legge 12 Dicembre 2016, n. 238. (21A01168). Ventunesima Revisione Dell’elenco dei Prodotti Agroalimentari Tradizionali 2021, Serie Generale n.48 del 26-02-2021—Suppl. Ordinario n. 15. Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/398 (accessed on 20 February 2024).
- Iobbi, V.; Santoro, V.; Maggi, N.; Giacomini, M.; Lanteri, A.P.; Minuto, G.; Minuto, A.; Fossa, P.; De Tommasi, N.; Bisio, A.; et al. Characterization of sulfur compounds and antiviral activity against Tomato brown rugose fruit virus (ToBRFV) of Italian “Vessalico” garlic compared to other cultivars and landrace. LWT 2023, 174, 114411. [Google Scholar] [CrossRef]
- GEVES. Le Catalogue Officiel des Espèces et Variétés de Plantes Cultivées en France; GEVES: Beaucouzé, France, 2015. [Google Scholar]
- Martins, N.; Petropoulos, S.; Ferreira, I.C.F.R. Chemical composition and bioactive compounds of garlic (Allium sativum L.) as affected by pre- and post-harvest conditions: A review. Food Chem. 2016, 211, 41–50. [Google Scholar] [CrossRef]
- Ellis, N.; Hattori, C.; Cheema, J.; Donarski, J.; Charlton, A.; Dickinson, M.; Venditti, G.; Kaló, P.; Szabó, Z.; Kiss, G.B.; et al. NMR metabolomics defining genetic variation in pea seed metabolites. Front. Plant Sci. 2018, 9, 1022. [Google Scholar] [CrossRef] [PubMed]
- Bueno, P.C.P.; Lopes, N.P. Metabolomics to characterize adaptive and signaling responses in legume crops under abiotic stresses. ACS Omega 2020, 5, 1752–1763. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Song, Y.; Jeong, J.-H.; Hwang, J.; Kim, Y. Geographical discrimination of Allium species (garlic and onion) using 1H NMR spectroscopy with multivariate analysis. Int. J. Food Prop. 2020, 23, 241–254. [Google Scholar] [CrossRef]
- Ritota, M.; Casciani, L.; Han, B.-Z.; Cozzolino, S.; Leita, L.; Sequi, P.; Valentini, M. Traceability of Italian garlic (Allium sativum L.) by means of HRMAS-NMR spectroscopy and multivariate data analysis. Food Chem. 2012, 135, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Pacholczyk-Sienicka, B.; Modranka, J.; Ciepielowski, G. Comparative analysis of bioactive compounds in garlic owing to the cultivar and origin. Food Chem. 2024, 439, 138141. [Google Scholar] [CrossRef] [PubMed]
- Muhie, S.H. Novel approaches and practices to sustainable agriculture. J. Agric. Food Res. 2022, 10, 100446. [Google Scholar] [CrossRef]
- Fenibo, E.O.; Ijoma, G.N.; Matambo, T. Biopesticides in sustainable agriculture: A critical sustainable development driver governed by green chemistry principles. Front. Sustain. Food Syst. 2021, 5, 619058. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Datta, S.; Dhanjal, D.S.; Singh, J. Plant disease management by bioactive natural products. In Natural Bioactive Products in Sustainable Agriculture; Singh, J., Yadav, A.N., Eds.; Springer: Singapore, 2020; pp. 15–29. [Google Scholar]
- Iqbal, A.; Hamayun, M.; Shah, F.; Hussain, A. Role of plant bioactives in sustainable agriculture. In Environment, Climate, Plant and Vegetation Growth; Fahad, S., Hasanuzzaman, M., Alam, M., Ullah, H., Saeed, M., Ali Khan, I., Adnan, M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 591–605. [Google Scholar]
- Vicente, J.G.; Holub, E.B. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 2013, 14, 2–18. [Google Scholar] [CrossRef]
- Gazdik, F.; Magnus, S.; Roberts, S.J.; Baranski, R.; Cechova, J.; Pokluda, R.; Eichmeier, A.; Grzebelus, D.; Baranek, M. Persistence of Xanthomonas campestris pv. campestris in field soil in Central Europe. Microorganisms 2021, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Shigaki, T.; Nelson, S.C.; Alvarez, A.M. Symptomless spread of blight-inducing strains of Xanthomonas campestris pv. campestris on cabbage seedlings in misted seedbeds. Eur. J. Plant Pathol. 2000, 106, 339–346. [Google Scholar] [CrossRef]
- Siddiqui, N.; Mothana, R.; Alam, P. Quantitative determination of alliin in dried garlic cloves and products by high-performance thin-layer chromatography. Trop. J. Pharm. Res. 2016, 15, 1759. [Google Scholar] [CrossRef]
- EPPO. Guidelines on good plant protection practice—Vegetable brassicas. EPPO Bull. 1996, 26, 311–347. [Google Scholar]
- Hakalová, E.; Čechová, J.; Tekielska, D.A.; Eichmeier, A.; Pothier, J.F. Combined effect of thyme and clove phenolic compounds on Xanthomonas campestris pv. campestris and biocontrol of black rot disease on cabbage seeds. Front. Microbiol. 2022, 13, 1007988. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.; Caproni, A.; Sicurella, M.; Manfredini, S.; Baldisserotto, A.; Marconi, P. Effects of flavonoids and phenols from Moringa oleifera leaf extracts on biofilm processes in Xanthomonas campestris pv. campestris. Plants 2023, 12, 1508. [Google Scholar] [CrossRef] [PubMed]
- Velasco, P.; Lema, M.; Francisco, M.; Soengas, P.; Cartea, M.E. In vivo and in vitro effects of secondary metabolites against Xanthomonas campestris pv. campestris. Molecules 2013, 18, 11131–11143. [Google Scholar] [CrossRef] [PubMed]
- da Silva, R.S.; de Oliveira, M.M.G.; de Melo, J.O.; Blank, A.F.; Corrêa, C.B.; Scher, R.; Fernandes, R.P.M. Antimicrobial activity of Lippia gracilis essential oils on the plant pathogen Xanthomonas campestris pv. campestris and their effect on membrane integrity. Pestic. Biochem. Physiol. 2019, 160, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Ahmad, H.; Hayat, S.; Ghani, M.I.; Amin, B.; Atif, M.J.; Wali, K.; Cheng, Z. Application of garlic allelochemicals improves growth and induces defense responses in eggplant (Solanum melongena) against Verticillium dahliae. Ecotoxicol. Environ. Saf. 2021, 215, 112132. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Ahmad, H.; Ali, M.; Hayat, K.; Khan, M.A.; Cheng, Z. Aqueous garlic extract as a plant biostimulant enhances physiology, improves crop quality and metabolite abundance, and primes the defense responses of receiver plants. Appl. Sci. 2018, 8, 1505. [Google Scholar] [CrossRef]
- Curtis, H.; Noll, U.; Störmann, J.; Slusarenko, A. Broad-spectrum activity of the volatile phytoanticipin allicin in extracts of garlic (Allium sativum L.) against plant pathogenic bacteria, fungi and Oomycetes. Physiol. Mol. Plant Pathol. 2004, 65, 79–89. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 889/2008 of 5 September 2008—Laying Down Detailed Rules for the Implementation of Council Regulation (EC) No 834/2007 on Organic Production and Labelling of Organic Products with Regard to Organic Production, Labelling and Control; Document 32008R0889; European Commission: Brussels, Belgium, 2008. [Google Scholar]
- European Commission. Commission Implementing Regulation (EU) 2018/1584 of 22 October 2018—Amending Regulation (EC) No 889/2008 Laying Down Detailed Rules for the Implementation of Council Regulation (EC) No 834/2007 on Organic Production and Labelling of Organic Products with Regard to Organic Production, Labelling and Control; Implementing Regulation (EU) 2018/1584; European Commission: Brussels, Belgium, 2018. [Google Scholar]
- European Commission. Commission Implementing Regulation (EU) 2021/129 of 3 February 2021—Renewing the Approval of the Active Substance Garlic Extract in Accordance with Regulation (EC) No 1107/2009 of the European Parliament and of the Council Concerning the Placing of Plant Protection Products on the Market, and Amending the Annex to Commission Implementing Regulation (EU) No 540/2011 (Text with EEA Relevance); Document 32021R0129, Implementing Regulation (EU) 2021/129; European Commission: Brussels, Belgium, 2021. [Google Scholar]
- European Parliament. Directive 2009/128/EC of the European Parliament and of the Council of 21 October 2009 Establishing a Framework for Community Action to Achieve the Sustainable Use of Pesticides; European Parliament: Strasbourg, France, 2009; pp. 71–84. [Google Scholar]
- European Parliament Council of the European Union. Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009—Concerning the Placing of Plant Protection Products on the Market and Repealing Council Directives 79/117/EEC and 91/414/EEC; Document 32009R1107, Regulation (EC) No 1107/2009; European Parliament Council of the European Union: Brussels, Belgium, 2009. [Google Scholar]
- Liang, T.; Wei, F.; Lu, Y.; Kodani, Y.; Nakada, M.; Miyakawa, T.; Tanokura, M. Comprehensive NMR Analysis of Compositional Changes of Black Garlic during Thermal Processing. J. Agric. Food. Chem. 2015, 63, 683–691. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, 1, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; Sarojini, V. Pseudomonas syringae pv. actinidiae: Chemical control, resistance mechanisms and possible alternatives. Plant Pathol. 2014, 63, 1–11. [Google Scholar] [CrossRef]
- Németh, J. Practice of applying streptomycin to control fireblight in Hungary. In Proceedings of the EPPO Conference on Fireblight, Budapest, Hungary, 7–9 October 2003; EPPO: Paris, France, 2004; pp. 381–382. [Google Scholar]
- Verhaegen, M.; Bergot, T.; Liebana, E.; Stancanelli, G.; Streissl, F.; Mingeot-Leclercq, M.-P.; Mahillon, J.; Bragard, C. On the use of antibiotics to control plant pathogenic bacteria: A genetic and genomic perspective. Front. Microbiol. 2023, 14, 1221478. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Ferreira, J.P.; LeJeune, J.T. Antimicrobial use and resistance in plant agriculture: A one health perspective. Agriculture 2022, 12, 289. [Google Scholar] [CrossRef]
- Pirela, M.; Peña, M.; Peña-Vera, M.; Sulbarán, M.; Perez, E.; Lárez Velásquez, C. Characterization and determination of antimicrobial and metal resistant profles of Xanthomonas strains isolated from natural environments. J. Anal. Pharm. Res. 2019, 8, 55–60. [Google Scholar] [CrossRef]
- Srivastava, V.; Deblais, L.; Kathayat, D.; Rotondo, F.; Helmy, Y.A.; Miller, S.A.; Rajashekara, G. Novel small molecule growth inhibitors of Xanthomonas spp. causing bacterial spot of tomato. Phytopathology 2021, 111, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Block, E. The chemistry of garlic and onions. Sci. Am. 1985, 252, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Amagase, H. Clarifying the real bioactive constituents of garlic. J. Nutr. 2006, 136, 716s–725s. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Ide, N.; Ono, K. Changes in organosulfur compounds in garlic cloves during storage. J. Agric. Food. Chem. 2006, 54, 4849–4854. [Google Scholar] [CrossRef]
- Beato, V.M.; Sánchez, A.H.; de Castro, A.; Montaño, A. Effect of processing and storage time on the contents of organosulfur compounds in pickled blanched garlic. J. Agric. Food. Chem. 2012, 60, 3485–3491. [Google Scholar] [CrossRef]
- Hornickova, J.; Kubec, R.; Cejpek, K.; Velisek, J.; Ovesna, J.; Stavelikova, H. Profiles of S-alk(en)ylcysteine sulfoxides in various garlic genotypes. Czech J. Food Sci. 2010, 28, 298–308. [Google Scholar] [CrossRef]
- Kodera, Y.; Kurita, M.; Nakamoto, M.; Matsutomo, T. Chemistry of aged garlic: Diversity of constituents in aged garlic extract and their production mechanisms via the combination of chemical and enzymatic reactions (Review). Exp. Ther. Med. 2020, 19, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Montaño, A.; Casado, F.J.; de Castro, A.; Sánchez, A.H.; Rejano, L. Vitamin content and amino acid composition of pickled garlic processed with and without fermentation. J. Agric. Food. Chem. 2004, 52, 7324–7330. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, L.; Guo, W.; Fu, M.; Yang, M.; Huang, S.; Zhang, F.; Liu, Y. A new methodology for sensory quality assessment of garlic based on metabolomics and an artificial neural network. RSC Adv. 2019, 9, 17754–17765. [Google Scholar] [CrossRef] [PubMed]
- Molina-Calle, M.; de Medina, V.S.; Priego-Capote, F.; de Castro, M.D.L. Establishing compositional differences between fresh and black garlic by a metabolomics approach based on LC–QTOF MS/MS analysis. J. Food Compos. Anal. 2017, 62, 155–163. [Google Scholar] [CrossRef]
- Hrbek, V.; Rektorisova, M.; Chmelarova, H.; Ovesna, J.; Hajslova, J. Authenticity assessment of garlic using a metabolomic approach based on high resolution mass spectrometry. J. Food Compos. Anal. 2018, 67, 19–28. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, M.; Wang, C.; Zhou, H.; Fan, L.; Huang, X. Thermolysis kinetics and thermal degradation compounds of alliin. Food Chem. 2017, 223, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zaini, A.S.; Putra, N.R.; Idham, Z.; Mohd Faizal, A.N.; Che Yunus, M.A.; Mamat, H.; Abdul Aziz, A.H. Comparison of alliin recovery from Allium sativum L. using soxhlet extraction and subcritical water extraction. Chem. Eng. 2022, 6, 73. [Google Scholar] [CrossRef]
- Bloem, E.; Haneklaus, S.; Schnug, E. Storage life of field-grown garlic bulbs (Allium sativum L.) as influenced by nitrogen and sulfur fertilization. J. Agric. Food. Chem. 2011, 59, 4442–4447. [Google Scholar] [CrossRef]
- Lawson, L.D.; Wood, S.G.; Hughes, B.G. HPLC analysis of allicin and other thiosulfinates in garlic clove homogenates. Planta Med. 1991, 57, 263–270. [Google Scholar] [CrossRef]
- Fujisawa, H.; Suma, K.; Origuchi, K.; Kumagai, H.; Seki, T.; Ariga, T. Biological and chemical stability of garlic-derived allicin. J. Agric. Food. Chem. 2008, 56, 4229–4235. [Google Scholar] [CrossRef] [PubMed]
- Rybak, M.E.; Calvey, E.M.; Harnly, J.M. Quantitative determination of allicin in garlic: supercritical fluid extraction and standard addition of alliin. J. Agric. Food. Chem. 2004, 52, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Kodera, Y.; Suzuki, A.; Imada, O.; Kasuga, S.; Sumioka, I.; Kanezawa, A.; Taru, N.; Fujikawa, M.; Nagae, S.; Masamoto, K.; et al. Physical, chemical, and biological properties of S-allylcysteine, an amino acid derived from garlic. J. Agric. Food. Chem. 2002, 50, 622–632. [Google Scholar] [CrossRef]
- Rais, N.; Ved, A.; Ahmad, R.; Kumar, M.; Deepak Barbhai, M.; Radha; Chandran, D.; Dey, A.; Dhumal, S.; Senapathy, M.; et al. S-Allyl-L-Cysteine—A garlic bioactive: Physicochemical nature, mechanism, pharmacokinetics, and health promoting activities. J. Funct. Foods 2023, 107, 105657. [Google Scholar] [CrossRef]
- Bae, S.E.; Cho, S.Y.; Won, Y.D.; Lee, S.H.; Park, H.J. A comparative study of the different analytical methods for analysis of S-allyl-cysteine in black garlic by HPLC. LWT 2012, 46, 532–535. [Google Scholar] [CrossRef]
- Yudhistira, B.; Punthi, F.; Lin, J.-A.; Sulaimana, A.S.; Chang, C.-K.; Hsieh, C.-W. S-Allyl cysteine in garlic (Allium sativum): Formation, biofunction, and resistance to food processing for value-added product development. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2665–2687. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, H.; Kim, H.S.; Kim, Y.-R.; Noh, S.H. Optimum conditions for S-allyl-(L)-cysteine accumulation in aged garlic by RSM. Food Sci. Biotechnol. 2014, 23, 717–722. [Google Scholar] [CrossRef]
- Bae, S.E.; Cho, S.Y.; Won, Y.D.; Lee, S.H.; Park, H.J. Changes in S-allyl cysteine contents and physicochemical properties of black garlic during heat treatment. LWT 2014, 55, 397–402. [Google Scholar] [CrossRef]
- Le Mao, I.; Martin-Pernier, J.; Bautista, C.; Lacampagne, S.; Richard, T.; Da Costa, G. 1H-NMR Metabolomics as a tool for winemaking monitoring. Molecules 2021, 26, 6771. [Google Scholar] [CrossRef]
- Wang, K.; Liao, X.; Xia, J.; Xiao, C.; Deng, J.; Xu, Z. Metabolomics: A promising technique for uncovering quality-attribute of fresh and processed fruits and vegetables. Trends Food Sci. Technol. 2023, 142, 104213. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic diversity, conservation, and utilization of plant genetic resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.M.F.; Lau, H.Y.; Abu-Bakar, N. Integration of advanced technologies for plant variety and cultivar identification. J. Biosci. 2021, 46, 91. [Google Scholar] [CrossRef]
- Lazaridi, E.; Kapazoglou, A.; Gerakari, M.; Kleftogianni, K.; Passa, K.; Sarri, E.; Papasotiropoulos, V.; Tani, E.; Bebeli, P.J. Crop landraces and indigenous varieties: A valuable source of genes for plant breeding. Plants 2024, 13, 758. [Google Scholar] [CrossRef] [PubMed]
- Jamali, S.H.; Cockram, J.; Hickey, L.T. Insights into deployment of DNA markers in plant variety protection and registration. Theor. Appl. Genet. 2019, 132, 1911–1929. [Google Scholar] [CrossRef]
- Reddy, P.P. Agro-Ecological Approaches to Pest Management for Sustainable Agriculture; Springer Nature: Singapore, 2017; p. 339. [Google Scholar]
- European Food Safety Authority. The 2017 European Union Report on Pesticide Residues in Food. EFSA J. 2019, 17, e05743. [Google Scholar]
- Directorate-General for Health and Food Safety. Pesticides; SANTE: Brussel, Belgium, 2023. [Google Scholar]
- Godlewska, K.; Ronga, D.; Michalak, I. Plant extracts—Importance in sustainable agriculture. Ital. J. Agron. 2021, 16, 1851. [Google Scholar] [CrossRef]
- Hayat, S.; Ahmad, A.; Ahmad, H.; Hayat, K.; Khan, M.A.; Runan, T. Garlic, from medicinal herb to possible plant bioprotectant: A review. Sci. Hortic. 2022, 304, 111296. [Google Scholar] [CrossRef]
- La Torre, A.; Iovino, V.; Caradonia, F. Copper in plant protection: Current situation and prospects. Phytopathol. Mediterr. 2018, 57, 201–236. [Google Scholar] [CrossRef]
- Tamm, L.; Thuerig, B.; Apostolov, S.; Blogg, H.; Borgo, E.; Corneo, P.E.; Fittje, S.; de Palma, M.; Donko, A.; Experton, C.; et al. Use of copper-based fungicides in organic agriculture in twelve European countries. Agronomy 2022, 12, 673. [Google Scholar] [CrossRef]
- Bonn, W.G.; Lesage, S. Control of bacterial speck of tomato by copper and ethylenebisuithiocarbamate fungicides: Their efficacy and residues on leaves. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 1984, 19, 29–38. [Google Scholar] [CrossRef]
- Roberts, J.M.; Bruce, T.J.A.; Monaghan, J.M.; Pope, T.W.; Leather, S.R.; Beacham, A.M. Vertical farming systems bring new considerations for pest and disease management. Ann. Appl. Biol. 2020, 176, 226–232. [Google Scholar] [CrossRef]
- Fontana, R.; Caproni, A.; Buzzi, R.; Sicurella, M.; Buratto, M.; Salvatori, F.; Pappadà, M.; Manfredini, S.; Baldisserotto, A.; Marconi, P. Effects of Moringa oleifera leaf extracts on Xanthomonas campestris pv. campestris. Microorganisms 2021, 9, 2244. [Google Scholar] [CrossRef] [PubMed]
- Darji, B.; Ratani, J.; Doshi, M.; Kothari, V. In vitro antimicrobial activity in certain plant products/seed extracts against selected phytopathogens. Res. Pharm. 2012, 2, 1–10. [Google Scholar]
- Guimarães, P.; Moreira, I.; Pedro Campos, P.; Ferraz, J.; Novaes, Q.; Batista, R. Antibacterial activity of Schinopsis brasiliensis against phytopathogens of agricultural interest. Rev. Fitos 2015, 9, 161–252. [Google Scholar] [CrossRef]
- Choo, S.; Chin, V.K.; Wong, E.H.; Madhavan, P.; Tay, S.T.; Yong, P.V.C.; Chong, P.P. Review: Antimicrobial properties of allicin used alone or in combination with other medications. Folia Microbiol. 2020, 65, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Khameneh, B.; Iranshahy, M.; Soheili, V.; Fazly Bazzaz, B.S. Review on plant antimicrobials: A mechanistic viewpoint. Antimicrob. Resist. Infect. Control 2019, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Bloem, E.; Haneklaus, S.; Schnug, E. Influence of fertilizer practices on S-containing metabolites in garlic (Allium sativum L.) under field conditions. J. Agric. Food. Chem. 2010, 58, 10690–10696. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, C.-H.; Cai, J.; Zhang, W.; Qi, W.-L.; Wang, Z.; Liu, Z.-B.; Yang, Y. Broad-spectrum antimicrobial activity, chemical composition and mechanism of action of garlic (Allium sativum) extracts. Food Control 2018, 86, 117–125. [Google Scholar] [CrossRef]
- Varympopi, A.; Dimopoulou, A.; Papafotis, D.; Avramidis, P.; Sarris, I.; Karamanidou, T.; Kerou, A.K.; Vlachou, A.; Vellis, E.; Giannopoulos, A.; et al. Antibacterial activity of copper nanoparticles against Xanthomonas campestris pv. vesicatoria in tomato plants. Int. J. Mol. Sci. 2022, 23, 4080. [Google Scholar] [CrossRef]
- Fan, Q.; Liao, Y.-Y.; Kunwar, S.; Da Silva, S.; Young, M.; Santra, S.; Minsavage, G.V.; Freeman, J.H.; Jones, J.B.; Paret, M.L. Antibacterial effect of copper composites against Xanthomonas euvesicatoria. Crop Prot. 2021, 139, 105366. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kim, K.-M.; Kang, S.-W.; Kim, J.-Y.; Kim, J.-Y.; Kim, J.-H. Synergistic antibacterial effect of geraniol, thymol and o-vanillin against Xanthomonas campestris pv. vesicatoria. Korean J. Pestic. Sci. 2023, 27, 318–323. [Google Scholar] [CrossRef]
- Hanelt, P. Taxonomy, evolution, and history. In Onions and Allied Crops; Brewster, J.L., Rabinowitch, H.D., Eds.; CRC Press: Boca Raton, FL, USA, 1989; Volume I: Botany Physiology and Genetics, pp. 1–26. [Google Scholar]
- Takagi, H. Garlic (Allium sativum). In Onions and Allied Crops; Brewster, J.L., Rabinowitch, H.D., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2019; Volume III: Biochemistry, Food Science, and Minor Crops, pp. 109–146. [Google Scholar]
- Tajidin, N.E.; Shaari, K.; Maulidiani, M.; Salleh, N.S.; Ketaren, B.R.; Mohamad, M. Metabolite profiling of Andrographis paniculata (Burm. f.) Nees. young and mature leaves at different harvest ages using 1H NMR-based metabolomics approach. Sci. Rep. 2019, 9, 16766. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Liu, X.; Shen, D.; Qiu, Y.; Zhang, X.; Song, J. Influence of pH, concentration and light on stability of allicin in garlic (Allium sativum L.) aqueous extract as measured by UPLC. J. Sci. Food Agric. 2015, 95, 1838–1844. [Google Scholar] [CrossRef]
- Monzón Daza, G.; Meneses Macías, C.; Forero, A.M.; Rodríguez, J.; Aragón, M.; Jiménez, C.; Ramos, F.A.; Castellanos, L. Identification of α-amylase and α-glucosidase inhibitors and ligularoside A, a new triterpenoid saponin from Passiflora ligularis Juss (Sweet Granadilla) leaves, by a nuclear magnetic resonance-based metabolomic study. J. Agric. Food. Chem. 2021, 69, 2919–2931. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-throughput metabolomics by 1D NMR. Angew. Chem. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Iobbi, V.; Donadio, G.; Lanteri, A.P.; Maggi, N.; Kirchmair, J.; Parisi, V.; Minuto, G.; Copetta, A.; Giacomini, M.; Bisio, A.; et al. Targeted metabolite profiling of Salvia rosmarinus Italian local ecotypes and cultivars and inhibitory activity against Pectobacterium carotovorum subsp. carotovorum. Front. Plant Sci. 2024, 15, 1164859. [Google Scholar] [CrossRef] [PubMed]
- Lanzotti, V. The analysis of onion and garlic. J. Chromatogr. A 2006, 1112, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Torres, C.; Huber, G.; Ichikawa, A.; Nishiyama, Y.; Wong, A. HR-μMAS NMR-based metabolomics: Localized metabolic profiling of a garlic clove with μg tissues. Anal. Chem. 2018, 90, 13736–13743. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef]
- Grimaldi, M.; Marino, C.; Buonocore, M.; Santoro, A.; Sommella, E.; Merciai, F.; Salviati, E.; De Rosa, A.; Nuzzo, T.; Errico, F.; et al. Prenatal and early postnatal cerebral D-aspartate depletion influences L-amino acid pathways, bioenergetic processes, and developmental brain metabolism. J. Proteome Res. 2021, 20, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Osmala, M.; Raiko, T.; Lagus, K.; Sysi-Aho, M.; Orešič, M.; Honkela, T.; Lähdesmäki, H. Self-organization and missing values in SOM and GTM. Neurocomputing 2015, 147, 60–70. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi- and Megavariate Data Analysis: Part I: Basic Principles and Applications, 2nd revised and enlarged ed.; Umetrics Umeå: Umeå, Sweden, 2006. [Google Scholar]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier Academic Press: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Ross, L.N.; Woodward, J.F. Koch’s postulates: An interventionist perspective. Stud. Hist. Philos. Biol. Biomed. Sci. 2016, 59, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Dogra, D.; Julka, J.M.; Kumar, A. Insight into antimicrobic evaluation techniques and their role in better assessment of antimicrobic agents: A review. Asian J. Pharm. Clin. Res. 2021, 15, 6–13. [Google Scholar] [CrossRef]
- M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2012.
- Ankri, S.; Mirelman, D. Antimicrobial properties of allicin from garlic. Microb. Infect. 1999, 1, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Kunimura, K.; Suzuki, J.I.; Kodera, Y. Antimicrobial properties of hydrophobic compounds in garlic: Allicin, vinyldithiin, ajoene and diallyl polysulfides. Exp. Ther. Med. 2020, 19, 1550–1553. [Google Scholar] [CrossRef] [PubMed]
- Slusarenko, A.J.; Patel, A.; Portz, D. Control of plant diseases by natural products: Allicin from garlic as a case study. Eur. J. Plant Pathol. 2008, 121, 313–322. [Google Scholar] [CrossRef]
- Bal, D.; Kraska-Dziadecka, A.; Gryff-Keller, A. Solution structure of succinylacetone, an unsymmetrical beta-diketone, as studied by 13C NMR and GIAO-DFT calculations. J. Org. Chem. 2009, 74, 8604–8609. [Google Scholar] [CrossRef]
- Bourafai-Aziez, A.; Jacob, D.; Charpentier, G.; Cassin, E.; Rousselot, G.; Moing, A.; Deborde, C. Development, validation, and use of 1H-NMR spectroscopy for evaluating the quality of acerola-based food supplements and quantifying ascorbic acid. Molecules 2022, 27, 5614. [Google Scholar] [CrossRef]
- Fulmer, G.R.; Miller, A.J.M.; Sherden, N.H.; Gottlieb, H.E.; Nudelman, A.; Stoltz, B.M.; Bercaw, J.E.; Goldberg, K.I. NMR chemical shifts of trace impurities: Common laboratory solvents, organics, and gases in deuterated solvents relevant to the organometallic chemist. Organometallics 2010, 29, 2176–2179. [Google Scholar] [CrossRef]
- Gaviglio, C.; Doctorovich, F. Hydrogen-free homogeneous catalytic reduction of olefins in aqueous solutions. J. Org. Chem. 2008, 73, 5379–5384. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Kawashima, T.; Yajima, H.; Smirnov, S.V.; Kodera, T.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; Ogawa, J. Enzymatic synthesis of chiral amino acid sulfoxides by Fe(II)/α-ketoglutarate-dependent dioxygenase. Tetrahedron Asymmetry 2013, 24, 990–994. [Google Scholar] [CrossRef]
- Higuchi, O.; Tateshita, K.; Nishimura, H. Antioxidative activity of sulfur-containing compounds in Allium species for human low-density lipoprotein (ldl) oxidation in vitro. J. Agric. Food. Chem. 2003, 51, 7208–7214. [Google Scholar] [CrossRef]
- Horton, D.; Wałaszek, Z.; Ekiel, I. Conformations of D-gluconic, D-mannonic, and D-galactonic acids in solution, as determined by N.M.R. spectroscopy. Carbohydr. Res. 1983, 119, 263–268. [Google Scholar] [CrossRef]
- Ingallina, C.; Di Matteo, G.; Spano, M.; Acciaro, E.; Campiglia, E.; Mannina, L.; Sobolev, A.P. Byproducts of globe artichoke and cauliflower production as a new source of bioactive compounds in the green economy perspective: An NMR study. Molecules 2023, 28, 1363. [Google Scholar] [CrossRef]
- Jamieson, A.G.; Boutard, N.; Beauregard, K.; Bodas, M.S.; Ong, H.; Quiniou, C.; Chemtob, S.; Lubell, W.D. Positional scanning for peptide secondary structure by systematic solid-phase synthesis of amino lactam peptides. J. Am. Chem. Soc. 2009, 131, 7917–7927. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Nava, R.A.; Zepeda-Vallejo, L.G.; Santoyo-Tepole, F.; Chávez-Camarillo, G.M.; Cristiani-Urbina, E. RP-HPLC separation and 1H NMR identification of a yellow fluorescent compound—Riboflavin (vitamin B2) produced by the yeast Hyphopichia wangnamkhiaoensis. Biomolecules 2023, 13, 1423. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Maldonado, P.D.; Alvarez-Idaboy, J.R.; Aguilar-González, A.; Lira-Rocha, A.; Jung-Cook, H.; Medina-Campos, O.N.; Pedraza-Chaverrí, J.; Galano, A. Role of Allyl group in the hydroxyl and peroxyl radical scavenging activity of s-allylcysteine. J. Phys. Chem. B 2011, 115, 13408–13417. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, A.; Isogawa, T.; Morioka, Y.; Knappett, B.R.; Wheatley, A.E.H.; Saito, S.; Naka, H. Hydration of nitriles to amides by a chitin-supported ruthenium catalyst. RSC Adv. 2015, 5, 12152–12160. [Google Scholar] [CrossRef]
- Moing, A.; Maucourt, M.; Renaud, C.; Gaudillère, M.; Brouquisse, R.; Lebouteiller, B.; Gousset-Dupont, A.; Vidal, J.; Granot, D.; Denoyes-Rothan, B.; et al. Quantitative metabolic profiling by 1-dimensional 1H-NMR analyses: Application to plant genetics and functional genomics. Funct. Plant Biol. 2004, 31, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Duarte, I.F.; Almeida, C.; Delgadillo, I.; Goodfellow, B.J.; Gil, A.M.; Morris, G.A. High-resolution NMR and diffusion-ordered spectroscopy of port wine. J. Agric. Food. Chem. 2004, 52, 3736–3743. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.M.; Caldeira, M.M.; Gil, V.M. NMR study of the complexation of D-gulonic acid with tungsten(VI) and molybdenum(VI). Carbohydr. Res. 2000, 329, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Sabino, A.R.; Tavares, S.; Riffel, A.; Li, J.V.; Oliveira, D.J.; Feres, C.; Henrique, L.; Oliveira, J.S.; Correia, G.D.S.; Sabino, A.S.; et al. Short communication 1 H NMR metabolomic approach reveals chlorogenic acid as a response of sugarcane induced by exposure to Diatraea saccharalis. Ind. Crop. Prod. 2019, 140, 111651. [Google Scholar] [CrossRef]
- Verpoorte, R.; Choi, Y.H.; Kim, H.K. NMR-based metabolomics at work in phytochemistry. Phytochem. Rev. 2007, 6, 3–14. [Google Scholar] [CrossRef]
- Wang, M.-C.; Zhang, Q.-J.; Zhao, W.-X.; Wang, X.-D.; Ding, X.; Jing, T.-T.; Song, M.-P. Evaluation of enantiopure N-(ferrocenylmethyl)azetidin-2-yl(diphenyl)methanol for catalytic asymmetric addition of organozinc reagents to aldehydes. J. Org. Chem. 2008, 73, 168–176. [Google Scholar] [CrossRef]




| Treatment | MIC | |||
|---|---|---|---|---|
| (μg/mL) | μM | |||
| Strain 1 | Strain 2 | Strain 1 | Strain 2 | |
| S-allyl-L-cysteine | 500 | 500 | 3101.3 | 3101.3 |
| S-methyl-L-cysteine | 500 | 500 | 3698.77 | 3698.77 |
| L-alliin | 500 | 500 | 2821.35 | 2821.35 |
| Methiin | 500 | 500 | 3307.10 | 3307.10 |
| Allicin | 31.25 | 31.25 | 192.57 | 192.57 |
| Crude extract | 125 | 125 | - | - |
| Ampicillin | 0.25 | 0.5 | 0.72 | 1.43 |
| Streptomycin sulphate | 0.5 | 1 | 0.34 | 0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iobbi, V.; Parisi, V.; Lanteri, A.P.; Maggi, N.; Giacomini, M.; Drava, G.; Minuto, G.; Minuto, A.; Tommasi, N.D.; Bisio, A. NMR Metabolite Profiling for the Characterization of Vessalico Garlic Ecotype and Bioactivity against Xanthomonas campestris pv. campestris. Plants 2024, 13, 1170. https://doi.org/10.3390/plants13091170
Iobbi V, Parisi V, Lanteri AP, Maggi N, Giacomini M, Drava G, Minuto G, Minuto A, Tommasi ND, Bisio A. NMR Metabolite Profiling for the Characterization of Vessalico Garlic Ecotype and Bioactivity against Xanthomonas campestris pv. campestris. Plants. 2024; 13(9):1170. https://doi.org/10.3390/plants13091170
Chicago/Turabian StyleIobbi, Valeria, Valentina Parisi, Anna Paola Lanteri, Norbert Maggi, Mauro Giacomini, Giuliana Drava, Giovanni Minuto, Andrea Minuto, Nunziatina De Tommasi, and Angela Bisio. 2024. "NMR Metabolite Profiling for the Characterization of Vessalico Garlic Ecotype and Bioactivity against Xanthomonas campestris pv. campestris" Plants 13, no. 9: 1170. https://doi.org/10.3390/plants13091170







