First Clarification of the Involvement of Glycosyltransferase MdUGT73CG22 in the Detoxification Metabolism of Nicosulfuron in Apple
Abstract
:1. Introduction
2. Results
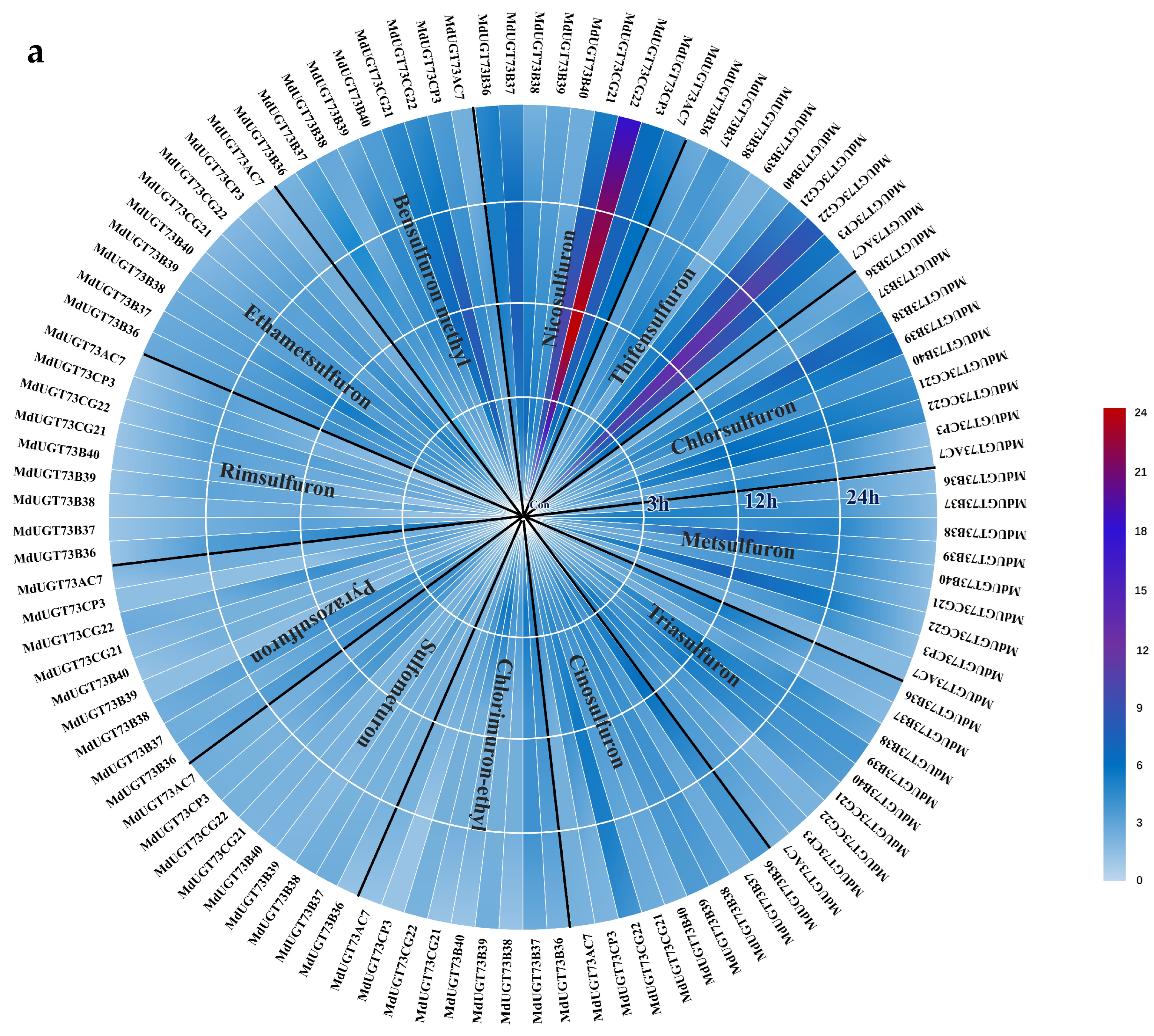
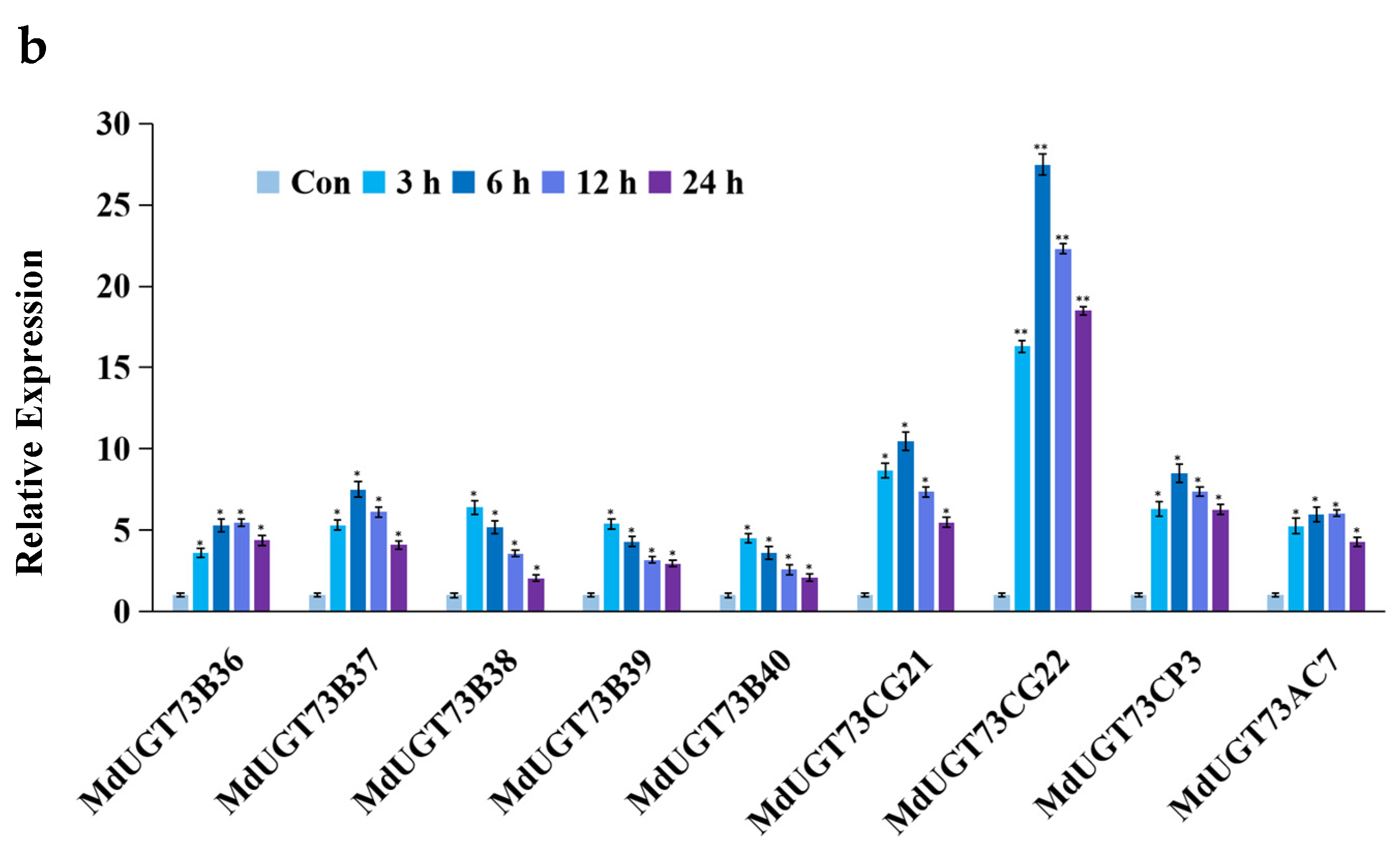
2.1. Candidate Gene MdUGT73CG22 Identified
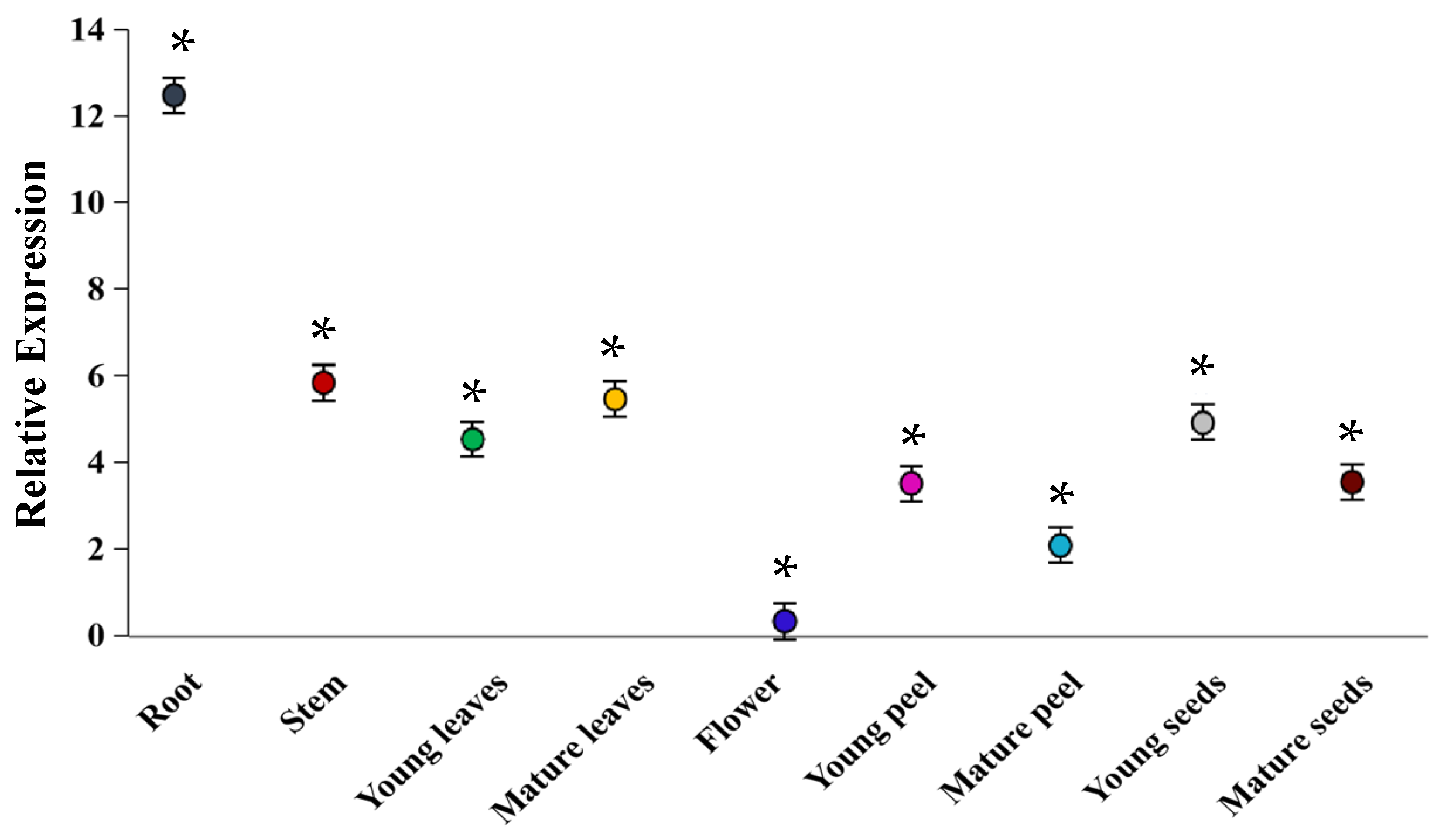
2.2. Analysis of Apple Glycosyltransferase MdUGT73CG22 Tissue Expression Pattern
2.3. Identification of Substrates for Glycosylation by Apple Glycosyltransferase MdUGT73CG22
2.4. Acquisition of the MdUGT73CG22 Transgenic Apple Line and Identification of the Detoxification Function
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Treatment of Apple Seedlings with ALS-Inhibiting Herbicides
4.3. Real-Time PCR
4.4. Expression Vector Construction
4.5. Protein Purification and In Vitro Enzymatic Reactions
4.6. Transgenic Apple Seedling Screening
4.7. Endogenous Glycoside Extraction from Transgenic Apple Seedlings
4.8. High Performance Liquid Chromatography (HPLC) Analysis of Target Products
4.9. Data Statistics and Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choudri, B.S.; Charabi, Y.; Al-Nasiri, N. Pesticides and herbicides. Water Environ. Res. 2020, 92, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef] [PubMed]
- Milesi, M.M.; Lorenz, V.; Durando, M.; Rossetti, M.F.; Varayoud, J. Glyphosate Herbicide: Reproductive Outcomes and Multigenerational Effects. Front. Endocrinol. 2021, 12, 672532. [Google Scholar] [CrossRef] [PubMed]
- Widianto, R.; Kurniadie, D.; Widayat, D.; Umiyati, U.; Nasahi, C.; Sari, S.; Juraimi, A.S.; Kato-Noguchi, H. Acetolactate Synthase-Inhibitor Resistance in Monochoria vaginalis (Burm. f.) C. Presl from Indonesia. Plants 2022, 11, 400. [Google Scholar] [CrossRef] [PubMed]
- Low, Y.S.; Garcia, M.D.; Lonhienne, T.; Fraser, J.A.; Schenk, G.; Guddat, L.W. Triazolopyrimidine herbicides are potent inhibitors of Aspergillus fumigatus acetohydroxyacid synthase and potential antifungal drug leads. Sci. Rep. 2021, 11, 21055. [Google Scholar] [CrossRef] [PubMed]
- Cheron, M.; Costantini, D.; Brischoux, F. Nicosulfuron, a sulfonylurea herbicide, alters embryonic development and oxidative status of hatchlings at environmental concentrations in an amphibian species. Ecotoxicol. Environ. Saf. 2022, 232, 113277. [Google Scholar] [CrossRef] [PubMed]
- Petric, I.; Karpouzas, D.G.; Bru, D.; Udikovic-Kolic, N.; Kandeler, E.; Djuric, S.; Martin-Laurent, F. Nicosulfuron application in agricultural soils drives the selection towards NS-tolerant microorganisms harboring various levels of sensitivity to nicosulfuron. Environ. Sci. Pollut. Res. Int. 2016, 23, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Ganai, B.A. Deciphering the recent trends in pesticide bioremediation using genome editing and multi-omics approaches: A review. World J. Microbiol. Biotechnol. 2023, 39, 151. [Google Scholar] [CrossRef]
- Márquez-López, R.E.; Loyola-Vargas, V.M.; Santiago-García, P.A. Interaction between fructan metabolism and plant growth regulators. Planta 2022, 255, 49. [Google Scholar] [CrossRef]
- Biswas, A.; Thattai, M. Promiscuity and specificity of eukaryotic glycosyltransferases. Biochem. Soc. Trans. 2020, 48, 891–900. [Google Scholar] [CrossRef]
- Dimunová, D.; Matoušková, P.; Podlipná, R.; Boušová, I.; Skálová, L. The role of UDP-glycosyltransferases in xenobioticresistance. Drug Metab. Rev. 2022, 54, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Kurze, E.; Wüst, M.; Liao, J.; McGraphery, K.; Hoffmann, T.; Song, C.; Schwab, W. Structure-function relationship of terpenoid glycosyltransferases from plants. Nat. Prod. Rep. 2022, 39, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Taylor, V.L.; Cummins, I.; Brazier-Hicks, M.; Edwards, R. Protective responses induced by herbicide safeners in wheat. Environ. Exp. Bot. 2013, 88, 93–99. [Google Scholar] [CrossRef]
- Messner, B.; Thulke, O.; Schäffner, A.R. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 2003, 217, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Loutre, C.; Dixon, D.P.; Brazier, M.; Slater, M.; Cole, D.J.; Edwards, R. Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant 3,4-dichloroaniline. Plant J. 2003, 34, 485–493. [Google Scholar] [CrossRef] [PubMed]
- von Rad, U.; Hüttl, R.; Lottspeich, F.; Gierl, A.; Frey, M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 2001, 28, 633–642. [Google Scholar] [CrossRef]
- Su, X.; Wang, W.; Xia, T.; Gao, L.; Shen, G.; Pang, Y. Characterization of a heat responsive UDP: Flavonoid glucosyltransferase gene in tea plant (Camellia sinensis). PLoS ONE 2018, 13, e0207212. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Zhang, L.; Shu, J.; Court, M.H.; Sun, Z.; Jiang, L.; Zheng, C.; Shu, H.; Ji, L.; et al. Genome-wide analysis of the apple family 1 glycosyltransferases identified a flavonoid-modifying UGT, MdUGT83L3, which is targeted by MdMYB88 and contributes to stress adaptation. Plant Sci. 2022, 321, 111314. [Google Scholar] [CrossRef]
- Singh, Y.; Saxena, M.K. Insights into the recent advances in nano-bioremediation of pesticides from the contaminated soil. Front. Microbiol. 2022, 13, 982611. [Google Scholar] [CrossRef]
- Gangola, S.; Bhatt, P.; Kumar, A.J.; Bhandari, G.; Joshi, S.; Punetha, A.; Bhatt, K.; Rene, E.R. Biotechnological tools to elucidate the mechanism of pesticide degradation in the environment. Chemosphere 2022, 296, 133916. [Google Scholar] [CrossRef]
- Mörtl, M.; Vehovszky, Á.; Klátyik, S.; Takács, E.; Győri, J.; Székács, A. Neonicotinoids: Spreading, Translocation and Aquatic Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 2006. [Google Scholar] [CrossRef] [PubMed]
- Guerrero Ramírez, J.R.; Ibarra Muñoz, L.A.; Balagurusamy, N.; Frías Ramírez, J.E.; Alfaro Hernández, L.; Carrillo Campos, J. Microbiology and Biochemistry of Pesticides Biodegradation. Int. J. Mol. Sci. 2023, 24, 15969. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, T.; Zhu, F.; Ouyang, G. In vivo monitoring and exposure potency assessment of phase I metabolism of fenthion in vegetables. J. Hazard. Mater. 2020, 399, 123013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, Z.; Li, R.; Tan, Y.; Lv, S.; McClements, D.J. Impact of pesticide polarity and lipid phase dimensions on the bioaccessibility of pesticides in agricultural produce consumed with model fatty foods. Food Funct. 2020, 11, 6028–6037. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Gill, J.P.K.; Datta, S.; Singh, S.; Dhaka, V.; Kapoor, D.; Wani, A.B.; Dhanjal, D.S.; Kumar, M.; et al. Herbicide Glyphosate: Toxicity and Microbial Degradation. Int. J. Environ. Res. Public Health 2020, 17, 7519. [Google Scholar] [CrossRef] [PubMed]
- Sirivarasai, J.; Chanprasertyothin, S.; Kongtip, P.; Woskie, S. Genetic Polymorphisms of Pesticide-Metabolizing Enzymes and Transporters in Agricultural Workers and Thyroid Hormone Levels. Risk Manag. Healthc. Policy 2021, 14, 3435–3451. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, S.; Chang, J.; Pan, X.; Dong, F.; Li, Z.; Zheng, Y.; Li, Y. Insight into the uptake and metabolism of a new insecticide cyetpyrafen in plants. Environ. Int. 2022, 169, 107522. [Google Scholar] [CrossRef] [PubMed]
- Tungu, P.K.; Rowland, M.W.; Messenger, L.A.; Small, G.J.; Bradley, J.; Snetselaar, J.; Kirby, M.J.; Mbewe, N.J. Large-scale (Phase III) evaluation of broflanilide 50WP (VECTRON T500) for indoor residual spraying for malaria vector control in Northeast Tanzania: Study protocol for a two-arm, non-inferiority, cluster-randomised community trial. BMC Infect. Dis. 2022, 22, 171. [Google Scholar] [CrossRef] [PubMed]
- Kawahigashi, H. Transgenic plants for phytoremediation of herbicides. Curr. Opin. Biotechnol. 2009, 20, 225–230. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Raulli, R.; Laney, J.W.; Coley, W.; Walker, R.; O’Rourke, A.W.; Raine, K.; Horwith, G.; Gao, Y.; Eisnor, D.L.; et al. Systemic Bioavailability of Sublingual Atropine Ophthalmic Solution: A Phase I Study in Healthy Volunteers with Implications for Use as a Contingency Medical Countermeasure. J. Med. Toxicol. 2022, 18, 187–197. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, A.P.; Ma, L.Y.; Zhang, N.; Liu, J.; Yang, H. An ABCG-type transporter intensifies ametryn catabolism by Phase III reaction mechanism in rice. J. Hazard. Mater. 2023, 457, 131804. [Google Scholar] [CrossRef] [PubMed]
- Jie, C.Z.; Yan, Z.X.; Liu, J.; Zhang, N.; Yang, H. Detoxification and catabolism of mesotrione and fomesafen facilitated by a Phase II reaction acetyltransferase in rice. J. Adv. Res. 2023, 51, 1–11. [Google Scholar]
- Zhang, J.J.; Gao, S.; Xu, J.Y.; Lu, Y.C.; Lu, F.F.; Ma, L.Y.; Su, X.N.; Yang, H. Degrading and Phytoextracting Atrazine Residues in Rice (Oryza sativa) and Growth Media Intensified by a Phase II Mechanism Modulator. Environ. Sci. Technol. 2017, 51, 11258–11268. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.X.; Zhao, S.M.; Zhang, Y.Y.; Li, Y.J.; Shen, H.N.; Li, X.; Hou, B.K. A novel UDP-glycosyltransferase 91C1 confers specific herbicide resistance through detoxification reaction in Arabidopsis. Plant Physiol. Biochem. 2021, 159, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Chen, Q.; Chen, F.; Liu, H.; Lin, J.; Chen, R.; Ren, C.; Wei, J.; Zhang, Y.; Yang, F.; et al. Glutathione Promotes Degradation and Metabolism of Residual Fungicides by Inducing UDP-Glycosyltransferase Genes in Tomato. Front. Plant Sci. 2022, 13, 893508. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Edwards, R. Functional importance of the family 1 glucosyltransferase UGT72B1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J. 2005, 42, 556–566. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.Z.; Cao, Y.F. Apple Germplasm Resources Description Specification and Data Criteria; China Agricultural Press: Beijing, China, 2005. [Google Scholar]



| Chemical Compound | Structure Name | Structural Formula | Manufacturer | CAS |
|---|---|---|---|---|
| Bensulfuron methyl | [3-(4,6-dimethoxypyrimidin-2-yl)-1-(2-methoxycarbonylbenzyl)sulfonylurea] | 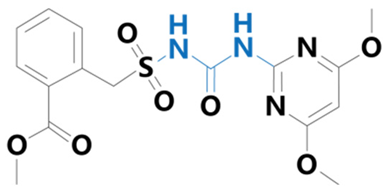 | Bermuda Biotechnology Co. (Nanjing, China) | 83055-99-6 |
| Nicosulfuron | [2-(4,6-dimethoxypyrimidin-2-ylcarbamoylsulfamoyl)-N,N-dimethylnicotinamide] |  | BioLabs Technology Ltd. (Beijing, China) | 111991-09-4 |
| Thifensulfuron | [3-(4-methoxy-6-methyl-1,3,5-triazin-2-ylaminocarbonylsulfamoyl)-thiophene-2-carboxylic acid] | 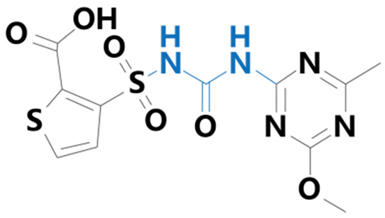 | Wittig Chemical Technology Co. (Zhengzhou, China) | 79277-67-1 |
| Chlorsulfuron | [1-(2-Chlorophenylsulfonyl)3-(4-methoxy-6-methyl-1,3,5-triazin-2-yl)urea] | 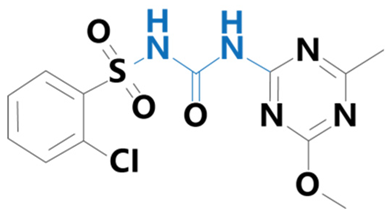 | BioLabs Technology Ltd. (Beijing, China) | 64902-72-3 |
| Metsulfuron | [3-(4-methoxy-6-methyl-1,3,5-triazin-2-yl)-1-(2-formylphenyl)sulfonylurea] |  | BioLabs Technology Ltd. (Beijing, China) | 142469-14-5 |
| Triasulfuron | [1-[2-(2-chloroethoxy)phenylsulfonyl]-3-(4-methoxy-6-methyl-1,3,5-triazin-2-yl)urea] | 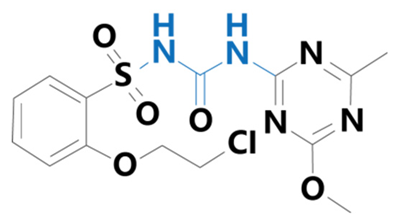 | Yuanye Biotechnology Co. (Shanghai, China) | 82097-50-5 |
| Cinosulfuron | [3-(4,6-dimethoxy-1,3,5-triazin-2-yl)-1-[2-(2-methoxyethoxy)phenyl]sulfonylurea] |  | BioLabs Technology Ltd. (Beijing, China) | 94593-91-6 |
| Chlorimuron-ethyl | [2-(4-chloro-6-methoxypyrimidin-2-ylcarbamoylsulfamoyl)benzoic acid] |  | BioLabs Technology Ltd. (Beijing, China) | 99283-00-8 |
| Sulfometuron | [2-(3-(4,6-dimethylpyrimidin-2-yl)ureasulfonyl)benzoic acid] |  | Eimage Technology Co. (Wuhan, China) | 74222-97-2 |
| Pyrazosulfuron | [5-(4,6-dimethoxypyrimidin-2-ylcarbamoylsulfamoyl)-1-methylpyrazole-4-carboxylic acid] | 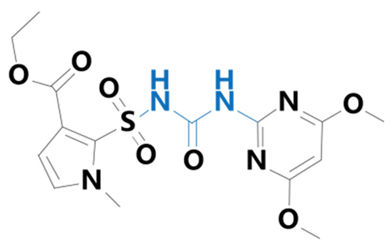 | Guyan Industry Co. (Shanghai, China) | 98389-04-9 |
| Rimsulfuron | [1-(4,6-dimethoxy-2-pyrimidinyl)-3-[3-(ethylsulfonyl)-2-pyridinesulfonyl]urea] |  | Yuanye Biotechnology Co. (Shanghai, China) | 122931-48-0 |
| Ethametsulfuron | [Methyl2-[(4-ethoxy-6-methylamino-1,3,5-triazin-2-yl)carbamoyl sulfamoyl]benzoate] | 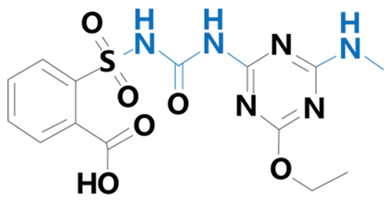 | Yuanye Biotechnology Co. (Shanghai, China) | 111353-84-5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhao, A.; Mu, L.; Teng, X.; Ma, Y.; Li, R.; Lei, K.; Ji, L.; Wang, X.; Li, P. First Clarification of the Involvement of Glycosyltransferase MdUGT73CG22 in the Detoxification Metabolism of Nicosulfuron in Apple. Plants 2024, 13, 1171. https://doi.org/10.3390/plants13091171
Zhang Y, Zhao A, Mu L, Teng X, Ma Y, Li R, Lei K, Ji L, Wang X, Li P. First Clarification of the Involvement of Glycosyltransferase MdUGT73CG22 in the Detoxification Metabolism of Nicosulfuron in Apple. Plants. 2024; 13(9):1171. https://doi.org/10.3390/plants13091171
Chicago/Turabian StyleZhang, Yuefeng, Aijuan Zhao, Lijun Mu, Xiao Teng, Yingxin Ma, Ru Li, Kang Lei, Lusha Ji, Xuekun Wang, and Pan Li. 2024. "First Clarification of the Involvement of Glycosyltransferase MdUGT73CG22 in the Detoxification Metabolism of Nicosulfuron in Apple" Plants 13, no. 9: 1171. https://doi.org/10.3390/plants13091171






