Genome-Wide Identification and Expression Analysis of Growth-Regulating Factors in Eucommia ulmoides Oliver (Du-Zhong)
Abstract
:1. Introduction
2. Materials and Methods
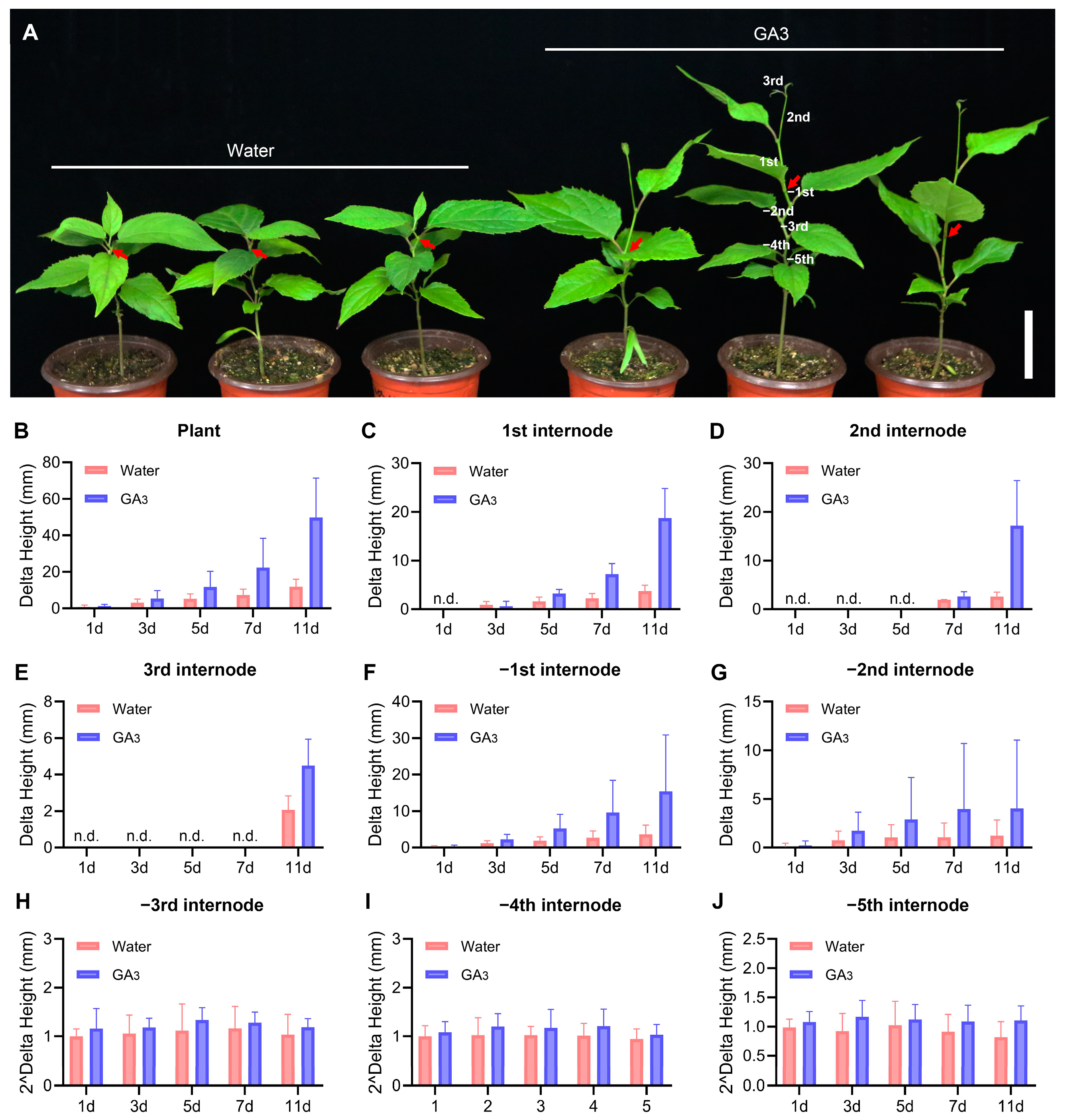
2.1. Plant Materials and Phytohormone Treatment
2.2. The Source of Original Data and Sequence Retrieval
2.3. The Identification of EuGRFs
2.4. The Identification of eul-miR396s
2.5. Bioinformatic Analysis of EuGRFs
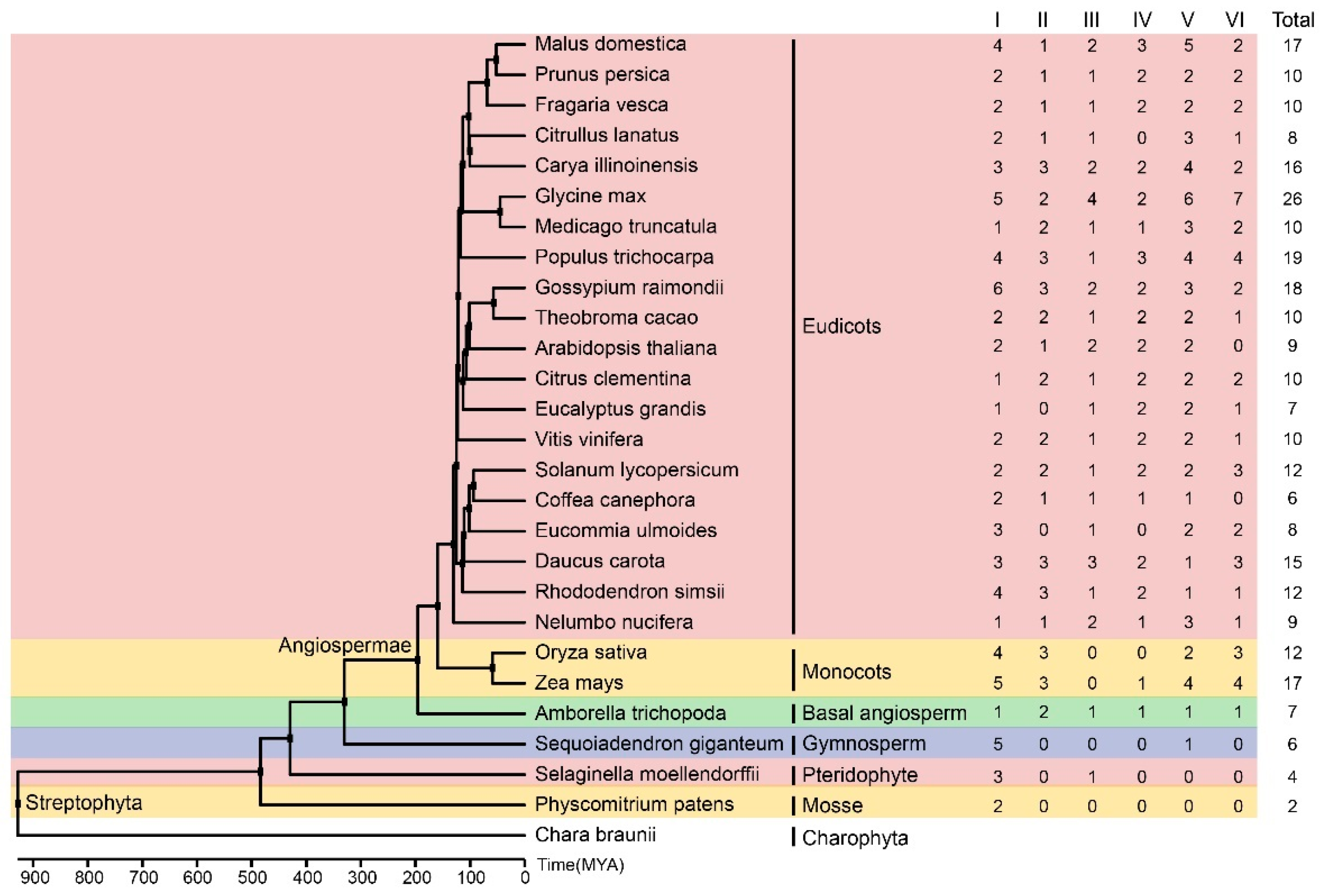
2.6. Phylogenetic Analysis of GRFs
2.7. Chromosomal Location and Collinearity Analysis
2.8. Total RNA Extraction and Real-Time Quantitative PCR
2.9. Construction of the Phylogenetic Tree of Plant Species
3. Results
3.1. The Characterization of EuGRFs in E. ulmoides
3.2. The Identification of miR396s in E. ulmoides and the Analysis of Their Target Binding Sites in the EuGRFs
3.3. Phylogenetic Analysis and the Classification of EuGRFs
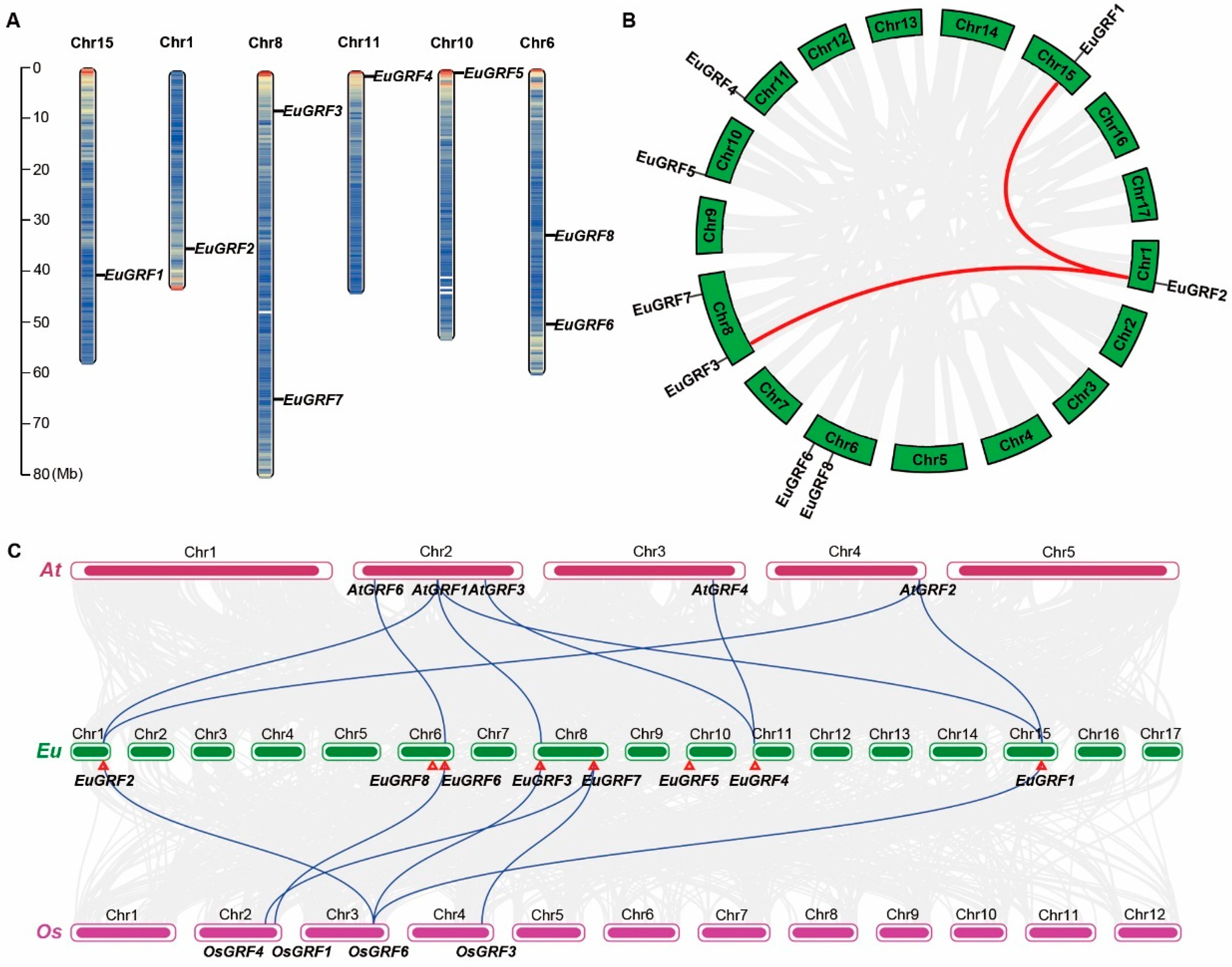
3.4. Chromosome Location and Gene Duplication of EuGRFs in E. ulmoides Genome
3.5. The Collinearity Analysis of GRFs in E. ulmoides, A. thaliana, and O. sativa
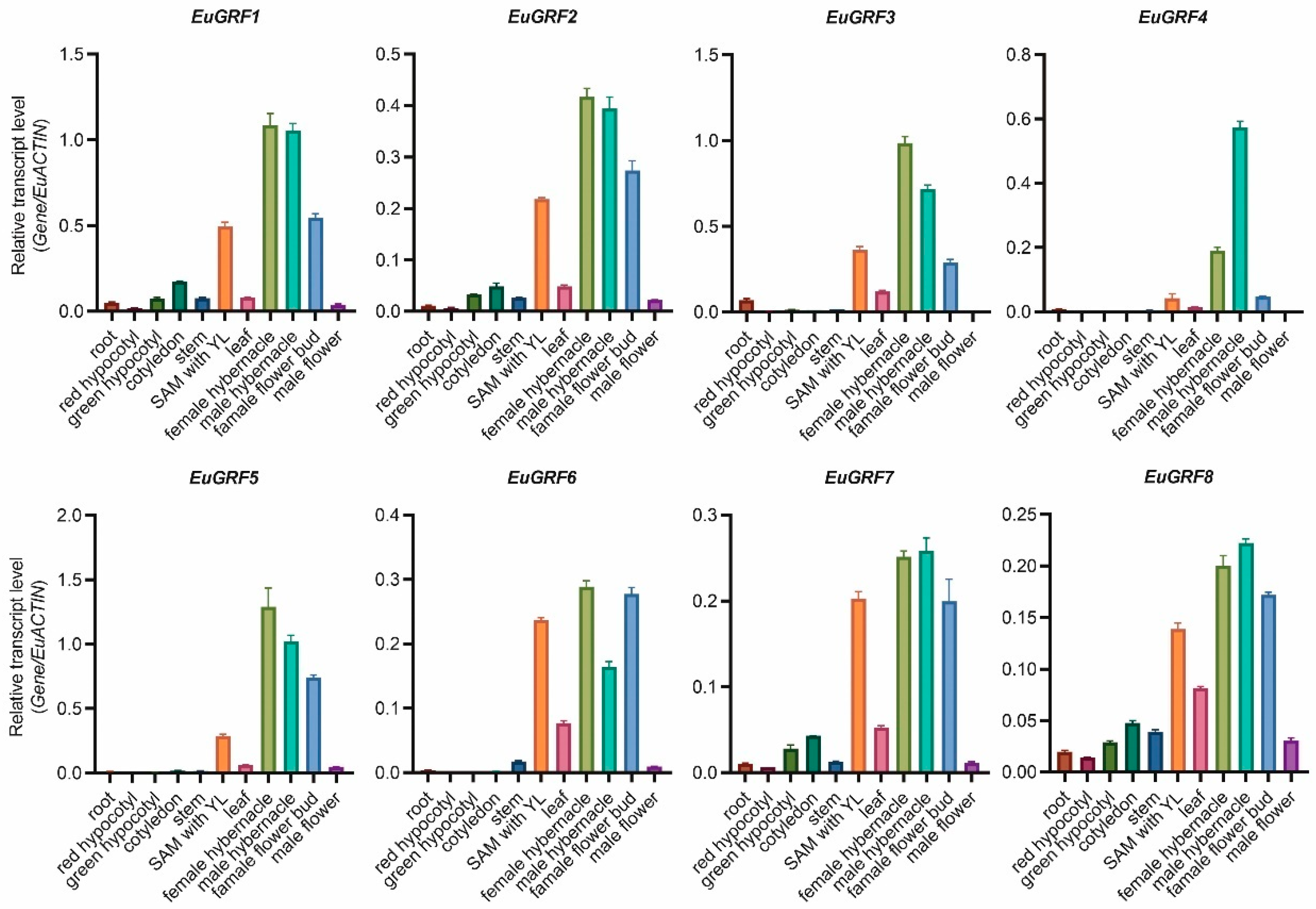
3.6. Expression Pattern Analysis of EuGRFs in Different Organs of E. ulmoides
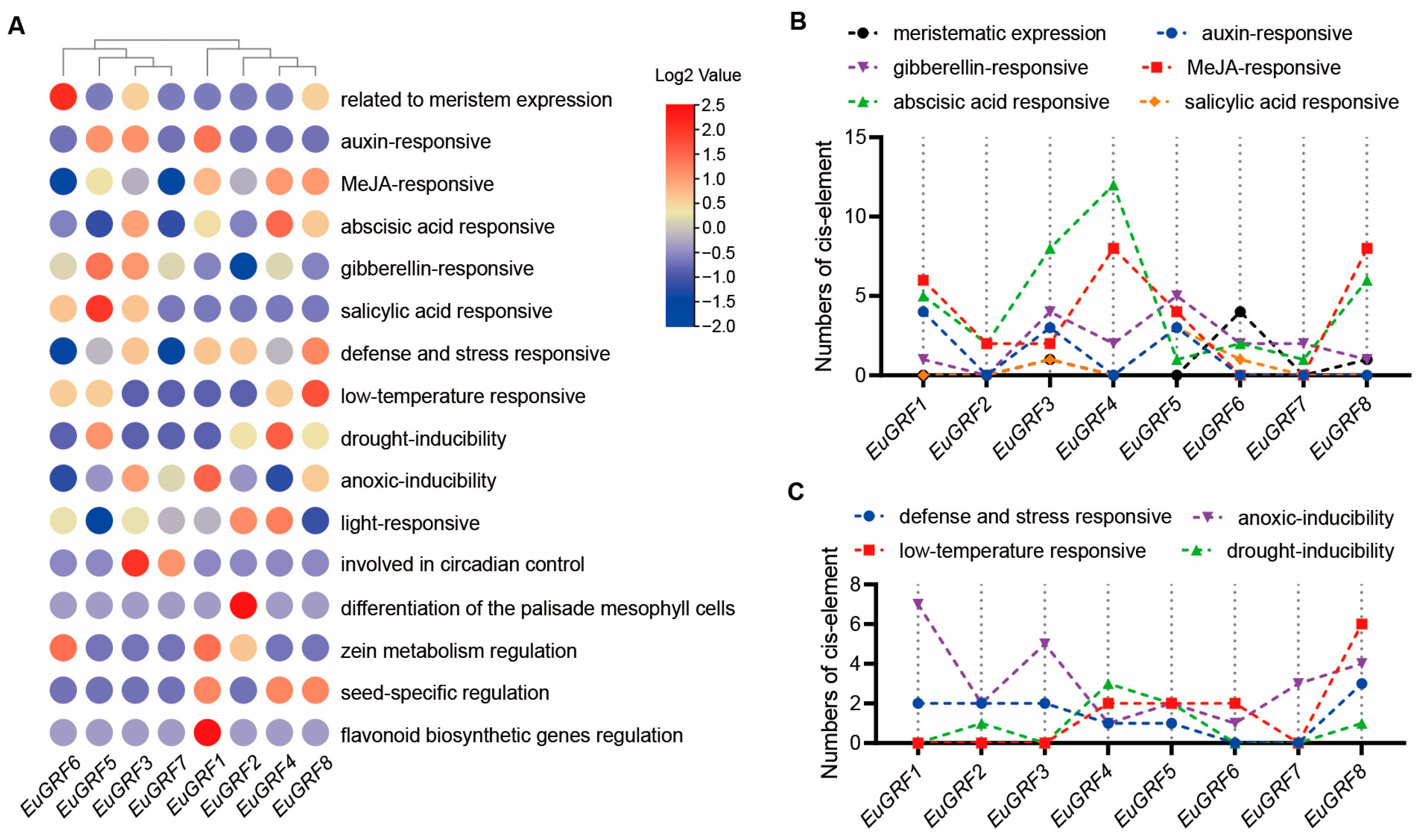
3.7. Cis-Element Analysis of EuGRFs
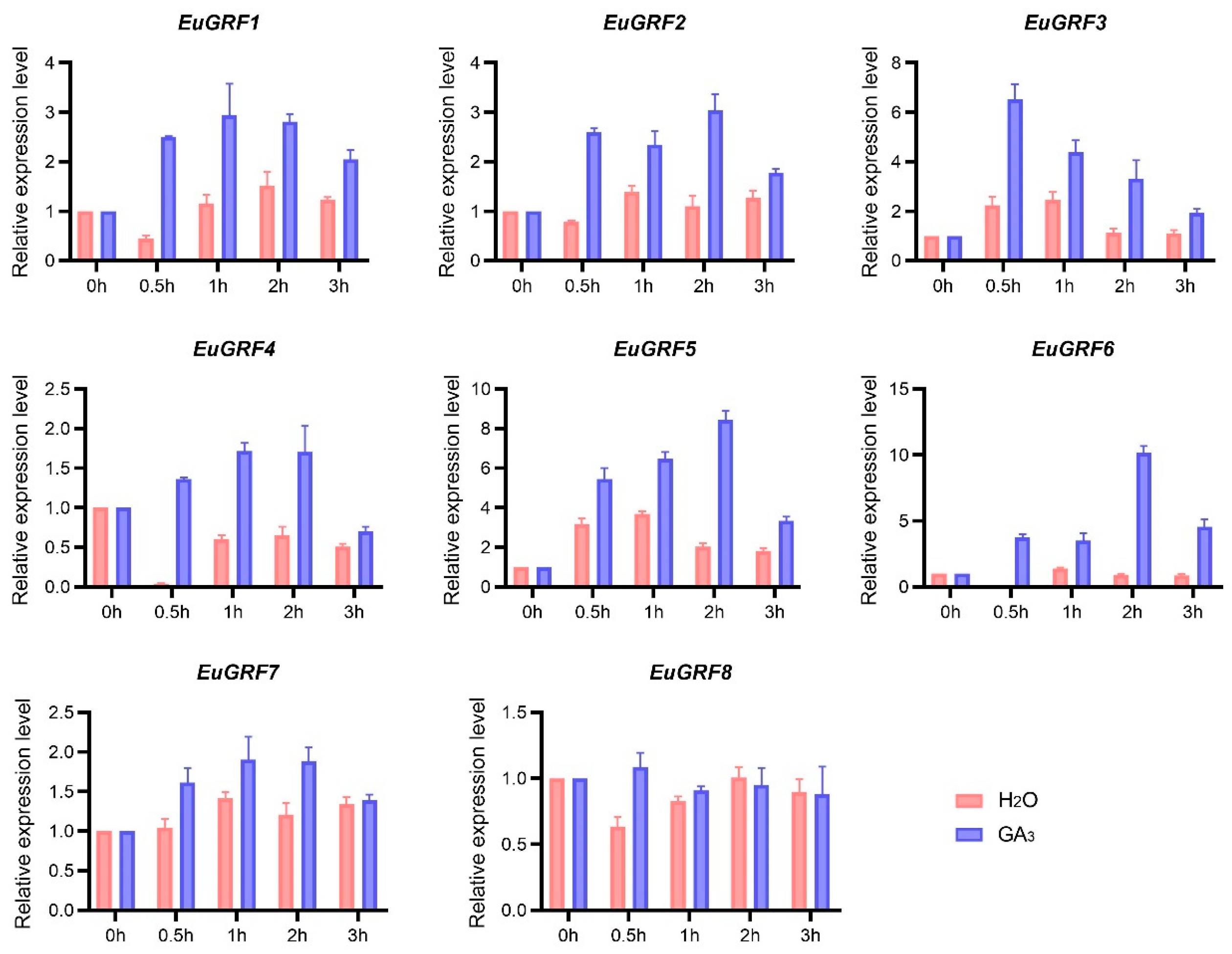
3.8. The Expression of EuGRFs in E. ulmoides Seedlings in Response to GA3 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Ercoli, M.F.; Debernardi, J.M.; Palatnik, J.F. Growth-Regulating Factors, A Transcription Factor Family Regulating More than Just Plant Growth. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016; pp. 269–280. [Google Scholar]
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tong, Y.; Luo, K.; Zhai, Z.; Liu, X.; Shi, Z.; Zhang, D.; Li, D. Identification of GROWTH-REGULATING FACTOR transcription factors in lettuce (Lactuca sativa) genome and functional analysis of LsaGRF5 in leaf size regulation. BMC Plant Biol. 2021, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, G.; Wang, Y.; Anwar, A.; He, B.; Zhang, J.; Chen, C.; Hao, Y.; Chen, R.; Song, S. Genome-wide identification of BcGRF genes in flowering Chinese cabbage and preliminary functional analysis of BcGRF8 in nitrogen metabolism. Front. Plant Sci. 2023, 14, 1144748. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gu, X.; Xu, D.; Wang, W.; Wang, H.; Zeng, M.; Chang, Z.; Huang, H.; Cui, X. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 2011, 62, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, S.; Xu, Y.; Li, C.; Zhang, Z.; Zhang, D.; Xu, S.; Zhang, C.; Chong, K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef]
- Gao, F.; Wang, K.; Liu, Y.; Chen, Y.; Chen, P.; Shi, Z.; Luo, J.; Jiang, D.; Fan, F.; Zhu, Y.; et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2015, 2, 15196. [Google Scholar] [CrossRef]
- Chen, F.; Yang, Y.; Luo, X.; Zhou, W.; Dai, Y.; Zheng, C.; Liu, W.; Yang, W.; Shu, K. Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qiu, N.; Ding, Q.; Li, J.; Zhang, Y.; Li, H.; Gao, J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2014, 15, 807. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Zhao, Y.; Sun, P.; Tang, F.; Song, X.; Lu, M.Z. Characterization of poplar growth-regulating factors and analysis of their function in leaf size control. BMC Plant Biol. 2020, 20, 509. [Google Scholar] [CrossRef]
- Wang, P.; Xiao, Y.; Yan, M.; Yan, Y.; Lei, X.; Di, P.; Wang, Y. Whole-genome identification and expression profiling of growth-regulating factor (GRF) and GRF-interacting factor (GIF) gene families in Panax ginseng. BMC Genom. 2023, 24, 334. [Google Scholar] [CrossRef]
- Du, F.; Guan, C.; Jiao, Y. Molecular Mechanisms of Leaf Morphogenesis. Mol Plant. 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.F.; Li, B.; Jia, G.Q.; Zhang, T.F.; Dai, J.R.; Li, J.S.; Wang, S.C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, B.H. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 2006, 49, 463–468. [Google Scholar] [CrossRef]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef]
- Nelissen, H.; Eeckhout, D.; Demuynck, K.; Persiau, G.; Walton, A.; van Bel, M.; Vervoort, M.; Candaele, J.; De Block, J.; Aesaert, S.; et al. Dynamic Changes in ANGUSTIFOLIA3 Complex Composition Reveal a Growth Regulatory Mechanism in the Maize Leaf. Plant Cell. 2015, 27, 1605–1619. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef]
- Pajoro, A.; Madrigal, P.; Muiño, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M.; et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, L.; Zheng, J.; Chen, F.; Wang, T.; Wang, M.; Tao, Y.; Wang, H.; Hong, Z.; Huang, Y.; et al. A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 2019, 70, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Zhao, Y.; Jiang, C.; Li, J.; Tang, F.; Liu, Y.; Zhao, S.; Hu, J.; Song, X.; et al. PagGRF12a interacts with PagGIF1b to regulate secondary xylem development through modulating PagXND1a expression in Populus alba × P. glandulosa. J. Integr. Plant Biol. 2021, 63, 1683–1694. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Wang, Q.; Lv, K.; Du, K.; Zhang, W.; Li, Q.; Kang, X.; Wei, H. Growth-regulating factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021, 230, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xie, J.; Wang, S.; Xu, W.; Chen, S.; Song, X.; Lu, M.; El-Kassaby, Y.A.; Zhang, D. Synonymous mutation in Growth Regulating Factor 15 of miR396a target sites enhances photosynthetic efficiency and heat tolerance in poplar. J. Exp. Bot. 2021, 72, 4502–4519. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Sun, Y.; El-Kassaby, Y.A.; Song, G.; Mi, Y.; Han, J.; Li, Y. PagGRF11 Overexpression Promotes Stem Development and Dwarfing in Populus. Int. J. Mol. Sci. 2022, 23, 7858. [Google Scholar] [CrossRef]
- Zuo, X.; Xiang, W.; Li, K.; Liu, Y.; Zheng, S.; Khan, A.; Zhang, D. MdGRF11, a growth-regulating factor, participates in the regulation of flowering time and interacts with MdTFL1/MdFT1 in apple. Plant Sci. 2022, 321, 111339. [Google Scholar] [CrossRef]
- Metcalfe, C.R. Distribution of latex in the plant kingdom. Econ. Bot. 1966, 21, 115–127. [Google Scholar] [CrossRef]
- Sando, T.; Hayashi, T.; Takeda, T.; Akiyama, Y.; Nakazawa, Y.; Fukusaki, E.; Kobayashi, A. Histochemical study of detailed laticifer structure and rubber biosynthesis-related protein localization in Hevea brasiliensis using spectral confocal laser scanning microscopy. Planta 2009, 230, 215–225. [Google Scholar] [CrossRef]
- Wuyun, T.N.; Wang, L.; Liu, H.; Wang, X.; Zhang, L.; Bennetzen, J.L.; Li, T.; Yang, L.; Liu, P.; Du, L.; et al. The Hardy Rubber Tree Genome Provides Insights into the Evolution of Polyisoprene Biosynthesis. Mol. Plant 2018, 11, 429–442. [Google Scholar] [CrossRef]
- Wei, X.; Peng, P.; Peng, F.; Dong, J. Natural Polymer Eucommia Ulmoides Rubber: A Novel Material. J. Agric. Food Chem. 2021, 69, 3797–3821. [Google Scholar] [CrossRef]
- Ran, X.; Liu, Y.; Zhao, D. The relationship between EuSRPP1 gene expression and rubber biosynthesis in Eucommia ulmoides Oliver (Du-zhong). Ind. Crops Prod. 2022, 175, 114246. [Google Scholar] [CrossRef]
- Zhou, H.; Song, X.; Wei, K.; Zhao, Y.; Jiang, C.; Wang, J.; Tang, F.; Lu, M. Growth-regulating factor 15 is required for leaf size control in Populus. Tree Physiol. 2019, 39, 381–390. [Google Scholar] [CrossRef]
- Van Bel, M.; Silvestri, F.; Weitz, E.M.; Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K. PLAZA 5.0: Extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res. 2022, 50, D1468–D1474. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Millar, A.A.; Waterhouse, P.M. Plant and animal microRNAs: Similarities and differences. Funct. Integr. Genom. 2005, 5, 129–135. [Google Scholar] [CrossRef]
- Li, Y.; Wei, H.; Yang, J.; Du, K.; Li, J.; Zhang, Y.; Qiu, T.; Liu, Z.; Ren, Y.; Song, L.; et al. High-quality de novo assembly of the Eucommia ulmoides haploid genome provides new insights into evolution and rubber biosynthesis. Hortic. Res. 2020, 7, 183. [Google Scholar] [CrossRef]
- Zhong, B.; Xi, Z.; Goremykin, V.V.; Fong, R.; McLenachan, P.A.; Novis, P.M.; Davis, C.C.; Penny, D. Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol. Biol. Evol. 2014, 31, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304.e15. [Google Scholar] [CrossRef]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of Subaerial Zygnematophyceae Provide Insights into Land Plant Evolution. Cell 2019, 179, 1057–1067.e14. [Google Scholar] [CrossRef]
- Liang, Z.; Geng, Y.; Ji, C.; Du, H.; Wong, C.E.; Zhang, Q.; Zhang, Y.; Zhang, P.; Riaz, A.; Chachar, S.; et al. Mesostigma viride Genome and Transcriptome Provide Insights into the Origin and Evolution of Streptophyta. Adv. Sci. 2019, 7, 1901850. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Li, H.; Sahu, S.K.; Wang, H.; Xu, Y.; Xian, W.; Song, B.; Liang, H.; Cheng, S.; et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2019, 6, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef]
- Qin, L.; Chen, H.; Wu, Q.; Wang, X. Identification and exploration of the GRF and GIF families in maize and foxtail millet. Physiol. Mol. Biol. Plants 2022, 28, 1717–1735. [Google Scholar] [CrossRef]


| Protein ID | Arabidopsis Homolog | Protein Length | Molecular Weight | Theoretical pI | Subcellular Localization | |
|---|---|---|---|---|---|---|
| EuGRF1 | GWHPBISF008846 | AtGRF1, 2 | 498 | 53,478.65 | 8.14 | Nucleus |
| EuGRF2 | GWHPBISF000663 | AtGRF1, 2 | 402 | 44,614.49 | 9.31 | Nucleus |
| EuGRF3 | GWHPBISF020960 | AtGRF1, 2 | 422 | 45,306.59 | 8.27 | Nucleus |
| EuGRF4 | GWHPBISF002516 | AtGRF3, 4 | 324 | 34,645.58 | 8.78 | Nucleus |
| EuGRF5 | GWHPBISF001116 | AtGRF5, 6 | 304 | 33,933.86 | 8.24 | Nucleus |
| EuGRF6 | GWHPBISF018630 | AtGRF5, 6 | 351 | 39,978.65 | 8.42 | Nucleus |
| EuGRF7 | GWHPBISF022031 | — | 312 | 34,787.05 | 8.65 | Nucleus |
| EuGRF8 | GWHPBISF018381 | — | 344 | 38,022.43 | 8.34 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Zhu, Y.; Zhao, D. Genome-Wide Identification and Expression Analysis of Growth-Regulating Factors in Eucommia ulmoides Oliver (Du-Zhong). Plants 2024, 13, 1185. https://doi.org/10.3390/plants13091185
Wang R, Zhu Y, Zhao D. Genome-Wide Identification and Expression Analysis of Growth-Regulating Factors in Eucommia ulmoides Oliver (Du-Zhong). Plants. 2024; 13(9):1185. https://doi.org/10.3390/plants13091185
Chicago/Turabian StyleWang, Ruoruo, Ying Zhu, and Degang Zhao. 2024. "Genome-Wide Identification and Expression Analysis of Growth-Regulating Factors in Eucommia ulmoides Oliver (Du-Zhong)" Plants 13, no. 9: 1185. https://doi.org/10.3390/plants13091185





