Synthesis and Functions of Jasmonates in Maize
Abstract
:1. Importance of Maize as a Crop and a Genetic Model
2. Jasmonates Belong to Oxylipins, a Group of Signals Better Understood in Mammals
3. Plant Oxylipins
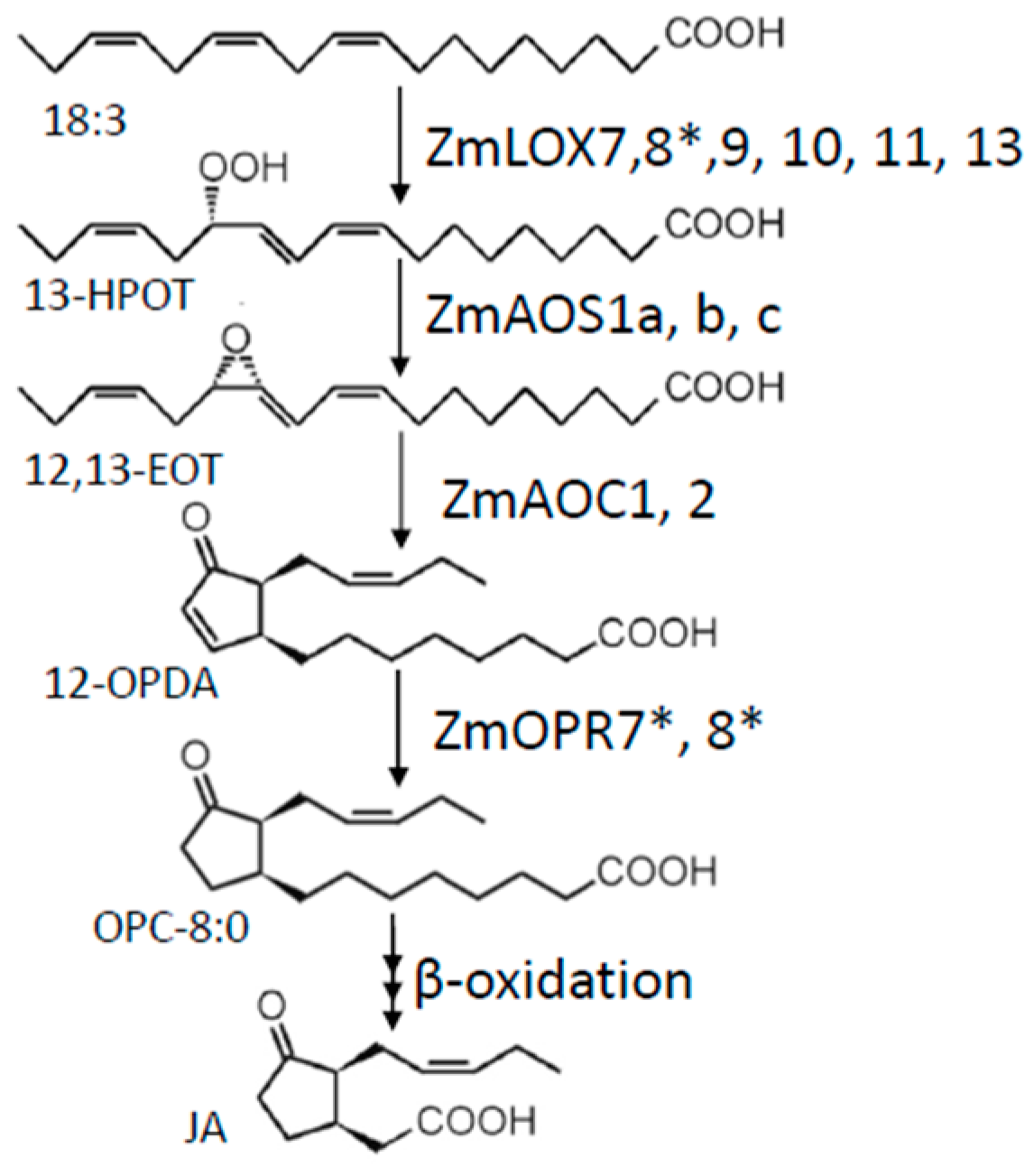
4. Jasmonate Biosynthesis in Maize
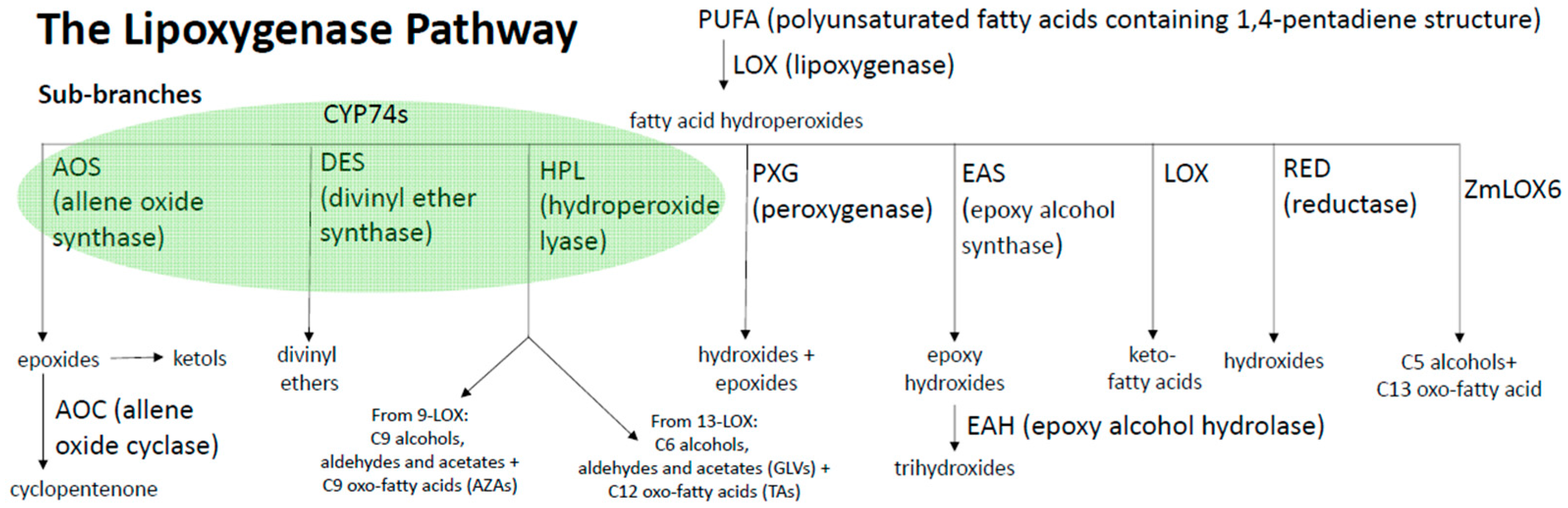
5. 13-Lipoxygenases
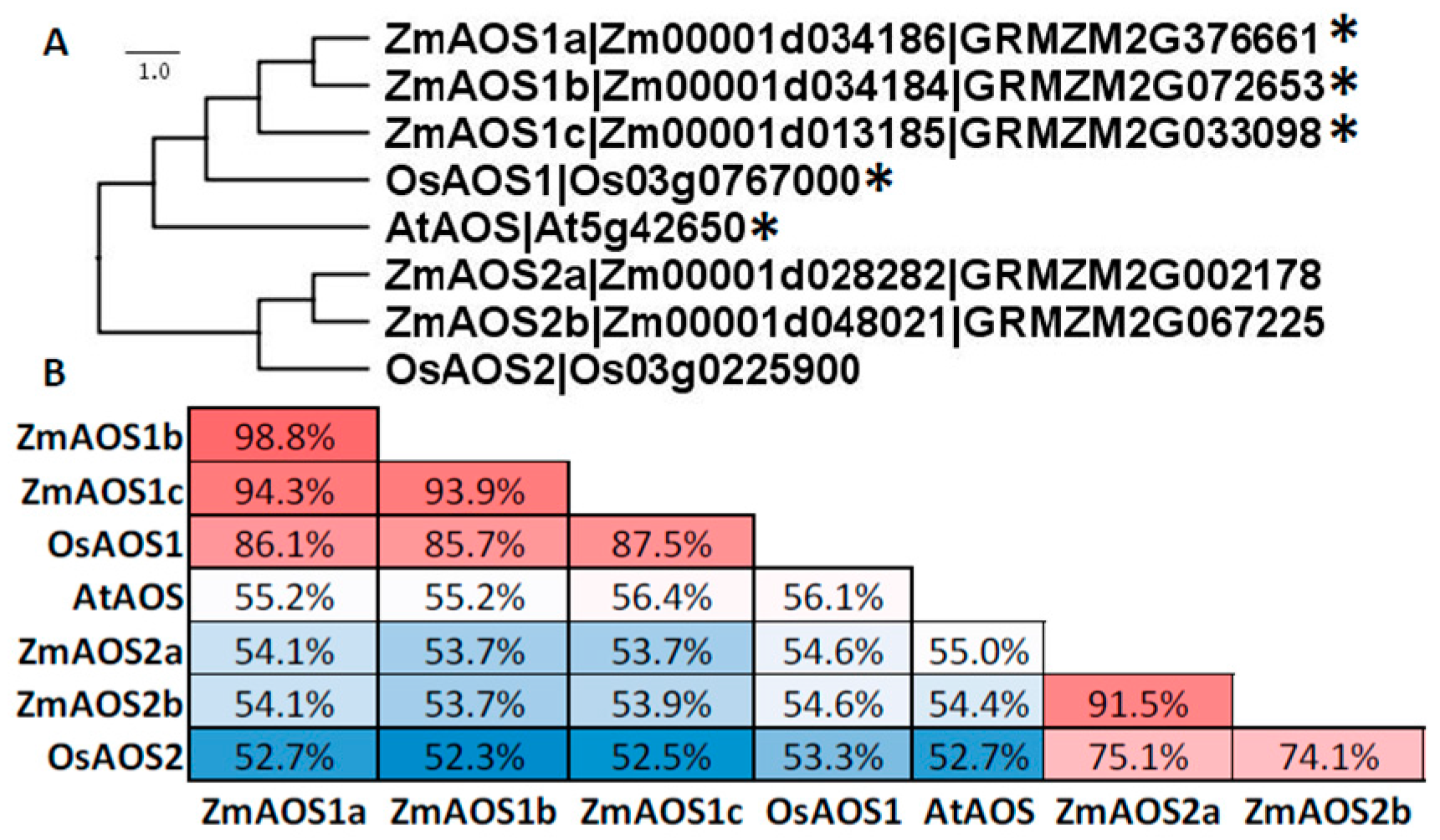
6. Allene Oxide Synthase
7. Allene Oxide Cyclase
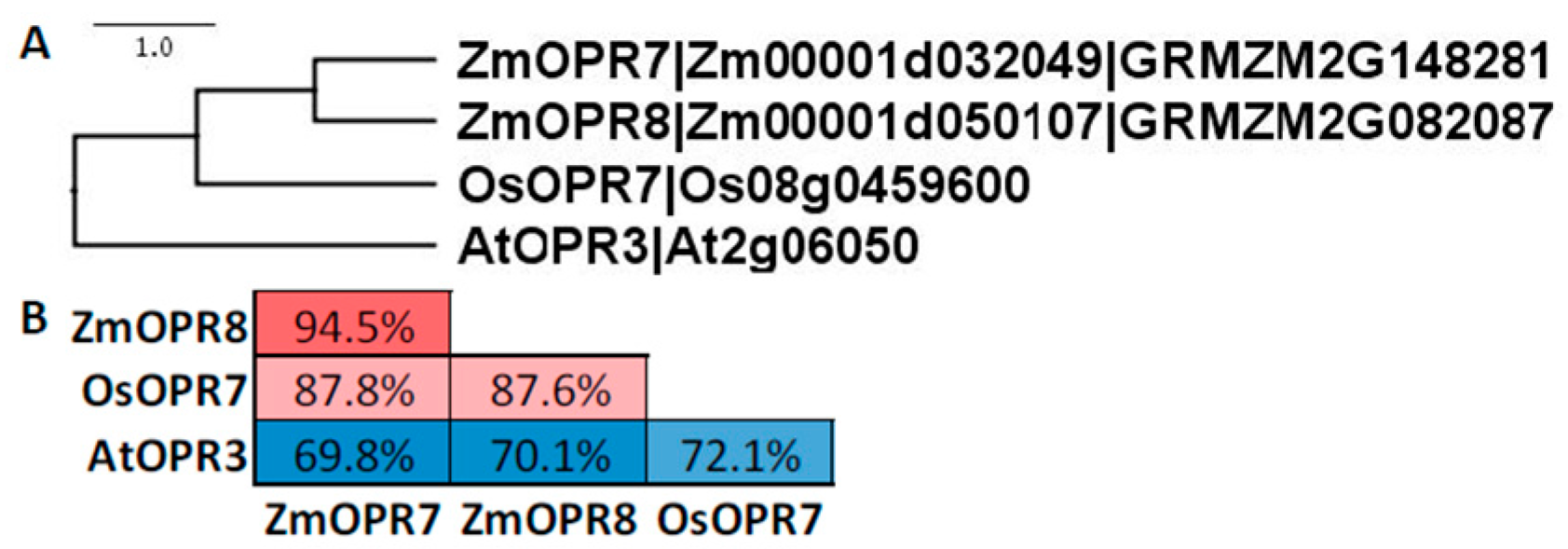
8. Oxo-Phytodienoic Acid Reductase and Beta-Oxidation
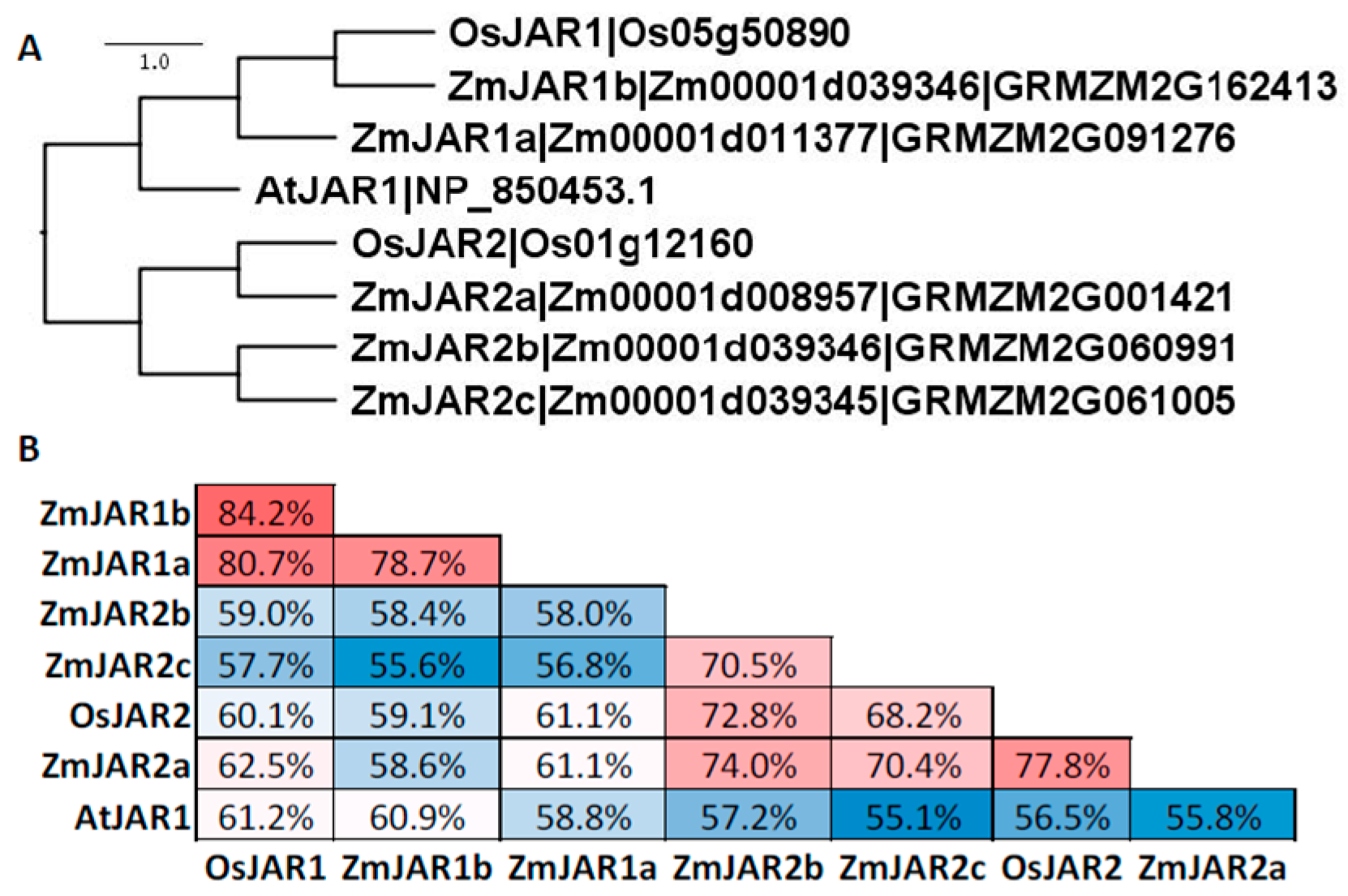
9. JA-Amino Acid Conjugation
10. 9-Oxylipin Jasmonate Analogs (10-OPEA, 10-OPDA and Derivatives)
11. Regulation of JA Production by the Non-JA Producing Branches of the LOX Pathway
11.1. The Role of GLV-Mediated Signaling on JA Production
11.2. The Role of 9-LOX-Mediated Signaling on JA Production
12. JA in Maize Defense against Insects
12.1. Plant Defenses, Genetic Evidence and Insect Elicitors
12.2. Direct Defenses
12.3. Indirect Defenses
13. JA in Maize Pathogen and Nematode Defense
14. JA in Maize Symbiosis
15. JA in Maize Growth, Development and Senescence
16. JA in Maize Anthocyanin Pigmentation and Photomorphogenesis
17. Maize Genes Regulated by JA
18. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Awika, J.M. Major cereal grains production and use around the world. Adv. Cereal Sci. Implic. Food Proc. Health Promot. 2011, 1089, 1–13. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Food Outlook: Biannual Report of Global Food Markets; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Vielle-Calzada, J.P.; de la Vega, O.M.; Hernandez-Guzman, G.; Ibarra-Laclette, E.; Alvarez-Mejia, C.; Vega-Arreguin, J.C.; Jimenez-Moraila, B.; Fernandez-Cortes, A.; Corona-Armenta, G.; Herrera-Estrella, L.; et al. The palomero genome suggests metal effects on domestication. Science 2009. [Google Scholar] [CrossRef] [PubMed]
- Andorf, C.M.; Cannon, E.K.; Portwood, J.L., 2nd; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Braun, B.L.; Campbell, D.A.; Vinnakota, A.G.; Sribalusu, V.V.; et al. Maizegdb update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016, 44, D1195–D1201. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.A. Evolutionary history of the grasses. Plant Physiol. 2001, 125, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Gowik, U.; Westhoff, P. The path from C3 to C4 photosynthesis. Plant Physiol. 2011, 155, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome sequencing and analysis of the model grass brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Brutnell, T.P.; Bennetzen, J.L.; Vogel, J.P. Brachypodium distachyon and Setaria viridis: Model genetic systems for the grasses. Annu. Rev. Plant Biol. 2015, 66, 465–485. [Google Scholar] [CrossRef] [PubMed]
- Estep, M.C.; McKain, M.R.; Vela Diaz, D.; Zhong, J.; Hodge, J.G.; Hodkinson, T.R.; Layton, D.J.; Malcomber, S.T.; Pasquet, R.; Kellogg, E.A. Allopolyploidy, diversification, and the miocene grassland expansion. Proc. Natl. Acad. Sci. USA 2014, 111, 15149–15154. [Google Scholar] [CrossRef] [PubMed]
- Buczynski, M.W.; Dumlao, D.S.; Dennis, E.A. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009, 50, 1505. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 2001, 294, 1871–1875. [Google Scholar] [CrossRef] [PubMed]
- Prost, I.; Dhondt, S.; Rothe, G.; Vicente, J.; Rodriguez, M.J.; Kift, N.; Carbonne, F.; Griffiths, G.; Esquerre-Tugaye, M.T.; Rosahl, S.; et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005, 139, 1902–1913. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.; Luan, S.; et al. Cyclophilin 20–3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [PubMed]
- v. Euler, U.S. Über die spezifische blutdrucksenkende substanz des menschlichen prostata-und samenblasensekretes. J. Mol. Med. 1935, 14, 1182–1183. [Google Scholar] [CrossRef]
- Goldblatt, M.W. Properties of human seminal plasma. J. Physiol. 1935, 84, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. How jasmonates earned their laurels: Past and present. Plant Growth Regul. 2015, 34, 761–794. [Google Scholar] [CrossRef]
- Corey, E.J.; Weinshenker, N.M.; Schaaf, T.K.; Huber, W. Stereo-controlled synthesis of DL-prostaglandins F2α and E2. J. Am. Chem. Soc 1969, 91, 5675–5677. [Google Scholar] [CrossRef] [PubMed]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Pountos, I.; Georgouli, T.; Bird, H.; Giannoudis, P.V. Nonsteroidal anti-inflammatory drugs: Prostaglandins, indications, and side effects. Int. J. Infereron Cytokine Mediator Res. 2011, 3, 19–27. [Google Scholar] [CrossRef]
- Vane, J.R. Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat.-New Biol. 1971, 231, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Meirer, K.; Steinhilber, D.; Proschak, E. Inhibitors of the arachidonic acid cascade: Interfering with multiple pathways. Basic Clin. Pharmacol. Toxicol. 2014, 114, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Goppelt-Struebe, M.; Wolter, D.; Resch, K. Glucocorticoids inhibit prostaglandin synthesis not only at the level of phospholipase A2 but also at the level of cyclo-oxygenase/pge isomerase. Br. J. Pharmacol. 1989, 98, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Laine, L. Approaches to nonsteroidal anti-inflammatory drug use in the high-risk patient. Gastroenterology 2001, 120, 594–606. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. National Drug Code Directory. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ucm142438.htm (accessed on13 October 2016).
- Andreou, A.; Brodhun, F.; Feussner, I. Biosynthesis of oxylipins in non-mammals. Prog. Lipid Res. 2009, 48, 148–170. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, M.; Ponce de Leon, I.; Sanz, A.; Castresana, C. Fatty acid alpha-dioxygenases. Prostaglandins Other Lipid Mediat. 2002, 68–69, 363–374. [Google Scholar] [CrossRef]
- Mueller, M.J. Archetype signals in plants: The phytoprostanes. Curr. Opin. Plant Biol. 2004, 7, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Cuyamendous, C.; Leung, K.S.; Durand, T.; Lee, J.C.; Oger, C.; Galano, J.M. Synthesis and discovery of phytofurans: Metabolites of alpha-linolenic acid peroxidation. Chem. Commun. (Camb.) 2015, 51, 15696–15699. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, A. Oxylipin Profiling Database. Available online: http://www.oxylipins.uni-goettingen.de/path.php?path=Pathway&caption=Oxylipin%20Pathway (accessed on 15 November 2016).
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in annals of botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Miersch, O.; Bohlmann, H.; Wasternack, C. Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 1999, 50, 517–523. [Google Scholar] [CrossRef]
- Miersch, O.; Bruckner, B.; Schmidt, J.; Sembdner, G. Cyclopentane fatty-acids from Gibberella fujikuroi. Phytochemistry 1992, 31, 3835–3837. [Google Scholar] [CrossRef]
- Miersch, O.; Günther, T.; Fritsche, W.; Sembdner, G. Jasmonates from different fungal species. Nat. Prod. Lett. 1993, 2, 293–299. [Google Scholar] [CrossRef]
- Tsukada, K.; Takahashi, K.; Nabeta, K. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry 2010, 71, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Borrego, E.J.; Kolomiets, M.V. Lipid-mediated signaling between fungi and plants. In Biocommunication of Fungi; Springer: New York, NY, USA, 2012; pp. 249–260. [Google Scholar]
- Russell, W. Registration of B70 and B73 parental lines of maize (reg. Nos. Pl16 and pl17). Crop Sci. 1972, 12, 721. [Google Scholar] [CrossRef]
- De La Fuente, G.N.; Murray, S.C.; Isakeit, T.; Park, Y.S.; Yan, Y.; Warburton, M.L.; Kolomiets, M.V. Characterization of genetic diversity and linkage disequilibrium of ZmLOX4 and ZmLOX5 loci in maize. PLoS ONE 2013, 8, e53973. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Borrego, E.; Kolomiets, M.V. Jasmonate Biosynthesis, Perception and Function in Plant Development and Stress Responses; InTech Open Access Publisher: Rijeka, Croatia, 2013. [Google Scholar]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.; Kolomiets, M.V. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, M.E.; Ghars, M.A.; Frettinger, P.; Perez-Alfocea, F.; Lutts, S.; Wathelet, J.P.; du Jardin, P.; Fauconnier, M.L. Organ-dependent oxylipin signature in leaves and roots of salinized tomato plants (Solanum lycopersicum). J. Plant Physiol. 2012, 169, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Stumpe, M.; Feussner, I.; Kolomiets, M. A novel plastidial lipoxygenase of maize (Zea mays) ZmLOX6 encodes for a fatty acid hydroperoxide lyase and is uniquely regulated by phytohormones and pathogen infection. Planta 2008, 227, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Kolomiets, M.V. The lipid language of plant-fungal interactions. Fungal Genet. Biol. 2011, 48, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Kunze, S.; Ni, X.; Feussner, I.; Kolomiets, M.V. Comparative molecular and biochemical characterization of segmentally duplicated 9-lipoxygenase genes ZmLOX4 and ZmLOX5 of maize. Planta 2010, 231, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Nemchenko, A.; Borrego, E.; Murray, I.; Sobhy, I.S.; Bosak, L.; DeBlasio, S.; Erb, M.; Robert, C.A.; Vaughn, K.A.; et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013, 74, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Umate, P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011, 6, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Nemchenko, A.; Kunze, S.; Feussner, I.; Kolomiets, M. Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 2006, 57, 3767–3779. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Luo, A.; Zadrozny, T.; Sylvester, A.; Jackson, D. Fluorescent protein marker lines in maize: Generation and applications. Int. J. Dev. Biol. 2013, 57, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Schiller, D.; Contreras, C.; Vogt, J.; Dunemann, F.; Defilippi, B.G.; Beaudry, R.; Schwab, W. A dual positional specific lipoxygenase functions in the generation of flavor compounds during climacteric ripening of apple. Hortic. Res. 2015, 2, Article 15003. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.K.; West, S.I.; Hornostaj, A.R.; Lawson, D.M.; Fairhurst, S.A.; Sanchez, R.O.; Hough, P.; Robinson, B.H.; Casey, R. Probing a novel potato lipoxygenase with dual positional specificity reveals primary determinants of substrate binding and requirements for a surface hydrophobic loop and has implications for the role of lipoxygenases in tubers. Biochem. J. 2001, 353, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Palmieri-Thiers, C.; Canaan, S.; Brunini, V.; Lorenzi, V.; Tomi, F.; Desseyn, J.L.; Garscha, U.; Oliw, E.H.; Berti, L.; Maury, J. A lipoxygenase with dual positional specificity is expressed in olives (Olea europaea L.) during ripening. Biochim. Biophys. Acta 2009, 1791, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Shen, W.; Liu, L.; Jiang, L.; Liu, Y.; Su, N.; Wan, J. A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol. Biol. 2008, 66, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Choi, E.; Kim, Y.; Cho, K.; Lee, A.; Shim, J.; Rakwal, R.; Agrawal, G.K.; Han, O. Dual positional specificity and expression of non-traditional lipoxygenase induced by wounding and methyl jasmonate in maize seedlings. Plant Mol. Biol. 2003, 52, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Von Heijne, G. Chlorop, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Han, Y.; Woo, J.C.; Baudisch, B.; Klosgen, R.B.; Oh, S.; Han, J.; Han, O. Cellular localization of dual positional specific maize lipoxygenase-1 in transgenic rice and calcium-mediated membrane association. Plant Sci. 2011, 181, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using CAS9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Itoh, A.; Schilmiller, A.L.; McCaig, B.C.; Howe, G.A. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. J. Biol. Chem. 2002, 277, 46051–46058. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Nioche, P.; Hamberg, M.; Raman, C.S. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 2008, 455, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T. Maize death acids, 9-lipoxygenase–derived cyclopente (a) nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef] [PubMed]
- Maucher, H.; Hause, B.; Feussner, I.; Ziegler, J.; Wasternack, C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome): Tissue specific regulation in seedling development. Plant J. 2000, 21, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Iino, M. Phytochrome-mediated transcriptional up-regulation of allene oxide synthase in rice seedlings. Plant Cell Physiol. 2004, 45, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Takano, M.; Neumann, R.; Iino, M. The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 2005, 17, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Raman, G.; Walley, J.W.; Perea, J.V.; Banu, G.; Theg, S.; Dehesh, K. Rice hydroperoxide lyases with unique expression patterns generate distinct aldehyde signatures in Arabidopsis. Plant Physiol. 2006, 141, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Yoeun, S.; Rakwal, R.; Han, O. Dual positional substrate specificity of rice allene oxide synthase-1: Insight into mechanism of inhibition by type П ligand imidazole. BMB Rep. 2013, 46, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Ogorodnikova, A.V.; Gorina, S.S.; Mukhtarova, L.S.; Mukhitova, F.K.; Toporkova, Y.Y.; Hamberg, M.; Grechkin, A.N. Stereospecific biosynthesis of (9S,13S)-10-oxophytoenoic acid in young maize roots. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 185, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.K.; Kang, H.G.; Kim, G.J.; Eu, H.J.; Oh, S.A.; Song, J.T.; Chung, I.K.; Eun, M.Y.; Park, S.K. Transgenic rice plants carrying RNA interference constructs of AOS (allene oxide synthase) genes show severe male sterility. Plant Breed. 2010, 129, 647–651. [Google Scholar] [CrossRef]
- Christensen, S.A.; Nemchenko, A.; Park, Y.S.; Borrego, E.; Huang, P.C.; Schmelz, E.A.; Kunze, S.; Feussner, I.; Yalpani, N.; Meeley, R.; et al. The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against Fusarium verticillioides in maize. Mol. Plant Microbe Interact. 2014, 27, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Starr, J.; Gobel, C.; Engelberth, J.; Feussner, I.; Tumlinson, J.; Kolomiets, M. Maize 9-lipoxygenase ZMLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant Microbe Interact. 2008, 21, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Engelberth, J.; Seidl-Adams, I.; Schultz, J.C.; Tumlinson, J.H. Insect elicitors and exposure to green leafy volatiles differentially upregulate major octadecanoids and transcripts of 12-oxo phytodienoic acid reductases in Zea mays. Mol. Plant Microbe Interact. 2007, 20, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Dafoe, N.J.; Thomas, J.D.; Shirk, P.D.; Legaspi, M.E.; Vaughan, M.M.; Huffaker, A.; Teal, P.E.; Schmelz, E.A. European corn borer (Ostrinia nubilalis) induced responses enhance susceptibility in maize. PLoS ONE 2013, 8, e73394. [Google Scholar] [CrossRef] [PubMed]
- Engelberth, J.; Contreras, C.F.; Viswanathan, S. Transcriptional analysis of distant signaling induced by insect elicitors and mechanical wounding in Zea mays. PLoS ONE 2012, 7, e34855. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Fokar, M.; Abd, H.; Zhang, H.; Allen, R.D.; Pare, P.W. (Z)-3-hexenol induces defense genes and downstream metabolites in maize. Planta 2005, 220, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.; Zerbe, P.; Schaller, F. The crystal structure of Arabidopsis thaliana allene oxide cyclase: Insights into the oxylipin cyclization reaction. Plant Cell 2006, 18, 3201–3217. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Kombrink, E. Jasmonates: Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 2010, 5, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. Allene oxide cyclase (AOC) gene family members of Arabidopsis thaliana: Tissue- and organ-specific promoter activities and in vivo heteromerization. J. Exp. Bot. 2012, 63, 6125–6138. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, J.; Wasternack, C.; Hamberg, M. On the specificity of allene oxide cyclase. Lipids 1999, 34, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Muller, A.; Korte, A.; Furuya, M.; Weiler, E.W.; Nick, P. Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 2003, 133, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M.G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S.Y.; Nadal, M.; et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Trengove, R.D. A compound from smoke that promotes seed germination. Science 2004, 305, 977. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, K.J.; Albani, M.C.; James, G.V.; Gutjahr, C.; Hartwig, B.; Turck, F.; Paszkowski, U.; Coupland, G.; Schneeberger, K. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 2013, 31, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased tolerance to salt stress in OPDA-deficient rice allene oxide cyclase mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 2015, 66, 3339–3352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Simmons, C.; Yalpani, N.; Crane, V.; Wilkinson, H.; Kolomiets, M. Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol. Biol. 2005, 59, 323–343. [Google Scholar] [CrossRef] [PubMed]
- Tani, T.; Sobajima, H.; Okada, K.; Chujo, T.; Arimura, S.; Tsutsumi, N.; Nishimura, M.; Seto, H.; Nojiri, H.; Yamane, H. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 2008, 227, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Schaller, F.; Biesgen, C.; Mussig, C.; Altmann, T.; Weiler, E.W. 12-oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 2000, 210, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, M.; Gardner, H.W. Oxylipin pathway to jasmonates: Biochemistry and biological significance. Biochim. Biophys. Acta 1992, 1165, 1–18. [Google Scholar] [CrossRef]
- Miersch, O.; Kramell, R.; Parthier, B.; Wasternack, C. Structure-activity relations of substituted, deleted or stereospecifically altered jasmonic acid in gene expression of barley leaves. Phytochemistry 1999, 50, 353–361. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Wakuta, S.; Suzuki, E.; Saburi, W.; Matsuura, H.; Nabeta, K.; Imai, R.; Matsui, H. OsJAR1 and OsJAR2 are jasmonyl-l-isoleucine synthases involved in wound- and pathogen-induced jasmonic acid signalling. Biochem. Biophys. Res. Commun. 2011, 409, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Riemann, M.; Riemann, M.; Takano, M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant Cell Environ. 2008, 31, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chen, Y.; Charnikhova, T.; Mulder, P.P.; Heijmans, J.; Hoogenboom, A.; Agalou, A.; Michel, C.; Morel, J.B.; Dreni, L.; et al. OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 2014, 86, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Miyamoto, K.; Miyamoto, K.; Minami, E.; Nishizawa, Y.; Iino, M.; Nojiri, H.; Yamane, H.; Okada, K. OsJAR1 contributes mainly to biosynthesis of the stress-induced jasmonoyl-isoleucine involved in defense responses in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, M. New cyclopentenone fatty acids formed from linoleic and linolenic acids in potato. Lipids 2000, 35, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Huffaker, A.; Hunter, C.T.; Alborn, H.T.; Schmelz, E.A. A maize death acid, 10-oxo-11-phytoenoic acid, is the predominant cyclopentenone signal present during multiple stress and developmental conditions. Plant Signal. Behav. 2015, 12, e1120395. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K. Green leaf volatiles: Hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 2006, 9, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Sugimoto, K.; Mano, J.; Ozawa, R.; Takabayashi, J. Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE 2012, 7, e36433. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Schuman, M.C.; Baldwin, I.T. The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 2016, 61, 373–394. [Google Scholar] [CrossRef] [PubMed]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef] [PubMed]
- Turlings, T.C.J.; Alborn, H.T.; Loughrin, J.H.; Tumlinson, J.H. Volicitin, an elicitor of maize volatiles in oral secretion of Spodoptera exigua: Isolation and bioactivity. J. Chem. Ecol. 2000, 26, 189–202. [Google Scholar] [CrossRef]
- Lait, C.G.; Alborn, H.T.; Teal, P.E.; Tumlinson, J.H., 3rd. Rapid biosynthesis of N-linolenoyl-l-glutamine, an elicitor of plant volatiles, by membrane-associated enzyme(s) in Manduca sexta. Proc. Natl. Acad. Sci. USA 2003, 100, 7027–7032. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Engelberth, J.; Alborn, H.T.; Tumlinson, J.H., 3rd; Teal, P.E. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc. Natl. Acad. Sci. USA 2009, 106, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Maes, L.; Goossens, A. Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species and trichome-specific regulatory mechanisms. Plant Signal. Behav. 2010, 5, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Velez-Bermudez, I.C.; Salazar-Henao, J.E.; Fornale, S.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Grotewold, E.; Gray, J.; Solano, R.; Schmidt, W.; Pages, M.; et al. A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell 2015, 27, 3245–3259. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, H.M. Hydroxamic acids derived from 2-hydroxy-2 h-1, 4-benzoxazin-3 (4 h)-one: Key defense chemicals of cereals. Agric. Food Chem. 2009, 57, 1677–1696. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Spiteller, D.; Boland, W.; Gierl, A. Transcriptional activation of IGL, the gene for indole formation in Zea mays: A structure-activity study with elicitor-active n-acyl glutamines from insects. Phytochemistry 2004, 65, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Dafoe, N.J.; Huffaker, A.; Vaughan, M.M.; Duehl, A.J.; Teal, P.E.; Schmelz, E.A. Rapidly induced chemical defenses in maize stems and their effects on short-term growth of Ostrinia nubilalis. J. Chem. Ecol. 2011, 37, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Chang, F.-R.; Wu, C.-C.; Wang, W.-Y.; Wu, Y.-C. New ent-kaurane diterpenoids with anti-platelet aggregation activity from Annona quamosa. J. Nat. Prod. 2002, 65, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Kaplan, F.; Huffaker, A.; Dafoe, N.J.; Vaughan, M.M.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 5455–5460. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Ma, P.W.; Pechan, T.; Bassford, E.R.; Williams, W.P.; Luthe, D.S. Degradation of the S. frugiperda peritrophic matrix by an inducible maize cysteine protease. J. Insect Physiol. 2006, 52, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Lopez, L.; Camas, A.; Shivaji, R.; Ankala, A.; Williams, P.; Luthe, D. MIR1-CP, a novel defense cysteine protease accumulates in maize vascular tissues in response to herbivory. Planta 2007, 226, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Pechan, T.; Ye, L.; Chang, Y.; Mitra, A.; Lin, L.; Davis, F.M.; Williams, W.P.; Luthe, D.S. A unique 33-kd cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 2000, 12, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Ankala, A.; Luthe, D.S.; Williams, W.P.; Wilkinson, J.R. Integration of ethylene and jasmonic acid signaling pathways in the expression of maize defense protein MIR1-CP. Mol. Plant Microbe Interact. 2009, 22, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.; Basu, S.; Varsani, S.; Castano-Duque, L.; Jiang, V.; Williams, W.P.; Felton, G.W.; Luthe, D.S. Ethylene contributes to MAIZE INSECT RESISTANCE1-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Physiol. 2015, 169, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, M.C.; Rufat, M.; Bravo, J.M.; San Segundo, B. Accumulation of a maize proteinase inhibitor in response to wounding and insect feeding, and characterization of its activity toward digestive proteinases of Spodoptera littoralis larvae. Planta 2000, 211, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Zhu-Salzman, K.; Zeng, R. Insect response to plant defensive protease inhibitors. Annu. Rev. Entomol. 2015, 60, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Johnson, R.R.; Ryan, C.A. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 1992, 98, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Ryan, C.A. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 1992, 4, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Lipka, A.E.; Schmelz, E.A.; Buckler, E.S.; Jander, G. Accumulation of 5-hydroxynorvaline in maize (Zea mays) leaves is induced by insect feeding and abiotic stress. J. Exp. Bot. 2014, eru385. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, P.; Freitak, D.; Vogel, H.; Heckel, D.G.; Boland, W. The phytohormone precursor OPDA is isomerized in the insect gut by a single, specific glutathione transferase. Proc. Natl. Acad. Sci. USA 2009, 106, 16304–16309. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Khan, S.A.; Vogel, H.; Heckel, D.G.; Boland, W. Opda isomerase GST16 is involved in phytohormone detoxification and insect development. FEBS J. 2014, 281, 2769–2783. [Google Scholar] [CrossRef] [PubMed]
- Heil, M. Indirect defence via tritrophic interactions. New Phytol. 2008, 178, 41–61. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Alborn, H.T.; Banchio, E.; Tumlinson, J.H. Quantitative relationships between induced jasmonic acid levels and volatile emission in Zea mays during Spodoptera exigua herbivory. Planta 2003, 216, 665–673. [Google Scholar] [PubMed]
- Kollner, T.G.; Lenk, C.; Schnee, C.; Kopke, S.; Lindemann, P.; Gershenzon, J.; Degenhardt, J. Localization of sesquiterpene formation and emission in maize leaves after herbivore damage. BMC Plant Biol. 2013, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Alborn, H.T.; Tumlinson, J.H. Synergistic interactions between volicitin, jasmonic acid and ethylene mediate insect-induced volatile emission in Zea mays. Physiol. Plant. 2003, 117, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, R.; Shiojiri, K.; Sabelis, M.W.; Arimura, G.; Nishioka, T.; Takabayashi, J. Corn plants treated with jasmonic acid attract more specialist parasitoids, thereby increasing parasitization of the common armyworm. J. Chem. Ecol. 2004, 30, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, R.; Shiojiri, K.; Sabelis, M.W.; Takabayashi, J. Maize plants sprayed with either jasmonic acid or its precursor, methyl linolenate, attract armyworm parasitoids, but the composition of attractants differs. Entomol. Exp. Appl. 2008, 129, 189–199. [Google Scholar] [CrossRef]
- Von Mérey, G.E.; Veyrat, N.; de Lange, E.; Degen, T.; Mahuku, G.; Valdez, R.L.; Turlings, T.C.; D’Alessandro, M. Minor effects of two elicitors of insect and pathogen resistance on volatile emissions and parasitism of Spodoptera frugiperda in mexican maize fields. Biol. Control. 2012, 60, 7–15. [Google Scholar] [CrossRef]
- Kim, J.; Felton, G.W. Priming of antiherbivore defensive responses in plants. Insect Sci. 2013, 20, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Sugimoto, K.; Ramadan, A.; Arimura, G. Memory of plant communications for priming anti-herbivore responses. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Conrath, U.; Peterhansel, C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011, 12, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Engelberth, J.; Alborn, H.T.; Schmelz, E.A.; Tumlinson, J.H. Airborne signals prime plants against insect herbivore attack. Proc. Natl. Acad. Sci. USA 2004, 101, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; D’Alessandro, M.; Jourdie, V.; Jakab, G.; Karlen, D.; Held, M.; Mauch-Mani, B.; Turlings, T.C. Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 2007, 49, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Veyrat, N.; Robert, C.A.; Xu, H.; Frey, M.; Ton, J.; Turlings, T.C. Indole is an essential herbivore-induced volatile priming signal in maize. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Oluwafemi, S.; Dewhirst, S.Y.; Veyrat, N.; Powers, S.; Bruce, T.J.; Caulfield, J.C.; Pickett, J.A.; Birkett, M.A. Priming of production in maize of volatile organic defence compounds by the natural plant activator cis-jasmone. PLoS ONE 2013, 8, e62299. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.M.; Huffaker, A.; Schmelz, E.A.; Dafoe, N.J.; Christensen, S.; Sims, J.; Martins, V.F.; Swerbilow, J.; Romero, M.; Alborn, H.T.; et al. Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides. Plant Cell Environ. 2014, 37, 2691–2706. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.D.; Perkins, A.; Williams, W.P.; Warburton, M.L. Using genome-wide associations to identify metabolic pathways involved in maize aflatoxin accumulation resistance. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Brodhagen, M.; Isakeit, T.; Brown, S.H.; Gobel, C.; Betran, J.; Feussner, I.; Keller, N.P.; Kolomiets, M.V. Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 2009, 22, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Dafoe, N.J.; Schmelz, E.A. Zmpep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 2011, 155, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, I.; Hause, B.; Maucher, H.; Pitzschke, A.; Miersch, O.; Ziegler, J.; Ryan, C.A.; Wasternack, C. Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato–amplification in wound signalling. Plant J. 2003, 33, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Constantino, N.N.; Mastouri, F.; Damarwinasis, R.; Borrego, E.J.; Moran-Diez, M.E.; Kenerley, C.M.; Gao, X.; Kolomiets, M.V. Root-expressed maize LIPOXYGENASE 3 negatively regulates induced systemic resistance to Colletotrichum graminicola in shoots. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Planchamp, C.; Glauser, G.; Mauch-Mani, B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Neal, A.L.; Ton, J. Systemic defense priming by Pseudomonas putida KT2440 in maize depends on benzoxazinoid exudation from the roots. Plant Signal. Behav. 2013, 8, e22655. [Google Scholar] [CrossRef] [PubMed]
- Yamane, H.; Sugawara, J.; Suzuki, Y.; Shimamura, E.; Takahashi, N. Syntheses of jasmonic acid related-compounds and their structure-activity-relationships on the growth of rice seedings. Agric. Biol. Chem. 1980, 44, 2857–2864. [Google Scholar] [CrossRef]
- Yan, Y.; Huang, P.-C.; Borrego, E.; Kolomiets, M. New perspectives into jasmonate roles in maize. Plant Signal. Behav. 2014, 9, e970442. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Urrea, A.; Dellaporta, S.L. Cell death and cell protection genes determine the fate of pistils in maize. Development 1999, 126, 435–441. [Google Scholar] [PubMed]
- DeLong, A.; Calderon-Urrea, A.; Dellaporta, S.L. Sex determination gene TASSELSEED 2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 1993, 74, 757–768. [Google Scholar] [CrossRef]
- Wu, X.; Knapp, S.; Stamp, A.; Stammers, D.K.; Jornvall, H.; Dellaporta, S.L.; Oppermann, U. Biochemical characterization of TASSELSEED 2, an essential plant short-chain dehydrogenase/reductase with broad spectrum activities. FEBS J. 2007, 274, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [CrossRef] [PubMed]




| Gene | Gene annotation | Gramene ID † | GRMZM ID † | Genbank † or TIGR ID † |
|---|---|---|---|---|
| α,β-Unsaturated carbonyl detoxification | ||||
| ZmOPR3 | 12-oxo-phytodienoic acid reductase 3 | Zm00001d037182 | GRMZM2G156712 | AY921640 |
| ZmOPR4 | 12-oxo-phytodienoic acid reductase 4 | Zm00001d013493 | N/A in RefGen_v3 | AY921641 |
| ZmOPR5 | 12-oxo-phytodienoic acid reductase 5 | Zm00001d003584 | GRMZM2G087192 | AY921642 |
| Direct defense | ||||
| ZmMIR1 | maize insect resistance 1 (mir1) cysteine proteinase | Zm00001d036542 | GRMZM2G150276 | NM_001112101 |
| ZmMPI | proteinase inhibitor | Zm00001d011080 | GRMZM2G028393 | X78988 |
| Ethylene associated | ||||
| ZmACS2 | 1-aminocyclopropane-1-carboxylate synthase 2 | Zm00001d002592 | GRMZM2G164405 | AY359569 |
| ZmERF2 | homologue to AtERF1 and 2 | Zm00001d002762 | GRMZM2G055180 | NM_001158578 |
| ZmERF6 | homologue to AtERF1 and 2 | Zm00001d034920 | GRMZM2G381441 | NM_001176924 |
| Flavonoid metabolism | ||||
| ZmF3H | flavanone 3-hydroxylase | Zm00001d001960 | GRMZM2G062396 | NM_001112225 |
| Lipase | ||||
| ZmPLC | phospholipase C | Zm00001d014903 | GRMZM5G889467 | NM_001111784 |
| Oxylipin biosynthesis | ||||
| ZmLOX3 | lipoxygenase 3 | Zm00001d033623 | GRMZM2G109130 | AF329371 |
| ZmLOX4 | lipoxygenase 4 | Zm00001d033624 | GRMZM2G109056 | DQ335762 |
| ZmLOX5 | lipoxygenase 5 | Zm00001d013493 | GRMZM2G102760 | DQ335763 |
| ZmLOX6 | lipoxygenase 6 | Zm00001d002000 | GRMZM2G040095 | DQ335764 |
| ZmLOX9 | lipoxygenase 9 | Zm00001d027893 | GRMZM2G017616 | DQ335767 |
| ZmLOX10 | lipoxygenase 10 | Zm00001d053675 | GRMZM2G015419 | DQ335768 |
| ZmLOX11 | lipoxygenase 11 | Zm00001d015852 | GRMZM2G009479 | DQ335769 |
| ZmAOS2a | allene oxide synthase 2a | Zm00001d028282 | GRMZM2G002178 | |
| ZmAOS2b | allene oxide synthase 2b | Zm00001d048021 | GRMZM2G067225 | AY488135 |
| Transcription factors | ||||
| ZmMYC2a | homologue to AtMYC2 | Zm00001d007536 | GRMZM2G303463 | |
| ZmWRKY14 | WRKY transcription factor 14 | Zm00001d043569 | GRMZM2G040298 | EU973705 |
| ZmWRKY46 | WRKY transcription factor 46 | Zm00001d052357 | GRMZM2G063216 | EU956406 |
| Repressors of JA signaling | ||||
| ZmJAZ1 | ZIM domain containing protein | Zm00001d026477 | GRMZM2G143402 | NM_001156069 |
| ZmJAZ3 | ZIM domain containing protein | Zm00001d048268 | GRMZM2G036288 | NM_001157436 |
| ZmJAZ4 | ZIM domain containing protein | Zm00001d027899 | GRMZM2G343157 | NM_001157673 |
| ZmJAZ5 | ZIM domain containing protein | Zm00001d027901 | GRMZM2G445634 | NM_001156053 |
| ZmJAZ6 | ZIM domain containing protein | Zm00001d048263 | GRMZM2G036351 | NM_001157328 |
| ZmJAZ7 | ZIM domain containing protein | Zm00001d014253 | GRMZM2G173596 | NM_001159100 |
| ZmJAZ8 | ZIM domain containing protein | Zm00001d033050 | GRMZM2G145412 | EU970040 |
| ZmJAZ10 | ZIM domain containing protein | Zm00001d020614 | GRMZM2G116614 | AZM5_3798 |
| ZmJAZ11 | ZIM domain containing protein | Zm00001d006860 | GRMZM2G101769 | AZM5_1941 |
| ZmJAZ12 | ZIM domain containing protein | Zm00001d027900 | GRMZM5G838098 | NM_001154831 |
| ZmJAZ19 | ZIM domain containing protein | Zm00001d036494 | GRMZM2G080509 | AZM5_87469 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borrego, E.J.; Kolomiets, M.V. Synthesis and Functions of Jasmonates in Maize. Plants 2016, 5, 41. https://doi.org/10.3390/plants5040041
Borrego EJ, Kolomiets MV. Synthesis and Functions of Jasmonates in Maize. Plants. 2016; 5(4):41. https://doi.org/10.3390/plants5040041
Chicago/Turabian StyleBorrego, Eli J., and Michael V. Kolomiets. 2016. "Synthesis and Functions of Jasmonates in Maize" Plants 5, no. 4: 41. https://doi.org/10.3390/plants5040041






