DRB1, DRB2 and DRB4 Are Required for Appropriate Regulation of the microRNA399/PHOSPHATE2 Expression Module in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
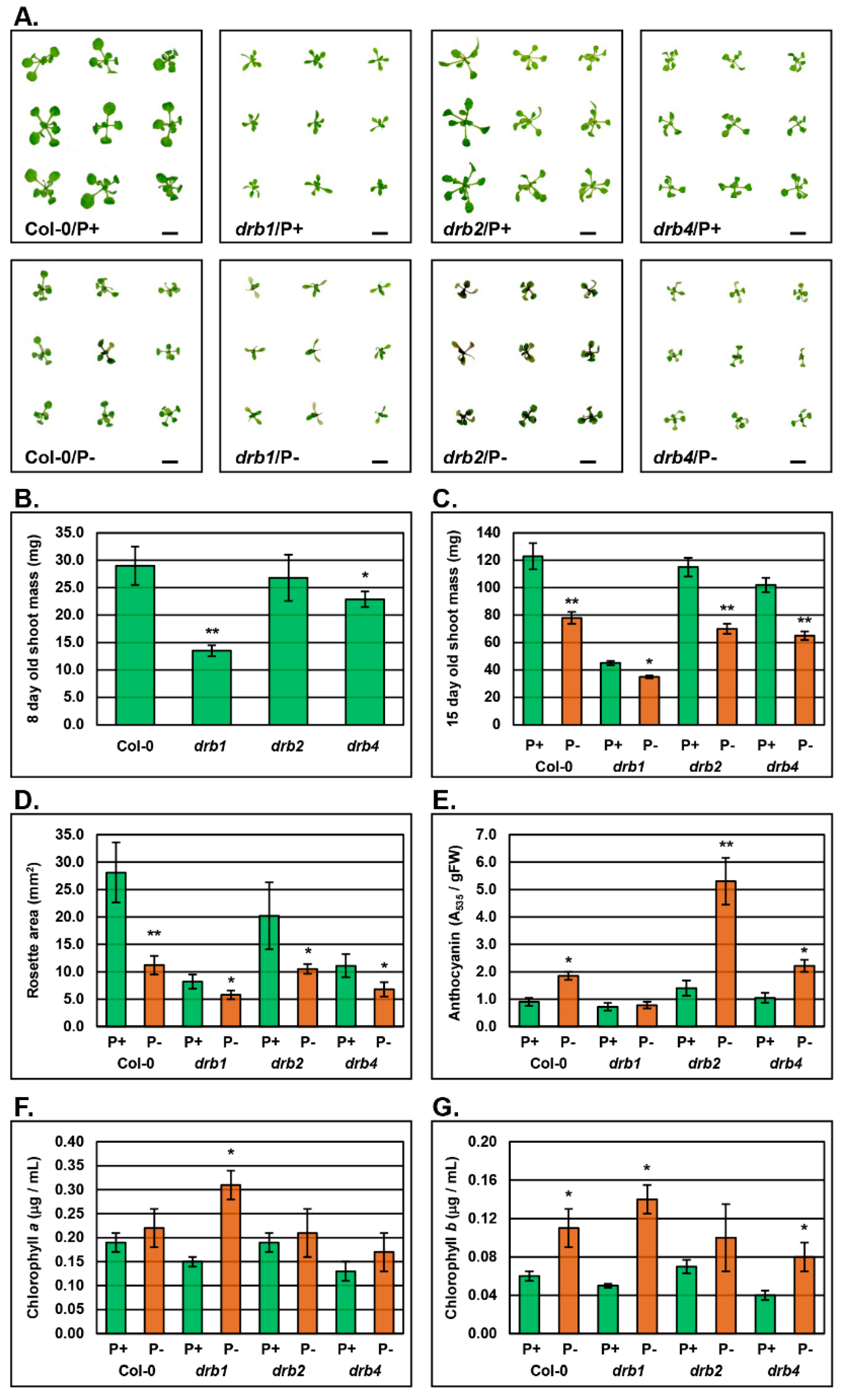
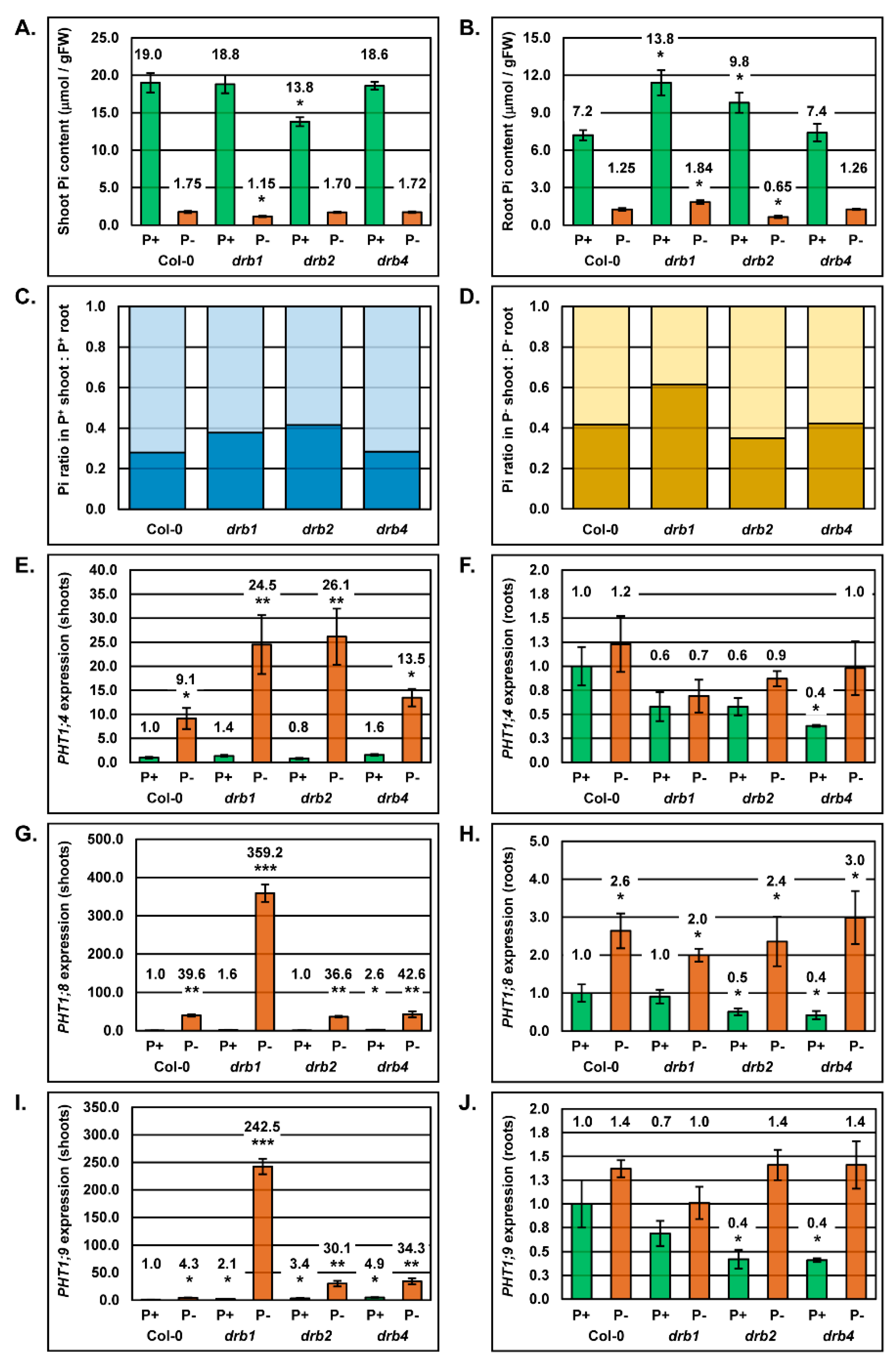
2.1. The Phenotypic and Physiological Response to PO4 Stress in the Shoot Tissues of Arabidopsis Plant Lines Defective in DRB Protein Activity
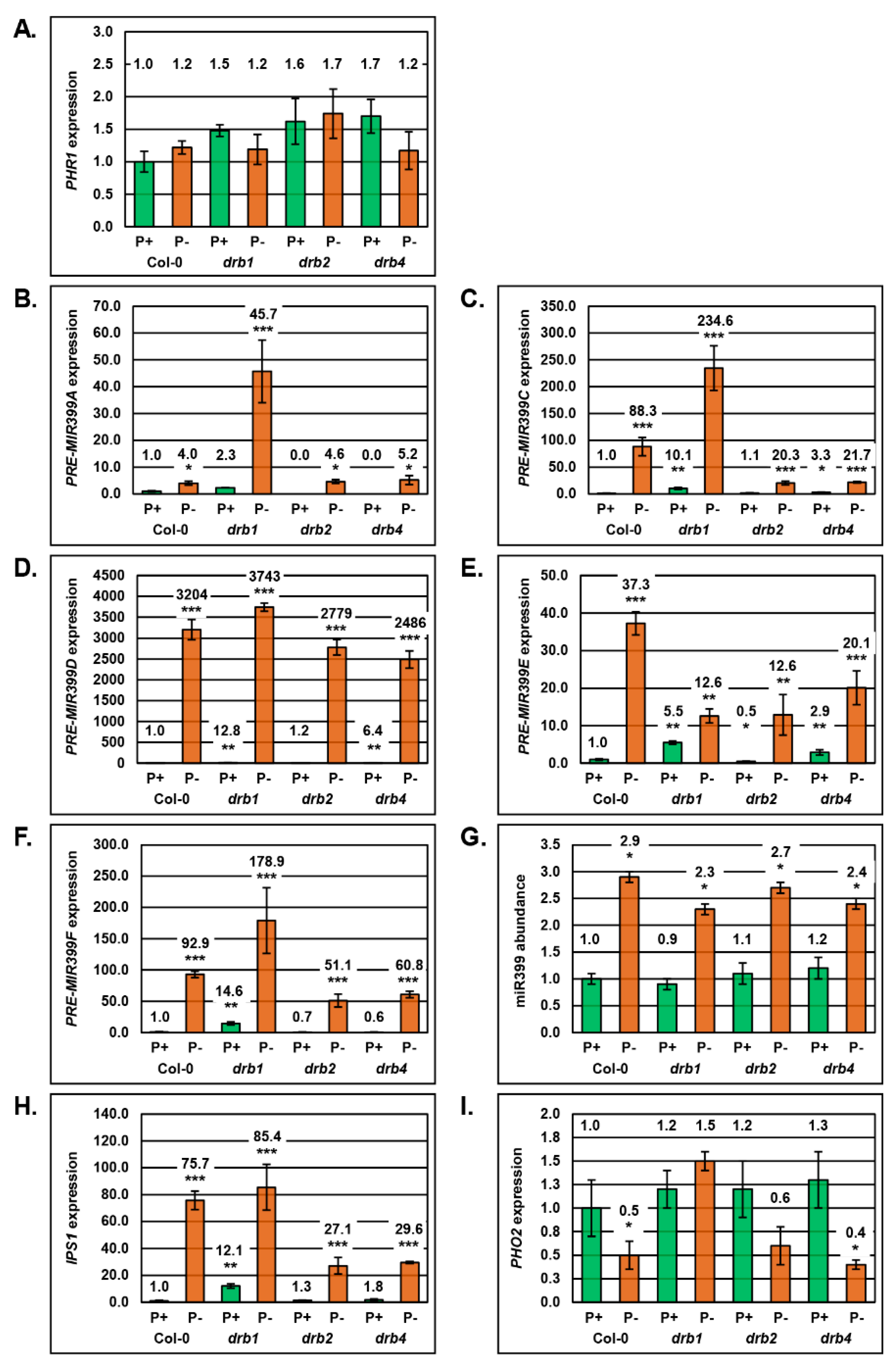
2.2. Molecular Profiling of the miR399/PHO2 Expression Module in the Shoot Tissues of Arabidopsis Plant Lines Defective in DRB Protein Activity
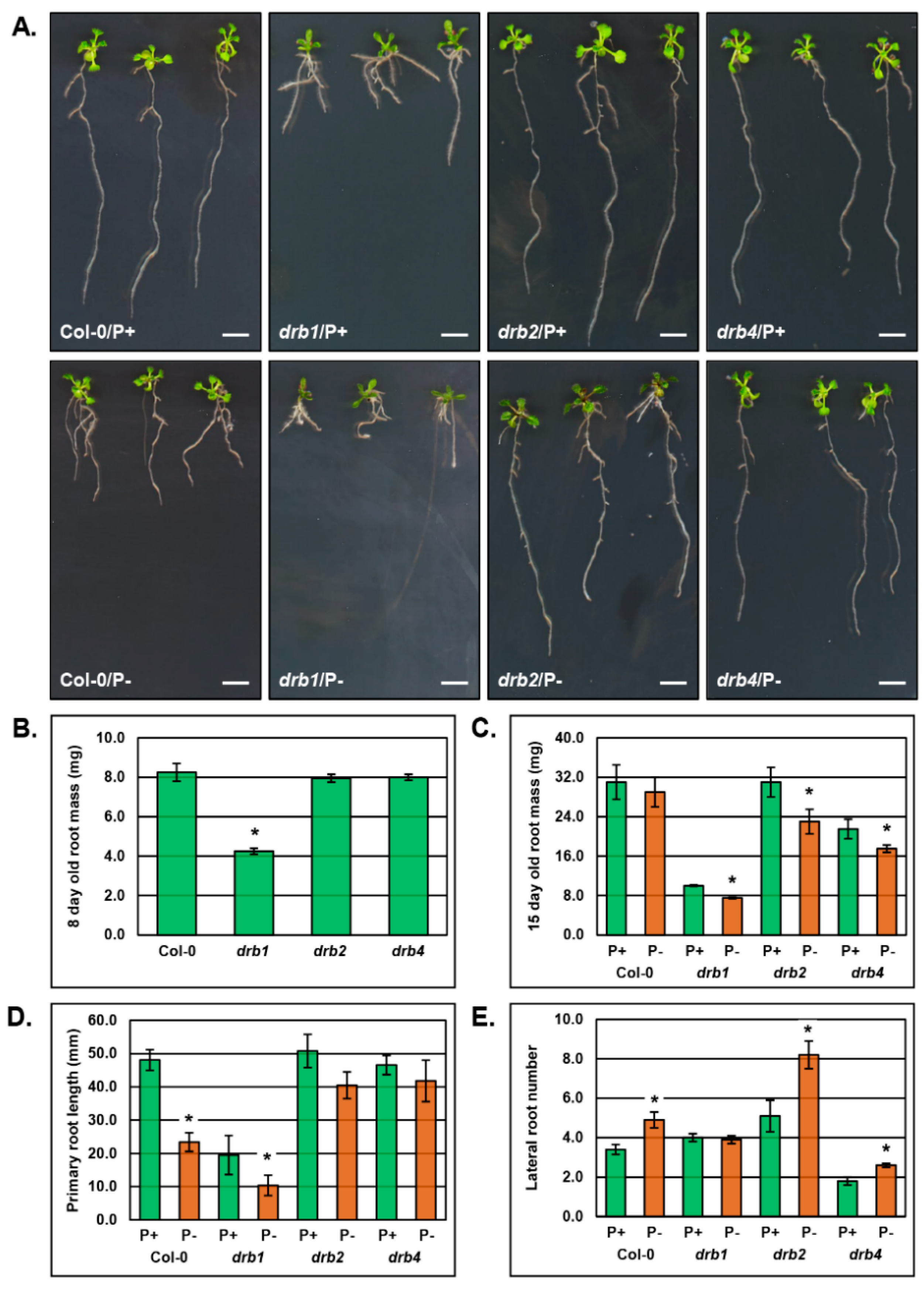
2.3. The Phenotypic and Physiological Response to PO4 Stress of the Root System of Arabidopsis Plant Lines Defective in DRB Protein Activity
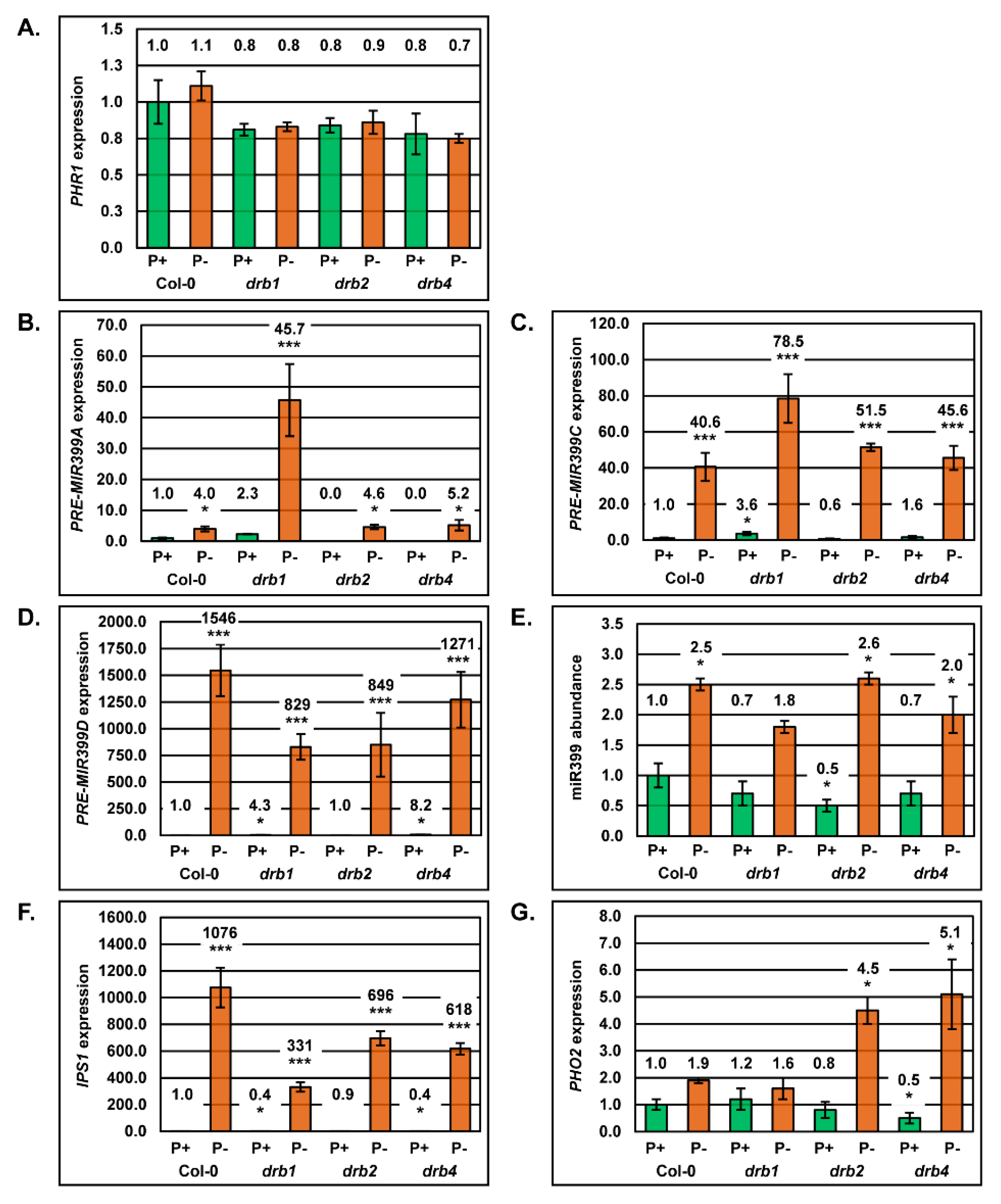
2.4. Molecular Profiling of the miR399/PHO2 Expression Module in the Root System of Arabidopsis Plant Lines Defective in DRB Protein Activity
2.5. Correct Inorganic Phosphate Partitioning Between the Shoot and Root Tissue of Arabidopsis Requires DRB1 and DRB2
3. Discussion
3.1. DRB1 is Required to Maintain Phosphorous Homeostasis in Arabidopsis
3.2. DRB2 is Required to Maintain Phosphate Homeostasis in Arabidopsis
3.3. DRB4 is Required For miR399 Production in Arabidopsis Roots
3.4. DRB1, DRB2 and DRB4 Are Required to Regulate the miR399/PHO2 Expression Module in Arabidopsis Shoots and Roots
4. Materials and Methods
4.1. Plant Material and Phosphate Stress Treatment
4.2. Phenotypic and Physiological Assessments
4.3. Total RNA Extraction for Quantitative Molecular Assessments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abel, S.; Ticconi, C.A.; Delatorre, C.A. Phosphate sensing in higher plants. Planta 2002, 115, 1–8. [Google Scholar] [CrossRef]
- Hammond, J.P.; Bennett, M.J.; Bowen, H.C.; Broadley, M.R.; Eastwood, D.C.; May, S.T.; Rahn, C.; Swarup, R.; Woolaway, K.E.; White, P.J. Changes in Gene Expression in Arabidopsis Shoot during Phosphate Starvation and the Potential for Developing Smart Plants. Society 2003, 132, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000, 3, 182–187. [Google Scholar] [CrossRef]
- Hackenberg, M.; Shi, B.J.; Gustafson, P.; Langridge, P. Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus- sufficient and phosphorus-deficient conditions. BMC Plant Biol. 2013, 13, 214. [Google Scholar] [CrossRef]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Ma, L.; Hou, X.; Wang, M.; Wu, Y.; Liu, F.; Deng, X.W. Phosphate Starvation Triggers Distinct Alterations of Genome Expression in Arabidopsis Roots and Leaves. Plant Physiol. 2003, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.I.; Chiang, S.F.; Lin, W.Y.; Chen, J.W.; Tseng, C.Y.; Wu, P.C.; Chiou, T.J. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 2008, 147, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L. Organic acids in the rhizosphere—A critical review. Plant Soil 1998, 205, 25–44. [Google Scholar] [CrossRef]
- Poirier, Y.; Thoma, S.; Somerville, C.; Schiefelbein, J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991, 97, 1087–1093. [Google Scholar] [CrossRef]
- Stefanovic, A.; Arpat, A.B.; Bligny, R.; Gout, E.; Vidoudez, C.; Bensimon, M.; Poirier, Y. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 2011, 66, 689–699. [Google Scholar] [CrossRef]
- Wang, Y.; Ribot, C.; Rezzonico, E.; Poirier, Y. Structure and Expression Profile of the Arabidopsis PHO1 Gene Family Indicates a Broad Role in Inorganic Phosphate Homeostasis 1. Plant Physiol. 2004, 135, 400–411. [Google Scholar] [CrossRef]
- Delhaize, E.; Randall, P.J. Characterization of a Phosphate-Accumulator Mutant of Arabidopsis thaliana. Plant Physiol. 1995, 107, 207–213. [Google Scholar] [CrossRef]
- Dong, B.; Rengel, Z.; Delhaize, E. Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 1998, 205, 251–256. [Google Scholar] [CrossRef]
- Park, B.S.; Seo, J.S.; Chua, N.H. Nitrogen Limitation Adaptation recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef]
- Aung, K. pho2, a Phosphate Overaccumulator, Is Caused by a Nonsense Mutation in a MicroRNA399 Target Gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef]
- Nilsson, L.; Müller, R.; Nielsen, T.H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 2007, 30, 1499–1512. [Google Scholar] [CrossRef]
- Rubio, V.; Linhares, F.; Solano, R.; Mart’in, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef]
- Buhtz, A.; Springer, F.; Chappell, L.; Baulcombe, D.C.; Kehr, J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008, 53, 739–749. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Garvin, D.F.; Kochian, L.V. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 2002, 130, 1361–1370. [Google Scholar] [CrossRef]
- Shin, R.; Berg, R.H.; Schachtman, D.P. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 2005, 46, 1350–1357. [Google Scholar] [CrossRef]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Yu, D. Identification of Nitrogen Starvation-Responsive MicroRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [PubMed]
- Doerner, P. Phosphate starvation signaling: A threesome controls systemic Pi homeostasis. Curr. Opin. Plant Biol. 2008, 11, 536–540. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Golm, S.P. PHO2, MicroRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, O.; Jost, R.; Kollehn, D.O.; Fenske, R.; Finnegan, P.M.; O’Brien, P.A.; Hardy, G.E.S.J.; Lambers, H. Acclimation responses of Arabidopsis thaliana to sustained phosphite treatments. J. Exp. Bot. 2013, 64, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.K.; Han, C.L.; Lin, S.I.; Chen, Y.J.; Tsai, Y.C.; Chen, Y.R.; Chen, J.W.; Lin, W.Y.; Chen, P.M.; Liu, T.Y.; et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef]
- Rouached, H.; Arpat, A.B.; Poirier, Y. Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 2010, 3, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.C.; Del Pozo, J.C.; Iglesias, J.; Rubio, V.; Solano, R.; De La Peňa, A.; Leyva, A.; Paz-Ares, J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000, 24, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Bouché, N. MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci. 2008, 13, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Han, M.H.; Goud, S.; Song, L.; Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 2004, 101, 1093–1098. [Google Scholar] [CrossRef]
- Eamens, A.L.; Smith, N.A.; Curtin, S.J.; Wang, M.B.; Waterhouse, P.M. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 2009, 15, 2219–2235. [Google Scholar] [CrossRef]
- Eamens, A.L.; Kim, K.W.; Curtin, S.J.; Waterhouse, P.M. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS ONE 2012, 7, e35933. [Google Scholar] [CrossRef]
- Eamens, A.L.; Kim, K.W.; Waterhouse, P.M. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal. Behav. 2012, 7, 1224–1229. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Pélissier, T.; Clavel, M.; Chaparro, C.; Pouch-Pélissier, M.N.; Vaucheret, H.; Deragon, J.M. Double-stranded RNA binding proteins DRB2 and DRB4 have an antagonistic impact on polymerase IV-dependent siRNA levels in Arabidopsis. RNA 2011, 17, 1502–1510. [Google Scholar] [CrossRef]
- Lu, C.; Fedoroff, N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000, 12, 2351–2366. [Google Scholar] [CrossRef]
- Curtin, S.J.; Watson, J.M.; Smith, N.A.; Eamens, A.L.; Blanchard, C.L.; Waterhouse, P.M. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008, 582, 2753–2760. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.J.; Mittal, A.; Jia, F.; Rock, C.D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2012, 80, 117–129. [Google Scholar] [CrossRef]
- Williamson, L.C.; Ribrioux, S.; Fitter, A.H.; Leyser, H.M.O. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001, 126, 875–882. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.J.; Finnegan, P.M. Regulation of phosphate starvation responses in higher plants. Ann. Bot. 2010, 105, 513–526. [Google Scholar] [CrossRef] [Green Version]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.L.; Bartel, D.P.; Crété, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef]
- Kurihara, Y.; Takashi, Y.; Watanabe, Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 2006, 12, 206–212. [Google Scholar] [CrossRef]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Hart-Smith, G.; Eamens, A.L.; Wilkins, M.R.; Waterhouse, P.M. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat. Plants 2015, 1, 14027. [Google Scholar] [CrossRef]
- Yang, L.; Wu, G.; Poethig, R.S. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Adenot, X.; Elmayan, T.; Lauressergues, D.; Boutet, S.; Bouché, N.; Gasciolli, V.; Vaucheret, H. DRB4-Dependent TAS3 trans-Acting siRNAs Control Leaf Morphology through AGO7. Curr. Biol. 2006, 16, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Hiraguri, A.; Moriyama, H.; Fukuhara, T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol. Biol. 2007, 63, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. DRB1, DRB2 and DRB4 Are Required for Appropriate Regulation of the microRNA399/PHOSPHATE2 Expression Module in Arabidopsis thaliana. Plants 2019, 8, 124. https://doi.org/10.3390/plants8050124
Pegler JL, Oultram JMJ, Grof CPL, Eamens AL. DRB1, DRB2 and DRB4 Are Required for Appropriate Regulation of the microRNA399/PHOSPHATE2 Expression Module in Arabidopsis thaliana. Plants. 2019; 8(5):124. https://doi.org/10.3390/plants8050124
Chicago/Turabian StylePegler, Joseph L., Jackson M. J. Oultram, Christopher P. L. Grof, and Andrew L. Eamens. 2019. "DRB1, DRB2 and DRB4 Are Required for Appropriate Regulation of the microRNA399/PHOSPHATE2 Expression Module in Arabidopsis thaliana" Plants 8, no. 5: 124. https://doi.org/10.3390/plants8050124




