Different Roles of Heat Shock Proteins (70 kDa) During Abiotic Stresses in Barley (Hordeum vulgare) Genotypes
Abstract
:1. Introduction
2. Results
2.1. HSP70s Showed Peculiar Roles against Abiotic Stress in Barley
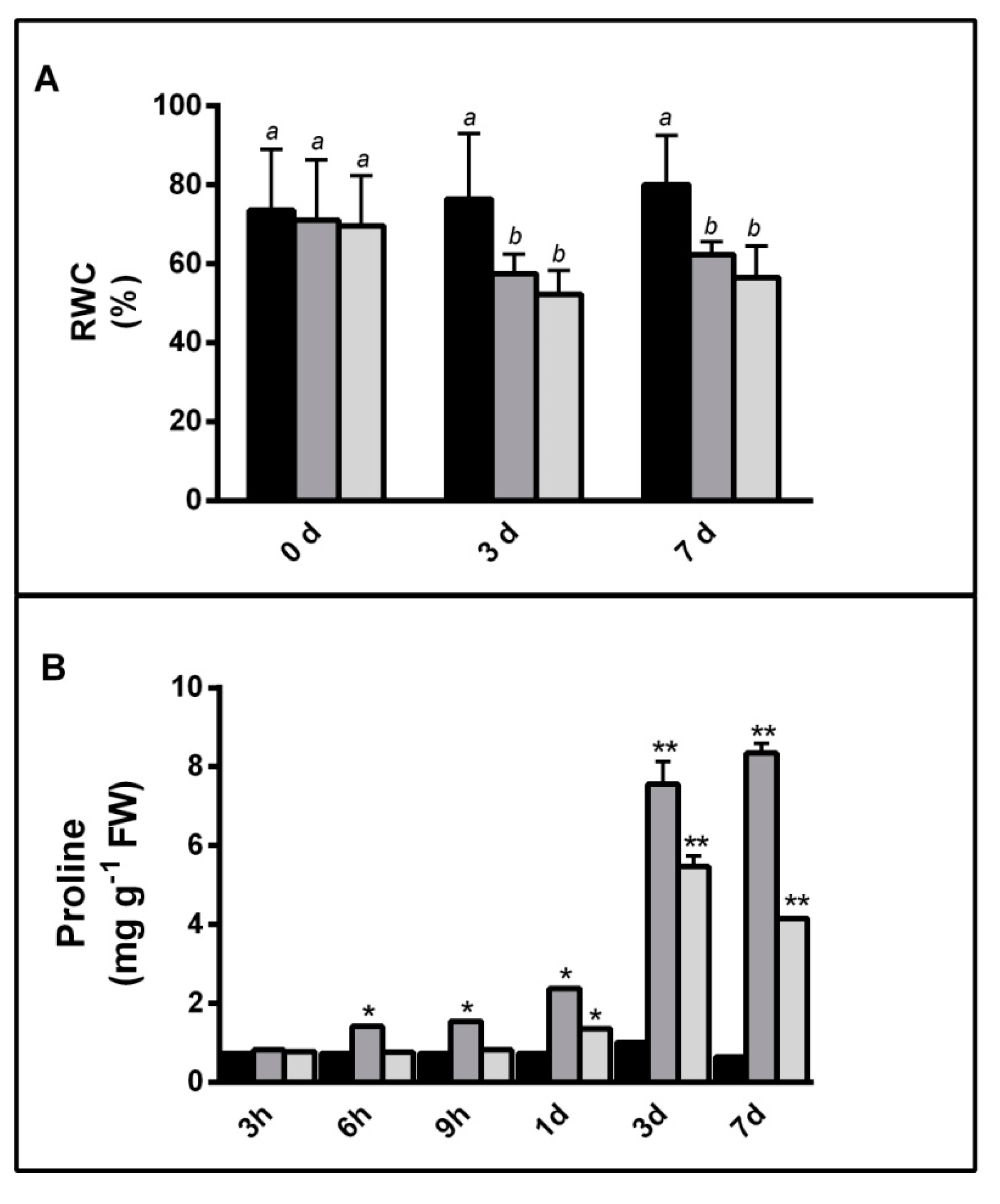
2.1.1. NaCl and PEG Effects on Barley Plants
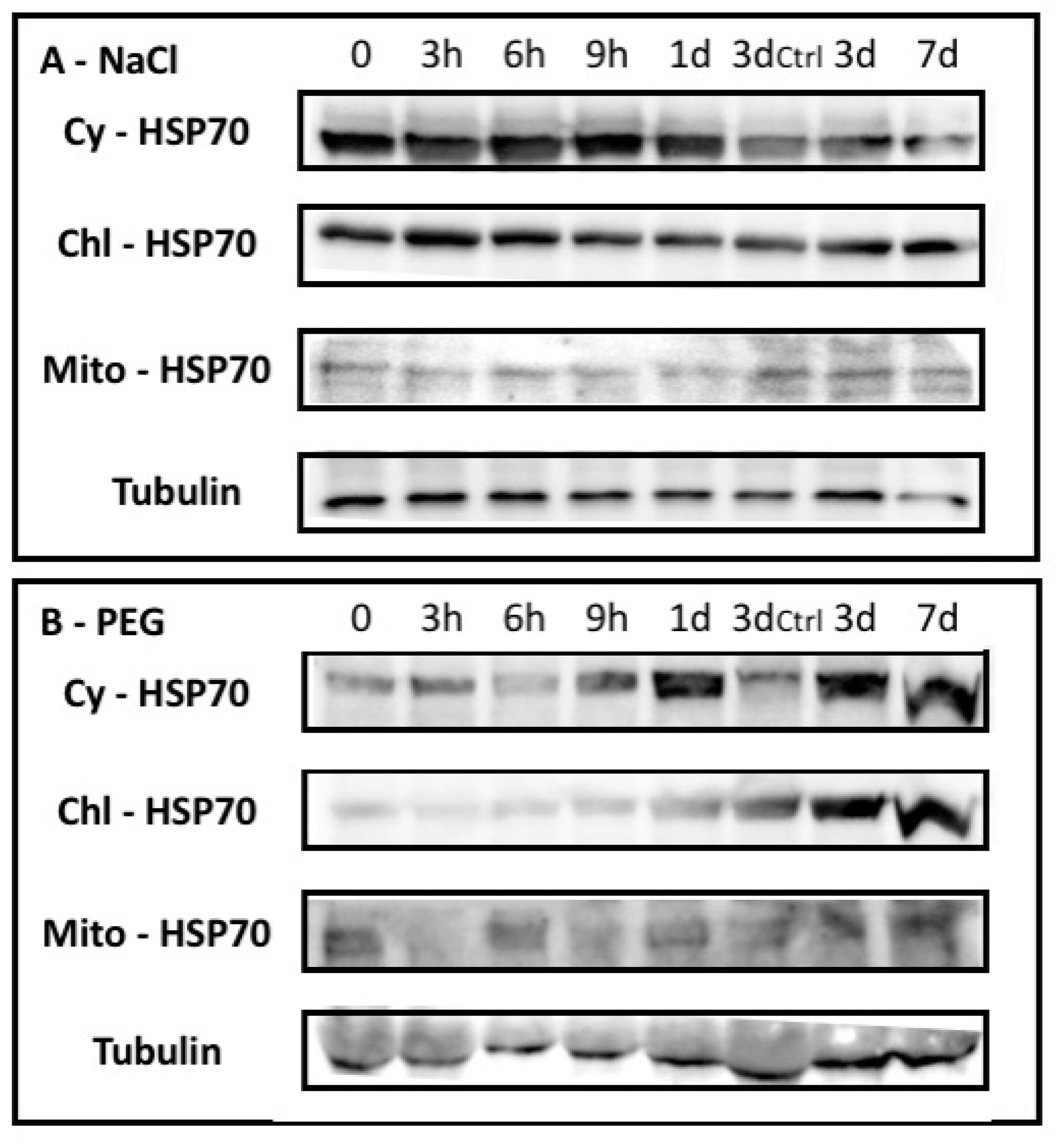
2.1.2. Barley HSP70 Isoforms Showed Specific Occurrence upon Abiotic Stresses
2.2. HSP70s in Barley: A Bioinformatic Overview
2.3. Analysis of cis-Acting Elements in the HSP70s Promoters
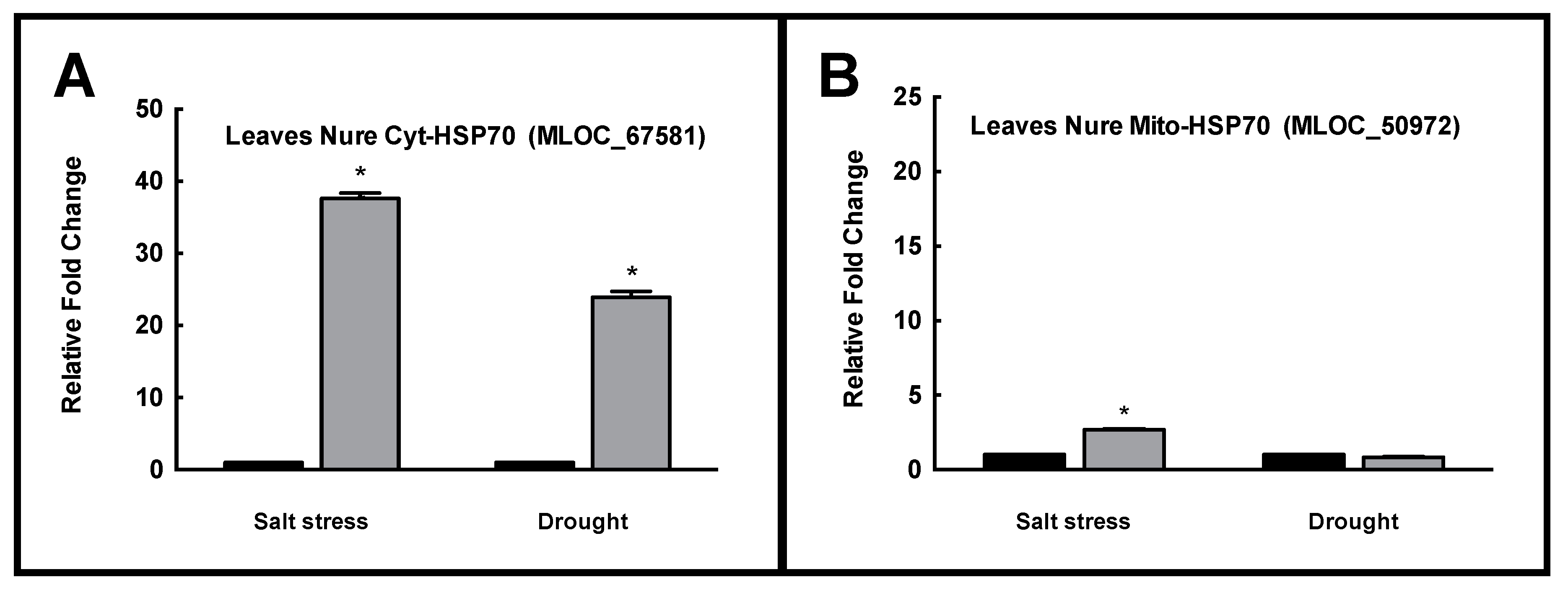
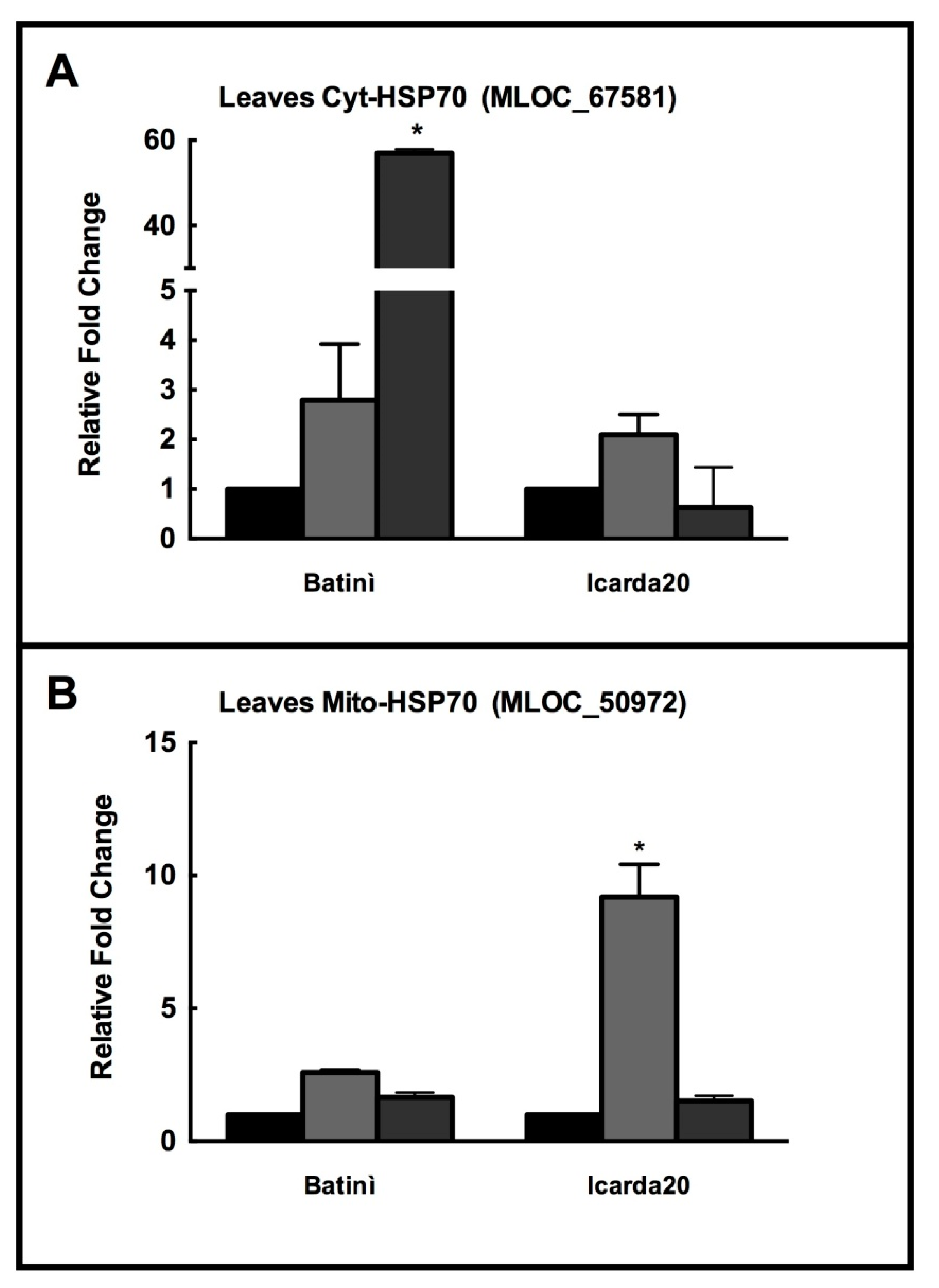
2.4. Real Time PCR of Selected HSP70 Isoforms
2.5. Effects of Abiotic Stress in in Different Barley Genotypes
2.6. HSP70s in Different Barley Genotypes
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Treatments
4.2. Growth Variation and Water Content Determination
4.3. Proline Content
4.4. Western Blotting
4.5. RNA Extraction and qRT-PCR
4.6. Bioinformatics Analysis
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Yu, A.; Li, P.; Tang, T.; Wang, J.; Chen, Y.; Liu, L. Roles of Hsp70s in Stress Responses of Microorganisms, Plants, and Animals. BioMed Res. Int. 2015, 2015, 510319. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, Z.; Broadley, S.A.; Shomura, Y.; Bracher, A.; Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of HSP70s. EMBO J. 2006, 25, 2519–2528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, H.K.; Dong, Q.L.; Zhang, Y.Y.; Wang, Y.M.; Li, H.Y.; Xing, G.J.; Li, Q.Y.; Dong, Y.S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesma, M.E.; Hendrickson, W.A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Kundnani, P.; Grover, A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperon 2013, 18, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.L.; Wang, J.S.; Liu, H.C.; Chen, R.W.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperon 2001, 6, 201–208. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.Y.; Martini, M.T.; Yusuff, O.A.; Ismail, R.M.; Miah, G. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 2017, 33, 26–39. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Su, P.H.; Li, H.M. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef]
- Leng, L.; Liang, Q.; Jiang, J.; Zhang, C.; Hao, Y.; Wang, X.; Su, W. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J. Plant Res. 2017, 130, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Wu, X.; Li, T.; Jia, M.; Liu, X.; Li, P.; Zhou, X.; Ji, X.; Yue, X. Genome-wide survey of heat shock factors and heat shock protein 70s and their regulatory network under abiotic stresses in Brachypodium distachyon. PLoS ONE 2017, 12, e0180352. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Lu, J.P.; Zhai, Y.F.; Chai, W.; Gong, Z.H.; Lu, M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Nurcato, R.; De Lillo, A.; Lentini, M.; Grillo, S.; Esposito, S. Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant Physiol. Biochem. 2016, 105, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Murata, N. Genetically engineered enhancement of salt tolerance in higher plants. In Stress Response of Photosynthetic Organisms: Molecular Mechanisms and Molecular Regulation; Sato, K., Murata, N., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 133–148. [Google Scholar]
- Reynolds, M.; Tuberosa, R. Translational research impacting on crop productivity in drought-prone environments. Curr. Opin. Plant Biol. 2008, 11, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; Aiese Cigliano, R.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 26, 1176. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Esposito, S. Nitrate uptake affects cell wall synthesis and modelling. Front. Plant Sci. 2017, 8, 1376. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 12, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Declerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Gill, B.S.; Appels, R.; Botha-Oberholster, A.M.; Buell, C.R.; Bennetzen, J.L.; Chalhoub, B.; Chumley, F.; Dvorák, J.; Iwanaga, M.; Keller, B.; et al. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 2004, 168, 1087–1096. [Google Scholar] [CrossRef]
- Cui, P.; Liu, H.; Islam, F.; Li, L.; Farooq, M.A.; Ruan, S.; Zhou, W. OsPEX11, a Peroxisomal Biogenesis Factor 11, Contributes to Salt Stress Tolerance in Oryza sativa. Front. Plant Sci. 2016, 7, 1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Fu, L.; Dai, F.; Jiang, L.; Zhang, G.; Wu, D. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genom. 2016, 17, 889. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Hausman, J.F.; Guerriero, G.; Esposito, S. Poaceae vs. abiotic stress: Focus on drought and salt stress, recent insight and perspectives. Front. Plant Sci. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Hammami, Z.; Gauffreteau, A.; BelhajFraj, M.; Sahli, A.; Jeuffroy, M.H.; Rezgui, S.; Bergaoui, K.; McDonnell, R.; Trifa, Y. Predicting yield reduction in improved barley (Hordeum vulgare L.) varieties and landraces under salinity using selected tolerance traits. Field Crops Res. 2017, 211, 10–18. [Google Scholar] [CrossRef]
- Ellis, R.P.; Forster, B.P.; Robinson, D.; Handley, L.L.; Gordon, D.C.; Russell, J.R.; Powell, W. Wild barley: A source of genes for crop improvement in the 21st century? J. Exp. Bot. 2000, 51, 9–17. [Google Scholar] [CrossRef]
- Ahmed, I.M.; Dai, H.; Zheng, W.; Cao, F.; Zhang, G.; Sun, D.; Wu, F. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2013, 63, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, S.; Karmous, C.; Chamekh, Z.; Hammami, Z.; Baraket, M.; Esposito, S.; Rezgui, S.; Trifa, Y. Effects of nitrogen rates on response of grain yield and nitrogen agronomic efficiency (NAE) of durum wheat cultivars under different environments. Ann. Appl. Biol. 2016, 168, 264–273. [Google Scholar] [CrossRef]
- Bouhaouel, I.; Gfeller, A.; Boudabbous, K.H.; Fauconnier, M.L.; Slim Amara, H.; du Jardin, P. Physiological and biochemical parameters: New tools to screen barley root exudate allelopathic potential (Hordeum vulgare L. subsp. vulgare). Acta Physiol. Plant 2018, 40, 38. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondellia, A.; Stanca, M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Challinor, A.J.; Watson, J.; Lobell, D.J.; Howden, S.M.; Smith, D.; Chetri, N. A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 2014, 4, 287–291. [Google Scholar] [CrossRef]
- Akar, T.; Avci, M.; Dusunceli, F. Barley: Post-Harvest Operations; FAO in PHO Post Harvest Compendium: Ankara, Turkey, 2004. [Google Scholar]
- Bhatty, R.S. Hulless barley: Development and Civilisation. In VII International Barley Genetics Symposium; University of Saskatoon: Saskatoon, SK, Canada, 1986; pp. 106–111. [Google Scholar]
- Mussatto, S.I. Brewer’s spent grain: A valuable feedstock for industrial applications. J. Sci. Food Agric. 2014, 94, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Moore, F.C.; Lobell, D.B. The fingerprint of climate trends on European crop yields. Proc. Nalt. Acad. Sci. USA 2015, 112, 2670–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, J.S. Drought decision-making. J. Exp. Bot. 2010, 61, 3493–3497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lentini, M.; De Lillo, A.; Paradisone, V.; Liberti, D.; Landi, S.; Esposito, S. Early responses to cadmium exposure in barley plants: Effects on biometric and physiological parameters. Acta Physiol. Plant 2018, 40, 178. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Yu, A.; Li, P.; Yang, H.; Liu, G.; Liu, Li. Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses. Sci. Rep. 2016, 6, 33650. [Google Scholar] [CrossRef] [PubMed]
- Iovieno, P.; Punzo, P.; Guida, G.; Mistretta, C.; Van Oosten, M.J.; Nurcato, R.; Bostan, H.; Colantuono, C.; Costa, A.; Bagnaresi, P.; et al. Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front. Plant Sci. 2011, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D846–D851. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Ceccarelli, S.; Blair, M.W.; Upadhyaya, H.D.; Are, A.K.; Ortiz, R. Landrace Germplasm for Improving Yield and Abiotic Stress Adaptation. Trends Plant Sci. 2016, 21, 31–42. [Google Scholar] [CrossRef]
- Lopes, S.M.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Landi, S.; De Lillo, A.; Nurcato, R.; Grillo, S.; Esposito, S. In-field study on traditional Italian tomato landraces: The constitutive activation of the ROS scavenging machinery reduces effects of drought stress. Plant Physiol. Biochem. 2017, 118, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Long, H.; Wang, Z.; Zhao, S.; Tang, Y.; Huang, Z.; Wang, Y.; Xu, Q.; Mao, L.; Deng, G.; et al. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc. Nalt. Acad. Sci. USA 2015, 112, 1095–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardi, M.; Castiglia, D.; Ferrara, M.; Guerriero, G.; Chiurazzi, M.; Esposito, S. The effects of salt stress cause a diversion of basal metabolism in barley roots: Possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 2015, 86, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Matin, M.; Brown, J.H.; Ferguson, H. Leaf water potential, relative water content, and diffusive resistance as screening techniques for drought resistance in barley. Agron. J. 1989, 81, 100–105. [Google Scholar] [CrossRef]
- Claussen, W. Proline as a measure of stress in tomato plants. Plant Sci. 2005, 168, 241–248. [Google Scholar] [CrossRef]
- Castiglia, D.; Cardi, M.; Landi, S.; Cafasso, D.; Esposito, S. Expression and characterization of a cytosolic glucose-6-phosphate dehydrogenase isoform from barley (Hordeum vulgare) roots. Protein Express Purif. 2015, 112, 8–14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. Plantcare, a data base of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Aoki, Y.; Okamura, Y.; Tadaka, S.; Kinoshita, K.; Obayashi, T. ATTED-II in 2016: A plant co-expression database towards lineage-specific co-expression. Plant Cell Physiol. 2016, 57, e5. [Google Scholar] [CrossRef] [PubMed]
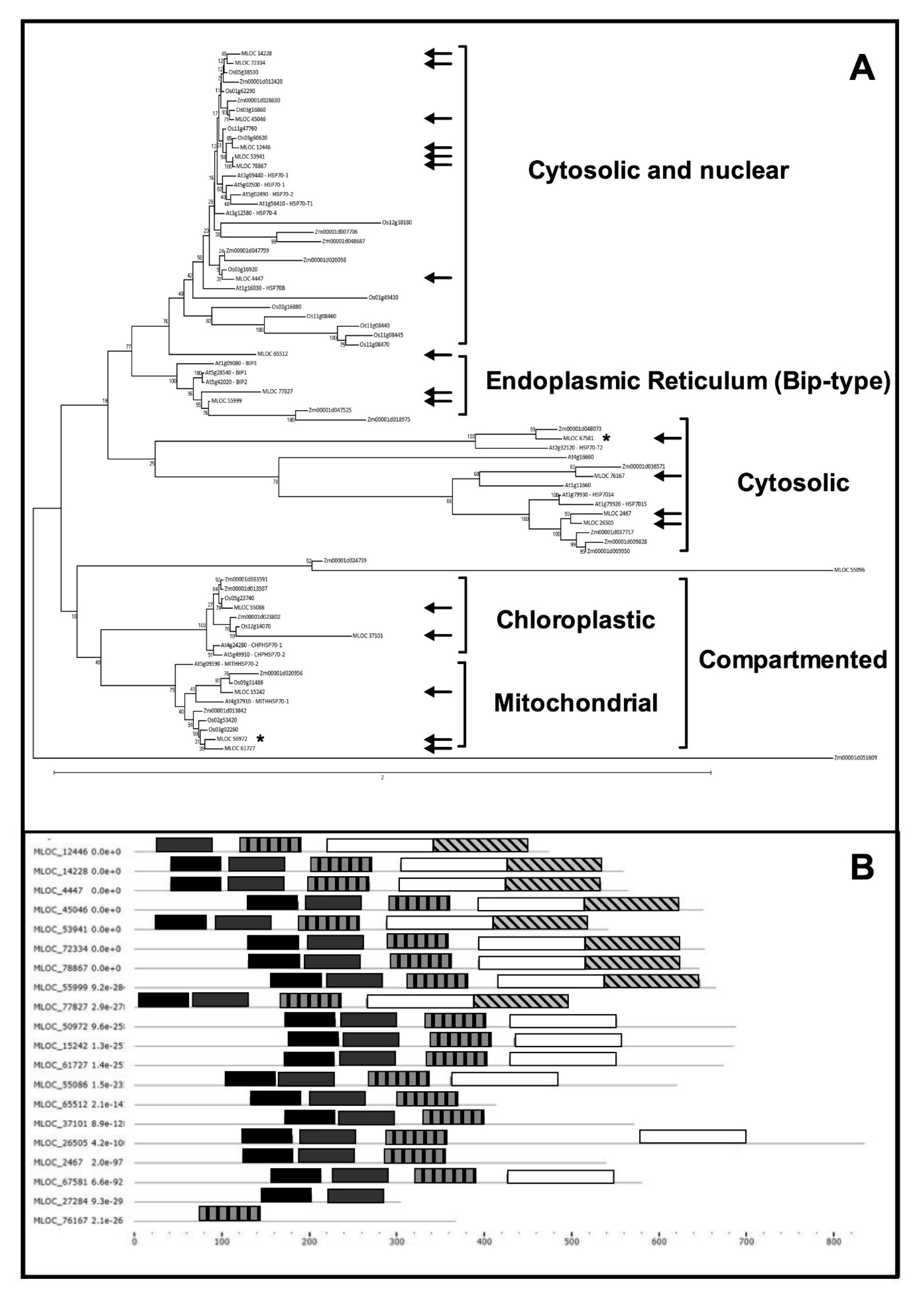


| Locus | Localization | Proposed Nomenclature | Pfam Domains IDs |
|---|---|---|---|
| MLOC_12446 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_14228 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_15242 | Mitochondrial | HvMithHSP70 | PF00012.20; PF06723.13 |
| MLOC_2467 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_26505 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_37101 | Chloroplast | HvCHPHSP70 | PF00012.20; PF06723.13 |
| MLOC_4447 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_45046 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_50972 | Mitochondrial | HvMithHSP70 | PF00012.20; PF06723.13; PF02782.16 |
| MLOC_53941 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_55086 | Chloroplast | HvCHPHSP70 | PF00012.20; PF06723.13 |
| MLOC_55096 | Cytoplasm | HvHSP70 | PF00685.27 |
| MLOC_55999 | Cytoplasm | HvBIP | PF00012.20; PF06723.13 |
| MLOC_61727 | Mitochondrial | HvMithHSP70 | PF00012.20; PF06723.13 |
| MLOC_65512 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_67581 | Cytoplasm | HvHSP70 | PF00012.20 |
| MLOC_72334 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| MLOC_76167 | Cytoplasm | HvHSP70 | PF00012.20 |
| MLOC_77827 | Cytoplasm | HvBIP | PF00012.20; PF06723.13 |
| MLOC_78867 | Cytoplasm | HvHSP70 | PF00012.20; PF06723.13 |
| Expression data (FPKM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| 4 Days Embryo | Seedling Shoots | Young Inflorescences | Developing Inflorescences | Seedling Roots | Developing Tillers | Developing Grains (5 DPA) | Developing Grains (15 DPA) | |
| MLOC_12446 | 928.19 | 674.36 | 1584.58 | 1909.81 | 1181.38 | 354.58 | 524.31 | 216.92 |
| MLOC_14228 | 39.44 | 202.24 | 1.21 | 3.92 | 418.49 | 4.72 | 72.14 | 220.23 |
| MLOC_15242 | 33.28 | 33.49 | 32.72 | 28.73 | 38.93 | 1.82 | 38.89 | 6.71 |
| MLOC_2467 | 224.4 | 139.3 | 166.2 | 182.7 | 203.6 | 308.2 | 193.2 | 123.5 |
| MLOC_26505 | 45.00 | 67.59 | 50.43 | 68.15 | 67.48 | 11.90 | 94.58 | 60.19 |
| MLOC_37101 | 135.86 | 237.33 | 139.46 | 98.02 | 60.39 | 71.76 | 116.04 | 45.66 |
| MLOC_4447 | 0.04 | 13.67 | 0.02 | 2.03 | 32.01 | 0.27 | 0.38 | 43.40 |
| MLOC_45046 | 101.50 | 336.15 | 3.47 | 2.63 | 226.17 | 210.66 | 77.81 | 20.03 |
| MLOC_50972 | 171.19 | 103.86 | 196.05 | 153.72 | 109.15 | 39.96 | 170.41 | 44.16 |
| MLOC_53941 | 162.85 | 134.29 | 322.87 | 549.16 | 175.32 | 92.78 | 302.49 | 79.18 |
| MLOC_55086 | 51.30 | 125.24 | 45.13 | 46.13 | 69.61 | 36.50 | 118.59 | 86.64 |
| MLOC_55096 | 1.68 | 0.13 | 0.00 | 0.00 | 2.58 | 0.53 | 0.04 | 0.00 |
| MLOC_55999 | 267.44 | 166.85 | 126.02 | 118.81 | 259.95 | 64.44 | 720.53 | 146.05 |
| MLOC_61727 | 3.53 | 11.46 | 0.92 | 12.78 | 23.20 | 0.23 | 13.54 | 26.91 |
| MLOC_65512 | 0.00 | 0.03 | 0.00 | 0.01 | 1.62 | 0.00 | 0.00 | 0.01 |
| MLOC_67581 | 0.80 | 6.53 | 0.40 | 0.58 | 17.17 | 0.01 | 2.02 | 44.56 |
| MLOC_72334 | 2.86 | 16.57 | 0.08 | 0.16 | 54.85 | 0.05 | 5.23 | 6.86 |
| MLOC_76167 | 36.80 | 27.78 | 52.17 | 54.60 | 27.87 | 46.08 | 44.69 | 13.62 |
| MLOC_77827 | 0.47 | 0.18 | 0.07 | 0.01 | 0.65 | 0.37 | 0.29 | 0.24 |
| MLOC_78867 | 336.38 | 242.84 | 726.65 | 1088.08 | 384.78 | 169.08 | 511.81 | 164.32 |
| Cis-Actig Elements in Promoter Region | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABRE | ARE | AUXRR-Core | BOX W1 | CCAAT | CGTCA | ERE | EIRE | GARE | GCN4 | HSE | Light | LTR | P-box | MBS | Motif IIB | Ry | SKN-1 | TATC | TC-rich | TCA | TGA | TGAGC | |
| MLOC_12446 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 2 | 8 | 0 | 0 | 1 | 0 | 0 | 3 | 1 | 3 | 0 | 0 | 0 |
| MLOC_14228 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| MLOC_15242 | No available data | ||||||||||||||||||||||
| MLOC_2467 | 0 | 0 | 2 | 0 | 1 | 4 | 0 | 1 | 3 | 0 | 1 | 18 | 4 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 4 |
| MLOC_26505 | 2 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 7 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 2 |
| MLOC_37101 | No available data | ||||||||||||||||||||||
| MLOC_4447 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 2 | 0 | 0 |
| MLOC_45046 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 17 | 2 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 |
| MLOC_50972 | 0 | 0 | 0 | 3 | 1 | 3 | 0 | 0 | 1 | 0 | 1 | 20 | 1 | 0 | 4 | 1 | 0 | 5 | 0 | 1 | 0 | 0 | 3 |
| MLOC_53941 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 10 | 1 | 1 | 3 | 0 | 1 | 5 | 0 | 1 | 1 | 0 | 1 |
| MLOC_55086 | No available data | ||||||||||||||||||||||
| MLOC_55096 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 27 | 0 | 0 | 14 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 1 |
| MLOC_55999 | 3 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 17 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 2 |
| MLOC_61727 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 18 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 4 |
| MLOC_65512 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 11 | 1 | 0 | 1 | 0 | 0 | 4 | 1 | 1 | 1 | 1 | 1 |
| MLOC_67581 | 4 | 2 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 19 | 2 | 0 | 1 | 0 | 2 | 8 | 0 | 0 | 1 | 0 | 2 |
| MLOC_72334 | 0 | 0 | 0 | 3 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 15 | 0 | 1 | 1 | 0 | 0 | 4 | 0 | 2 | 1 | 1 | 2 |
| MLOC_76167 | No available data | ||||||||||||||||||||||
| MLOC_77827 | No available data | ||||||||||||||||||||||
| MLOC_78867 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 15 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 1 | 2 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landi, S.; Capasso, G.; Ben Azaiez, F.E.; Jallouli, S.; Ayadi, S.; Trifa, Y.; Esposito, S. Different Roles of Heat Shock Proteins (70 kDa) During Abiotic Stresses in Barley (Hordeum vulgare) Genotypes. Plants 2019, 8, 248. https://doi.org/10.3390/plants8080248
Landi S, Capasso G, Ben Azaiez FE, Jallouli S, Ayadi S, Trifa Y, Esposito S. Different Roles of Heat Shock Proteins (70 kDa) During Abiotic Stresses in Barley (Hordeum vulgare) Genotypes. Plants. 2019; 8(8):248. https://doi.org/10.3390/plants8080248
Chicago/Turabian StyleLandi, Simone, Giorgia Capasso, Fatma Ezzahra Ben Azaiez, Salma Jallouli, Sawsen Ayadi, Youssef Trifa, and Sergio Esposito. 2019. "Different Roles of Heat Shock Proteins (70 kDa) During Abiotic Stresses in Barley (Hordeum vulgare) Genotypes" Plants 8, no. 8: 248. https://doi.org/10.3390/plants8080248





