High-Resolution Secretome Analysis of Chemical Hypoxia Treated Cells Identifies Putative Biomarkers of Chondrosarcoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. DMOG Treatment of SW 1353 Cells
2.2. Sample Processing and Mass Spectrometry Analysis
2.3. Proteomic Data Analysis
2.4. EVs Isolation and Characterization
2.5. Validation of GAPDH Levels by Western Blotting
2.6. Assessment of SW 1353 Cell Viability upon DMOG Treatment
3. Results
3.1. Quantitative Proteomics Identified Differences in the Secretome of Hypoxia-Induced Chondrosarcoma Cells
3.2. Enrichment Analysis Showed Hypoxia Enriched EVs and Exosomes Proteins Compartmentalization
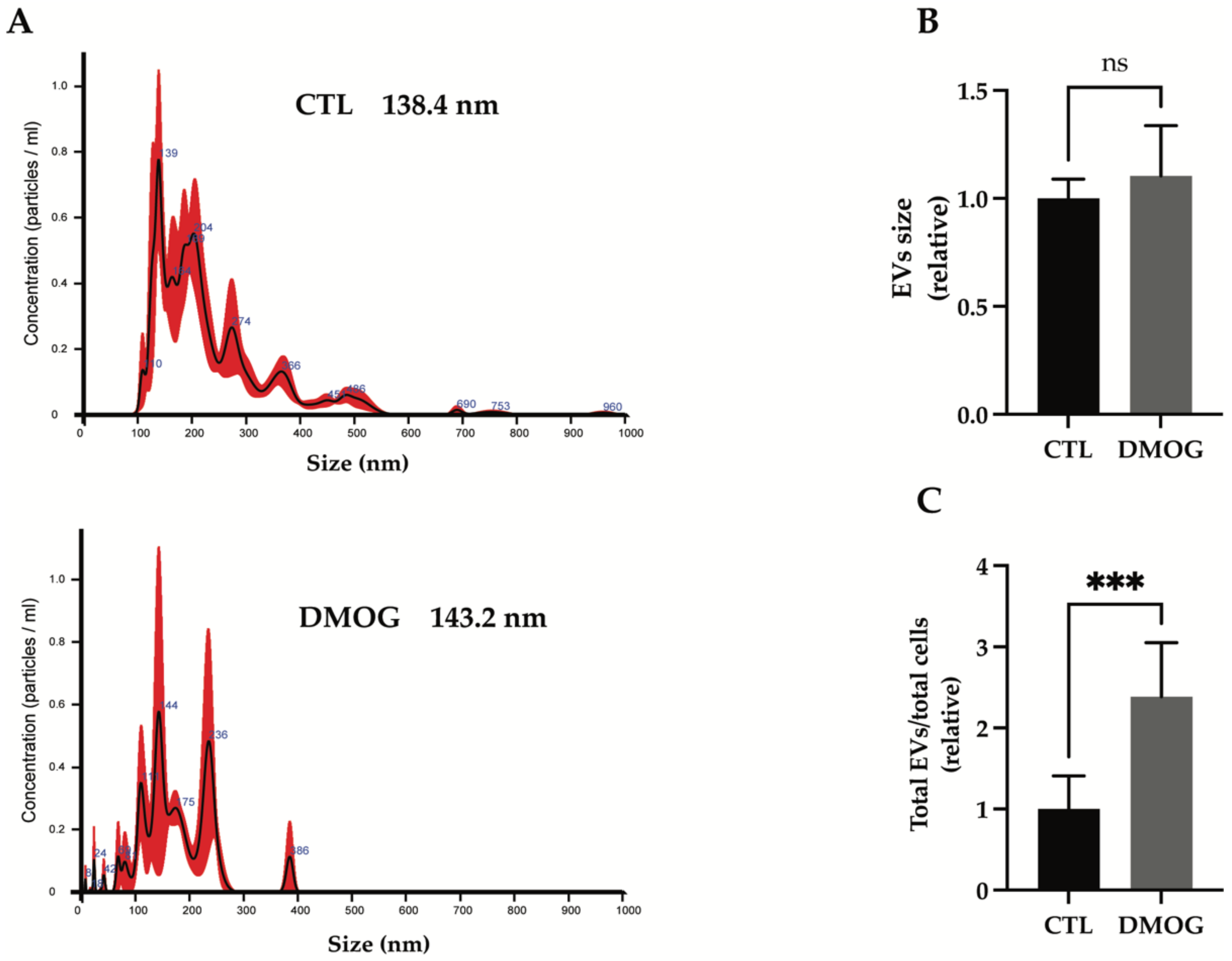
3.3. Hypoxia Increased Secretion of Extracellular Vesicles
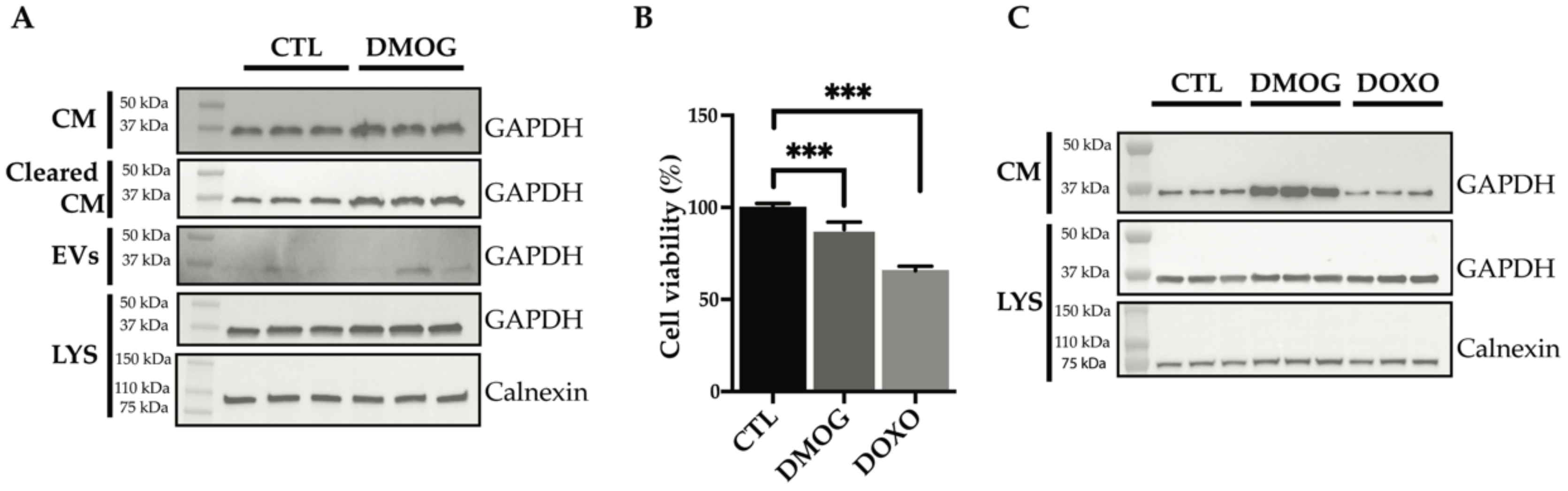
3.4. GAPDH: Validation of Secretome Analysis and Enrichment Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicolle, R.; Ayadi, M.; Gomez-Brouchet, A.; Armenoult, L.; Banneau, G.; Elarouci, N.; Tallegas, M.; Decouvelaere, A.-V.; Aubert, S.; Rédini, F.; et al. Integrated Molecular Characterization of Chondrosarcoma Reveals Critical Determinants of Disease Progression. Nat. Commun. 2019, 10, 4622. [Google Scholar] [CrossRef] [PubMed]
- van Praag, V.M.; Rueten-Budde, A.J.; Ho, V.; Dijkstra, P.D.S.; Fiocco, M.; van de Sande, M.A.J. Incidence, Outcomes and Prognostic Factors during 25 Years of Treatment of Chondrosarcomas. Surg. Oncol. 2018, 27, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nazeri, E.; Gouran Savadkoohi, M.; Majidzadeh-A, K.; Esmaeili, R. Chondrosarcoma: An Overview of Clinical Behavior, Molecular Mechanisms Mediated Drug Resistance and Potential Therapeutic Targets. Crit. Rev. Oncol. Hematol. 2018, 131, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zając, A.E.; Kopeć, S.; Szostakowski, B.; Spałek, M.J.; Fiedorowicz, M.; Bylina, E.; Filipowicz, P.; Szumera-Ciećkiewicz, A.; Tysarowski, A.; Czarnecka, A.M.; et al. Chondrosarcoma-from Molecular Pathology to Novel Therapies. Cancers 2021, 13, 2390. [Google Scholar] [CrossRef]
- Jeong, W.; Kim, H.-J. Biomarkers of Chondrosarcoma. J. Clin. Pathol. 2018, 71, 579–583. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; McGough, R.; Aswad, B.; Block, J.A.; Terek, R. Hypoxia Induces HIF-1α and VEGF Expression in Chondrosarcoma Cells and Chondrocytes. J. Orthop. Res. 2004, 22, 1175–1181. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The Hypoxic Tumour Microenvironment. Oncogenesis 2018, 7, 1175–1181. [Google Scholar] [CrossRef]
- Maes, C.; Carmeliet, G.; Schipani, E. Hypoxia-Driven Pathways in Bone Development, Regeneration and Disease. Nat. Rev. Rheumatol. 2012, 8, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Suo, C.; Zheng, C.; Zhang, H. Hypoxia and Metabolism in Metastasis. In Hypoxia and Cancer Metastasis; Gilkes, D.M., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; pp. 87–95. ISBN 978-3-030-12734-3. [Google Scholar]
- Sun, X.; Wei, L.; Chen, Q.; Terek, R.M. CXCR4/SDF1 Mediate Hypoxia Induced Chondrosarcoma Cell Invasion through ERK Signaling and Increased MMP1 Expression. Mol. Cancer 2010, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhou, H.; Wei, F.; Jiang, L.; Liu, X.; Liu, Z.; Ma, Q. Increased Levels of Hypoxia-Inducible Factor-1α Are Associated with Bcl-XL Expression, Tumor Apoptosis, and Clinical Outcome in Chondrosarcoma. J. Orthop. Res. 2011, 29, 143–151. [Google Scholar] [CrossRef]
- Nelson, B.S.; Kremer, D.M.; Lyssiotis, C.A. New Tricks for an Old Drug. Nat. Chem. Biol. 2018, 14, 990–991. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.D.; Pedersen, J.T.; Venning, F.A.; Singh, L.B.; Moeendarbary, E.; Charras, G.; Cox, T.R.; Sahai, E.; Erler, J.T. Hypoxia and Loss of PHD2 Inactivate Stromal Fibroblasts to Decrease Tumour Stiffness and Metastasis. EMBO Rep. 2015, 16, 1394–1408. [Google Scholar] [CrossRef] [PubMed]
- Bush, J.T.; Chan, M.C.; Mohammed, S.; Schofield, C.J. Quantitative MS-Based Proteomics: Comparing the MCF-7 Cellular Response to Hypoxia and a 2-Oxoglutarate Analogue. Chembiochem 2020, 21, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Meissner, F.; Scheltema, R.A.; Mollenkopf, H.-J.; Mann, M. Direct Proteomic Quantification of the Secretome of Activated Immune Cells. Science 2013, 340, 475–478. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Carreca, A.P.; Pravatà, V.M.; D’Apolito, D.; Bonelli, S.; Calligaris, M.; Monaca, E.; Müller, S.A.; Lichtenthaler, S.F.; Scilabra, S.D. Quantitative Proteomics Reveals Changes Induced by TIMP-3 on Cell Membrane Composition and Novel Metalloprotease Substrates. Int. J. Mol. Sci. 2021, 22, 2392. [Google Scholar] [CrossRef]
- Scilabra, S.D.; Pigoni, M.; Pravatá, V.; Schätzl, T.; Müller, S.A.; Troeberg, L.; Lichtenthaler, S.F. Increased TIMP-3 Expression Alters the Cellular Secretome through Dual Inhibition of the Metalloprotease ADAM10 and Ligand-Binding of the LRP-1 Receptor. Sci. Rep. 2018, 8, 14697. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E.; Chen, X.; Hehl, R.; Karas, H.; Liebich, I.; Matys, V.; Meinhardt, T.; Prüß, M.; Reuter, I.; Schacherer, F. TRANSFAC: An Integrated System for Gene Expression Regulation. Nucleic Acids Res. 2000, 28, 316–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingender, E. The TRANSFAC Project as an Example of Framework Technology That Supports the Analysis of Genomic Regulation. Brief. Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Binder, J.X.; Pletscher-Frankild, S.; Tsafou, K.; Stolte, C.; O’Donoghue, S.I.; Schneider, R.; Jensen, L.J. COMPARTMENTS: Unification and Visualization of Protein Subcellular Localization Evidence. Database 2014, 2014, bau012. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Ercolani, L.; Florence, B.; Denaro, M.; Alexander, M. Isolation and Complete Sequence of a Functional Human Glyceraldehyde-3-Phosphate Dehydrogenase Gene. J. Biol. Chem. 1988, 263, 15335–15341. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Zhang, F.; Hong, C.-Q.; Giuliano, A.E.; Cui, X.-J.; Zhou, G.-J.; Zhang, G.-J.; Cui, Y.-K. Critical Protein GAPDH and Its Regulatory Mechanisms in Cancer Cells. Cancer Biol. Med. 2015, 12, 10–22. [Google Scholar] [CrossRef]
- Sirover, M.A. GAPDH and Hypoxia. In Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH); Elsevier: Amsterdam, The Netherlands, 2017; pp. 155–165. ISBN 978-0-12-809852-3. [Google Scholar]
- Guo, C.; Liu, S.; Sun, M.-Z. Novel Insight into the Role of GAPDH Playing in Tumor. Clin. Transl. Oncol. 2013, 15, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, R.; Chatani, E.; Harada, N.; Sugimoto, K.; Inui, H.; Nakano, Y. Glyceraldehyde-3-Phosphate Dehydrogenase in the Extracellular Space Inhibits Cell Spreading. Biochim. Biophys. Acta 2005, 1726, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.S.; Rawat, P.; Malhotra, H.; Sheokand, N.; Kumar, M.; Patidar, A.; Chaudhary, S.; Jakhar, P.; Raje, C.I.; Raje, M. Secreted Multifunctional Glyceraldehyde-3-Phosphate Dehydrogenase Sequesters Lactoferrin and Iron into Cells via a Non-Canonical Pathway. Sci. Rep. 2015, 5, 18465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Desiderio, D.M. Proteomics Analysis of Prefractionated Human Lumbar Cerebrospinal Fluid. Proteomics 2005, 5, 541–550. [Google Scholar] [CrossRef]
- Jella, K.K.; Yu, L.; Yue, Q.; Friedman, D.; Duke, B.J.; Alli, A.A. Exosomal GAPDH from Proximal Tubule Cells Regulate ENaC Activity. PLoS ONE 2016, 11, e0165763. [Google Scholar] [CrossRef] [Green Version]
- Dar, G.H.; Mendes, C.C.; Kuan, W.-L.; Speciale, A.A.; Conceição, M.; Görgens, A.; Uliyakina, I.; Lobo, M.J.; Lim, W.F.; El Andaloussi, S.; et al. GAPDH Controls Extracellular Vesicle Biogenesis and Enhances the Therapeutic Potential of EV Mediated SiRNA Delivery to the Brain. Nat. Commun. 2021, 12, 6666. [Google Scholar] [CrossRef]
- Hu, X.; Li, C.; Wang, Q.; Wei, Z.; Chen, T.; Wang, Y.; Li, Y. Dimethyl Fumarate Ameliorates Doxorubicin-Induced Cardiotoxicity By Activating the Nrf2 Pathway. Front. Pharmacol. 2022, 13, 872057. [Google Scholar] [CrossRef]
- Aengenvoort, J.; Sekeres, M.; Proksch, P.; Fritz, G. Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve CAT-Triggered Tumor Cell Death. Molecules 2022, 27, 3567. [Google Scholar] [CrossRef]
- Wan-Ibrahim, W.I.; Singh, V.A.; Hashim, O.H.; Abdul-Rahman, P.S. Biomarkers for Bone Tumors: Discovery from Genomics and Proteomics Studies and Their Challenges. Mol. Med. 2016, 21, 861–872. [Google Scholar] [CrossRef]
- Stewart, P.A.; Parapatics, K.; Welsh, E.A.; Müller, A.C.; Cao, H.; Fang, B.; Koomen, J.M.; Eschrich, S.A.; Bennett, K.L.; Haura, E.B. A Pilot Proteogenomic Study with Data Integration Identifies MCT1 and GLUT1 as Prognostic Markers in Lung Adenocarcinoma. PLoS ONE 2015, 10, e0142162. [Google Scholar] [CrossRef] [Green Version]
- Corrêa, S.; Panis, C.; Binato, R.; Herrera, A.C.; Pizzatti, L.; Abdelhay, E. Identifying Potential Markers in Breast Cancer Subtypes Using Plasma Label-Free Proteomics. J. Proteom. 2017, 151, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Raffel, S.; Klimmeck, D.; Falcone, M.; Demir, A.; Pouya, A.; Zeisberger, P.; Lutz, C.; Tinelli, M.; Bischel, O.; Bullinger, L.; et al. Quantitative Proteomics Reveals Specific Metabolic Features of Acute Myeloid Leukemia Stem Cells. Blood 2020, 136, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.E.; Kitteringham, N.R.; Greenough, C.; Park, B.K. Albumin and Immunoglobulin Depletion of Human Plasma. Methods Mol. Biol. 2008, 425, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Baralla, A.; Fois, A.G.; Sotgiu, E.; Zinellu, E.; Mangoni, A.A.; Sotgia, S.; Zinellu, A.; Pirina, P.; Carru, C. Plasma Proteomic Signatures in Early Chronic Obstructive Pulmonary Disease. Proteom. Clin. Appl. 2018, 12, e1700088. [Google Scholar] [CrossRef] [PubMed]
- Sande, C.J.; Mutunga, M.; Muteti, J.; Berkley, J.A.; Nokes, D.J.; Njunge, J. Untargeted Analysis of the Airway Proteomes of Children with Respiratory Infections Using Mass Spectrometry Based Proteomics. Sci. Rep. 2018, 8, 13814. [Google Scholar] [CrossRef]
- Lai, X.; Chen, S. Identification of Novel Biomarker Candidates for Immunohistochemical Diagnosis to Distinguish Low-Grade Chondrosarcoma from Enchondroma. Proteomics 2015, 15, 2358–2368. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.W.; Ko, J.; Ju, C.; Eltzschig, H.K. Hypoxia Signaling in Human Diseases and Therapeutic Targets. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Singh, A.; Wilson, J.W.; Schofield, C.J.; Chen, R. Hypoxia-Inducible Factor (HIF) Prolyl Hydroxylase Inhibitors Induce Autophagy and Have a Protective Effect in an in-Vitro Ischaemia Model. Sci. Rep. 2020, 10, 1597. [Google Scholar] [CrossRef] [Green Version]
- Weidemann, A.; Breyer, J.; Rehm, M.; Eckardt, K.-U.; Daniel, C.; Cicha, I.; Giehl, K.; Goppelt-Struebe, M. HIF-1α Activation Results in Actin Cytoskeleton Reorganization and Modulation of Rac-1 Signaling in Endothelial Cells. Cell Commun. Signal. 2013, 11, 80. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.-Y.; Bonelli, S.; Calligaris, M.; Carreca, A.P.; Müller, S.A.; Lichtenthaler, S.F.; Troeberg, L.; Scilabra, S.D. Quantitative Proteomics Reveals That ADAM15 Can Have Proteolytic-Independent Functions in the Steady State. Membranes 2022, 12, 578. [Google Scholar] [CrossRef]
- Scilabra, S.D.; Troeberg, L.; Yamamoto, K.; Emonard, H.; Thøgersen, I.; Enghild, J.J.; Strickland, D.K.; Nagase, H. Differential Regulation of Extracellular Tissue Inhibitor of Metalloproteinases-3 Levels by Cell Membrane-Bound and Shed Low Density Lipoprotein Receptor-Related Protein 1. J. Biol. Chem. 2013, 288, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The Role of Collagen in Cancer: From Bench to Bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the Extracellular Matrix: Drivers of Tumour Metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-Inducible Factors and RAB22A Mediate Formation of Microvesicles That Stimulate Breast Cancer Invasion and Metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular Vesicles in Cancer—Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Shehzad, A.; Islam, S.U.; Shahzad, R.; Khan, S.; Lee, Y.S. Extracellular Vesicles in Cancer Diagnostics and Therapeutics. Pharmacol. Ther. 2021, 223, 107806. [Google Scholar] [CrossRef]
- Venturella, M.; Criscuoli, M.; Carraro, F.; Naldini, A.; Zocco, D. Interplay between Hypoxia and Extracellular Vesicles in Cancer and Inflammation. Biology 2021, 10, 606. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic Enhancement of Exosome Release by Breast Cancer Cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [Green Version]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes Secreted under Hypoxia Enhance Invasiveness and Stemness of Prostate Cancer Cells by Targeting Adherens Junction Molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Wysoczynski, M.; Ratajczak, M.Z. Lung Cancer Secreted Microvesicles: Underappreciated Modulators of Microenvironment in Expanding Tumors. Cancer Cell Biol. 2009, 125, 1595–1603. [Google Scholar] [CrossRef] [Green Version]
- Dorayappan, K.D.P.; Wanner, R.; Wallbillich, J.J.; Saini, U.; Zingarelli, R.; Suarez, A.A.; Cohn, D.E.; Selvendiran, K. Hypoxia-Induced Exosomes Contribute to a More Aggressive and Chemoresistant Ovarian Cancer Phenotype: A Novel Mechanism Linking STAT3/Rab Proteins. Oncogene 2018, 37, 3806–3821. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.S.; Kumar, M.; Chaudhary, S.; Dhiman, A.; Patidar, A.; Jakhar, P.; Jaswal, P.; Sharma, K.; Sheokand, N.; Malhotra, H.; et al. Trafficking of a Multifunctional Protein by Endosomal Microautophagy: Linking Two Independent Unconventional Secretory Pathways. FASEB J. 2019, 33, 5626–5640. [Google Scholar] [CrossRef]
- Sirover, M.A. Pleiotropic Effects of Moonlighting Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) in Cancer Progression, Invasiveness, and Metastases. Cancer Metastasis Rev. 2018, 37, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Lazarev, V.F.; Guzhova, I.V.; Margulis, B.A. Glyceraldehyde-3-Phosphate Dehydrogenase Is a Multifaceted Therapeutic Target. Pharmaceutics 2020, 12, E416. [Google Scholar] [CrossRef] [PubMed]
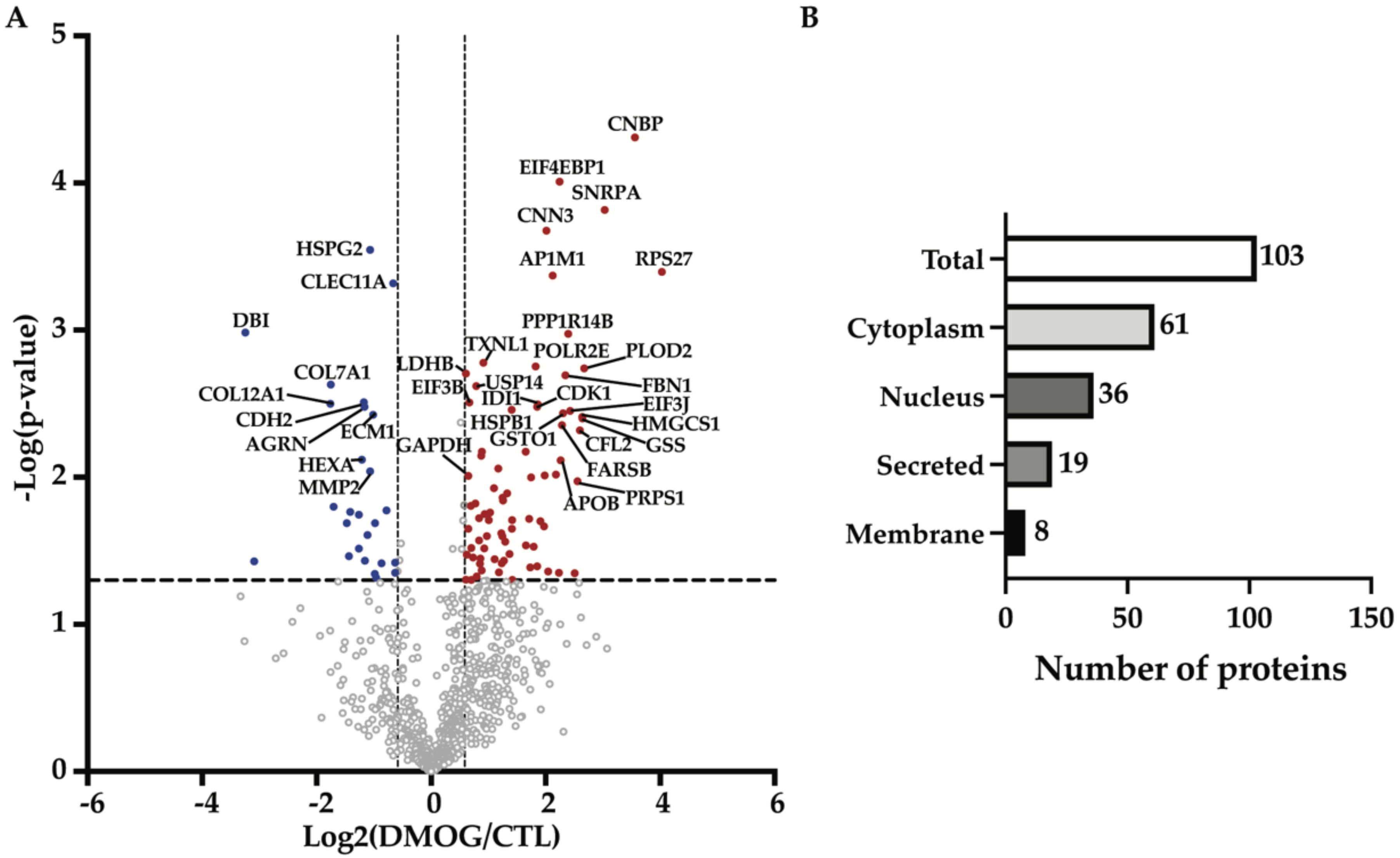
| Protein Name | Protein ID | Gene Name | p-Value | Fold Change | Ratio |
|---|---|---|---|---|---|
| Cellular nucleic acid-binding protein | P62633 | CNBP | 2.04 × 10−4 | 11.783 | 3.559 |
| Eukaryotic translation initiation factor 4E-binding protein 1 | Q13541 | EIF4EBP1 | 1.02 × 10−4 | 4.738 | 2.244 |
| U1 small nuclear ribonucleoprotein A | P09012 | SNRPA | 6.55 × 10−3 | 8.177 | 3.032 |
| Calponin-3 | Q15417 | CNN3 | 4.74 × 10−3 | 4.041 | 2.015 |
| 40S ribosomal protein S27 | P42677 | RPS27 | 2.48 × 10−3 | 16.358 | 4.032 |
| AP-1 complex subunit mu-1 | Q9BXS5 | AP1M1 | 2.35 × 10−3 | 4.356 | 2.123 |
| Protein phosphatase 1 regulatory subunit 14B | Q96C90 | PPP1R14B | 9.45 × 10−2 | 5.259 | 2.395 |
| Thioredoxin-like protein 1 | O43396 | TXNL1 | 5.99 × 10−2 | 1.883 | 0.913 |
| DNA-directed RNA polymerases I, II, and III subunit RPABC1 | P19388 | POLR2E | 5.66 × 10−2 | 3.545 | 1.826 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | O00469 | PLOD2 | 5.50 × 10−2 | 6.383 | 2.674 |
| L-lactate dehydrogenase B chain | P07195 | LDHB | 5.07 × 10−2 | 1.525 | 0.609 |
| Fibrillin-1 | P35555 | FBN1 | 4.93 × 10−2 | 5.079 | 2.345 |
| Ubiquitin carboxyl-terminal hydrolase 14 | P54578 | USP14 | 4.16 × 10−2 | 1.73 | 0.790 |
| Eukaryotic translation initiation factor 3 subunit B | P55884 | EIF3B | 3.22 × 10−2 | 1.590 | 0.669 |
| Isopentenyl-diphosphate Delta-isomerase 1 | Q13907 | IDI1 | 3.13 × 10−2 | 3.634 | 1.862 |
| Cyclin-dependent kinase 1 | P06493 | CDK1 | 3.01 × 10−2 | 3.611 | 1.852 |
| Heat shock protein b-1 | P04792 | HSPB1 | 2.87 × 10−2 | 2.650 | 1.406 |
| Eukaryotic translation initiation factor 3 subunit J | O75822 | EIF3J | 2.83 × 10−2 | 5.377 | 2.427 |
| Glutathione S-transferase omega-1 | P78417 | GSTO1 | 2.72 × 10−2 | 4.930 | 2.302 |
| Hydroxymethylglutaryl-CoA synthase, cytoplasmic | Q01581 | HMGCS1 | 2.57 × 10−2 | 6.231 | 2.639 |
| Glutathione synthetase | P48637 | GSS | 2.51 × 10−2 | 6.250 | 2.644 |
| Phenylalanine--tRNA ligase beta subunit | Q9NSD9 | FARSB | 2.26 × 10−2 | 4.877 | 2.286 |
| Cofilin-2 | Q9Y281 | CFL2 | 2.08 × 10−2 | 6.046 | 2.596 |
| Leucine-rich repeat flightless-interacting protein 1 | Q32MZ4 | LRRFIP1 | 1.49 × 10−2 | 1.847 | 0.886 |
| X-ray repair cross-complementing protein 5 | P13010 | XRCC5 | 1.49 × 10−2 | 3.137 | 1.649 |
| UMP-CMP kinase | P30085 | CMPK1 | 1.40 × 10−2 | 1.833 | 0.875 |
| Apolipoprotein B-100 | P04114 | APOB | 1.30 × 10−2 | 4.802 | 2.264 |
| Eukaryotic translation initiation factor 4 gamma 1 | Q04637 | EIF4G1 | 1.15 × 10−2 | 2.2550 | 1.173 |
| Actin-related protein 2/3 complex subunit 3 | O15145 | ARPC3 | 1.04 × 10−2 | 4.534 | 2.181 |
| 60S ribosomal protein L8 | P62917 | RPL8 | 1.03 × 10−2 | 3.955 | 1.984 |
| Glyceraldehyde-3-phosphate dehydrogenase | P04406 | GAPDH | 1.02 × 10−2 | 1.565 | 0.646 |
| Protein Name | Protein ID | Gene Name | p-Value | Fold Change | Ratio |
|---|---|---|---|---|---|
| Basement membrane-specific heparan sulfate proteoglycan core protein 2 | P98160 | HSPG2 | 3.51 × 10−3 | 0.477 | 1.069 |
| C-type lectin domain family 11 member A | Q9Y240 | CLEC11A | 2.08 × 10−3 | 0.632 | 0.663 |
| Collagen alpha-1(VII) chain | Q02388 | COL7A1 | 4.26 × 10−2 | 0.297 | 1.753 |
| Cadherin-2 | P19022 | CDH2 | 3.23 × 10−2 | 0.442 | 1.178 |
| Collagen alpha-1(XII) chain | Q99715 | COL12A1 | 3.16 × 10−2 | 0.295 | 1.760 |
| Agrin | O00468 | AGRN | 3.01 × 10−2 | 0.446 | 1.166 |
| Extracellular matrix protein 1 | Q16610 | ECM1 | 2.65 × 10−2 | 0.495 | 1.013 |
| Beta-hexosaminidase subunit alpha | P06865 | HEXA | 1.31 × 10−2 | 0.433 | 1.209 |
| 72 kDa type IV collagenase | P08253 | MMP2 | 1.10 × 10−2 | 0.478 | 1.066 |
| TRANSFAC | ||
|---|---|---|
| Term | p-Value | Proteins |
| HIF1A (human) | 1.50 × 10−4 | EIF5A, PRPS1, SNRPN, DBNL, RPS6, CSTF2, TXNL1, PLOD2, ACTN4, PPM1G, LDHB, TUBA1C, PSMC3, OAF, POLR2E, MAPK1, MCM6, EIF3A, EIF4G1, AP1M1 |
| GATA1 (human) | 3.76 × 10−4 | APP, IDI1, PRPS1, ECM1, CSTF2, PPM1G, SCRN1, CFL2, CAPN2, CCT8, PDLIM5, EIF5A, NUDC, SNRPN, DBNL, HMGCS1, XRCC5, NONO, MMP2, RPS6, TXNL1, CLEC11A, HSPG2, ACTA1, CDK1, CMPK1, EIF4G1 |
| GO Biological Process 2021 | ||
|---|---|---|
| Term | p-Value | Proteins |
| Extracellular matrix organization (GO:0030198) | 4.81 × 10−9 | APP, MMP2, COL12A1, TNC, FN1, PLOD2, NID1, HSPG2, COL7A1, CAPN2, AGRN, PRSS2, FBN1 |
| Cellular protein metabolic process (GO:0044267) | 3.01 × 10−8 | APP, MMP2, RPS6, TNC, FN1, PLOD2, PLAT, RPL8, RPS27, CDH2, APOB, EIF4G1, FARSB, FBN1 |
| GO Cellular Component 2021 | ||
|---|---|---|
| Term | p-Value | Proteins |
| Secretory granule lumen (GO:0034774) | 8.95 × 10−9 | LGALS3BP, APP, ECM1, DBNL, XRCC5, TUBB, FN1, ACTN4, PSMC3, IMPDH2, MAPK1, CCT8, PRSS2 |
| Intracellular organelle lumen (GO:0070013) | 5.79 × 10−8 | APP, DBNL, OAT, GSR, COL12A1, TNC, FN1, DBI, HSPG2, PSMC3, CDH2, COL7A1, IMPDH2, CDK1, MAPK1, CCT8, AGRN, APOB, FBN1 |
| Collagen-containing extracellular matrix (GO:0062023) | 7.84 × 10−8 | LGALS3BP, ECM1, MMP2, COL12A1, TNC, FN1, PLAT, NID1, HSPG2, CDH2, COL7A1, AGRN, FBN1 |
| Focal adhesion (GO:0005925) | 9.68 × 10−8 | TNC, HSPB1, ACTN4, RPL8, HSPG2, CNN3, PROCR, CDH2, ARPC3, CAPN2, ITGBL1, MAPK1, TLN1 |
| Jensen COMPARTMENT | ||
|---|---|---|
| Term | p-Value | Proteins |
| Extracellular exosome | 1.91 × 10−22 | LGALS3BP, APP, EIF4A1, ECM1, COL12A1, HEXA, HSPB1, PLOD2, PLAT, DBI, TUBB6, TUBA1A, CDH2, CFL2, CAPN2, EIF5A, DBNL, GSTO1, TUBB, EPDR1, ACTN4, HSPG2, SND1, DDB1, PROCR, ACTA1, MTHFD1, OAF, CMPK1, TLN1, GART, GAPDH, USP14, RAB1B, NID1, LDHB, PCBP1, MAPK1, CCT8, APOB, AP1M1, HSPA4, GSS, GSR, TXNL1, FN1, GGCT, ARPC3, IMPDH2, CDK1, ACO1, AGRN, RBMX, FBN1, AARS, EIF3B |
| Extracellular vesicle | 2.45 × 10−22 | LGALS3BP, APP, EIF4A1, ECM1, COL12A1, HEXA, HSPB1, PLOD2, PLAT, DBI, TUBB6, TUBA1A, CDH2, CFL2, CAPN2, EIF5A, DBNL, GSTO1, TUBB, EPDR1, ACTN4, HSPG2, SND1, DDB1, PROCR, ACTA1, MTHFD1, OAF, CMPK1, TLN1, GART, GAPDH, USP14, RAB1B, NID1, LDHB, PCBP1, MAPK1, CCT8, APOB, AP1M1, HSPA4, GSS, GSR, TXNL1, FN1, GGCT, ARPC3, IMPDH2, CDK1, ACO1, AGRN, RBMX, FBN1, AARS, EIF3B |
| Extracellular organelle | 2.49 × 10−22 | LGALS3BP, APP, EIF4A1, ECM1, COL12A1, HEXA, HSPB1, PLOD2, PLAT, DBI, TUBB6, TUBA1A, CDH2, CFL2, CAPN2, EIF5A, DBNL, GSTO1, TUBB, EPDR1, ACTN4, HSPG2, SND1, DDB1, PROCR, ACTA1, MTHFD1, OAF, CMPK1, TLN1, GART, GAPDH, USP14, RAB1B, NID1, LDHB, PCBP1, MAPK1, CCT8, APOB, AP1M1, HSPA4, GSS, GSR, TXNL1, FN1, GGCT, ARPC3, IMPDH2, CDK1, ACO1, AGRN, RBMX, FBN1, AARS, EIF3B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spanò, D.P.; Bonelli, S.; Calligaris, M.; Carreca, A.P.; Carcione, C.; Zito, G.; Nicosia, A.; Rizzo, S.; Scilabra, S.D. High-Resolution Secretome Analysis of Chemical Hypoxia Treated Cells Identifies Putative Biomarkers of Chondrosarcoma. Proteomes 2022, 10, 25. https://doi.org/10.3390/proteomes10030025
Spanò DP, Bonelli S, Calligaris M, Carreca AP, Carcione C, Zito G, Nicosia A, Rizzo S, Scilabra SD. High-Resolution Secretome Analysis of Chemical Hypoxia Treated Cells Identifies Putative Biomarkers of Chondrosarcoma. Proteomes. 2022; 10(3):25. https://doi.org/10.3390/proteomes10030025
Chicago/Turabian StyleSpanò, Donatella Pia, Simone Bonelli, Matteo Calligaris, Anna Paola Carreca, Claudia Carcione, Giovanni Zito, Aldo Nicosia, Sergio Rizzo, and Simone Dario Scilabra. 2022. "High-Resolution Secretome Analysis of Chemical Hypoxia Treated Cells Identifies Putative Biomarkers of Chondrosarcoma" Proteomes 10, no. 3: 25. https://doi.org/10.3390/proteomes10030025






