Enhanced Synthesis of Antioxidant Enzymes, Defense Proteins and Leghemoglobin in Rhizobium-Free Cowpea Roots after Challenging with Meloydogine incognita
Abstract
:1. Introduction
2. Experimental Section
2.1. Nematode Inoculums
2.2. Plant Material and Nematode Inoculation
2.3. Extraction of Proteins from the Cowpea Roots
2.4. Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-SDS-PAGE)
2.5. In-Gel Digestion
2.6. Mass Spectrometry Analysis
2.7. Gene Expression Analysis
3. Results
3.1. Proteomic Analysis of RKN-Inoculated and Non-Inoculated Cowpea Roots
| Target Gene (GenBank Accession Number) | Oligonucleotide Sequences 2 | Position 3 | Amplicon Size (bp) | Annealing Temperature (°C)/Cycle | Reference |
|---|---|---|---|---|---|
| Asparaginyl endopeptidase (D89971) | 5'-AACGGCTATTGGAACTAC-3' (f) | 217–234 | 876 | 52.5/35 | This work |
| 5'-GAGATCAGCATCCCTTTG-3' (r) | 1074–1092 | ||||
| ACC synthase (Z12135) | 5'-CAAATGGGTCTTGCTGAGAAT-3' (f) | 70–90 | 858 | 58.9/20 | This work |
| 5'-TCTCAGCCTCTCCCTGTT-3' (r) | 910–927 4 | ||||
| ARG 10 (AB012110) | 5'-CGAACACCATCGCCAAAG-3' (f) | 104–121 | 498 | 56.4/35 | This work |
| 5'-AGGGAAAGAAGCAAGCGA-3' (r) | 584–601 | ||||
| Chalcone-flavanone isomerase (AB073787) | 5'-GAGAGGGGTTGACGATT-3' (f) | 112–129 | 477 | 54.2/35 | This work |
| 5'-GCCAATCATCGTCTCCAA-3' (r) | 571–588 | ||||
| Cysteinyl endopeptidase (U49445) | 5'-TACGAGAGATGGAGGAGT-3' (f) | 119–136 | 765 | 51.2/25 | This work |
| 5'-TCCGACAATTGCTACACC-3' (r) | 866–883 | ||||
| Leghemoglobin (U33205) | 5'-ATGGTTGCTTTCTCTGACAAG-3' (f) | 46–66 | 375 | 64.7/35 | This work |
| 5'-TTCATCACTCCATTTGTCTCC-3' (r) | 400–420 | ||||
| CuZn-superoxide dismutase (AJ278668) | 5'-AAAGCGGTGGCGGTGCTGAAA-3' (f) | 195–215 | 393 | 61.0/35 | This work |
| 5'-GCTCAGTTCATGGCCGCCTTT-3' (r) | 567–587 | ||||
| nodC (AE006469) | 5'-TGATYGAYATGGARTAYTGGCT-3' (f) | 545–566 | 640 | 55.6/35 | Sarita et al. 2005 [21] |
| 5'-CGYGACARCCARTCGCTRTTG-3' (r) | 1164–1184 | ||||
| Chitinase I (X88800) | 5'-AGGATGATATGGAGCGTAGC-3' (f) | 14–33 | 972 | 58.0/28 | This work |
| 5'-GACAGGGTGAGATGTAGATC-3' (r) | 966–985 | ||||
| Chitinase IIIa (X88802) | 5'-CTATCAACAACTGCAACGTG-3' (f) | 152–171 | 576 | 55.0/28 | This work |
| 5'-ATTTGGAAGAACCCTTGATG-3' (r) | 708–727 | ||||
| Chitinase IIIb (X88801) | 5'-ACGTCAACATAGCTTTCCTC-3' (f) | 175–194 | 563 | 55.0/28 | This work |
| 5'-CTTCCCAGCAGGTACTGTAC-3' (r) | 718–737 | ||||
| Chitinase IV (X88803) | 5'-GCTCAGAACTGTGGTTGTGC-3' (f) | 8–27 | 743 | 57.0/28 | This work |
| 5'-TAGCAAGTAAGATTATCACC-3' (r) | 731–750 | ||||
| Actin (AF143208) | 5'-GCGTGATCTCACTGATGC-3' (f) | 669–686 | 530 | 59.0/35 | Costa et al., 2004 [23] |
| 5'-TCGCAATCCACATCTGTTGG-3' (r) | 1179–1198 |
|
|
|
|
|
|
|
|
|
|
|
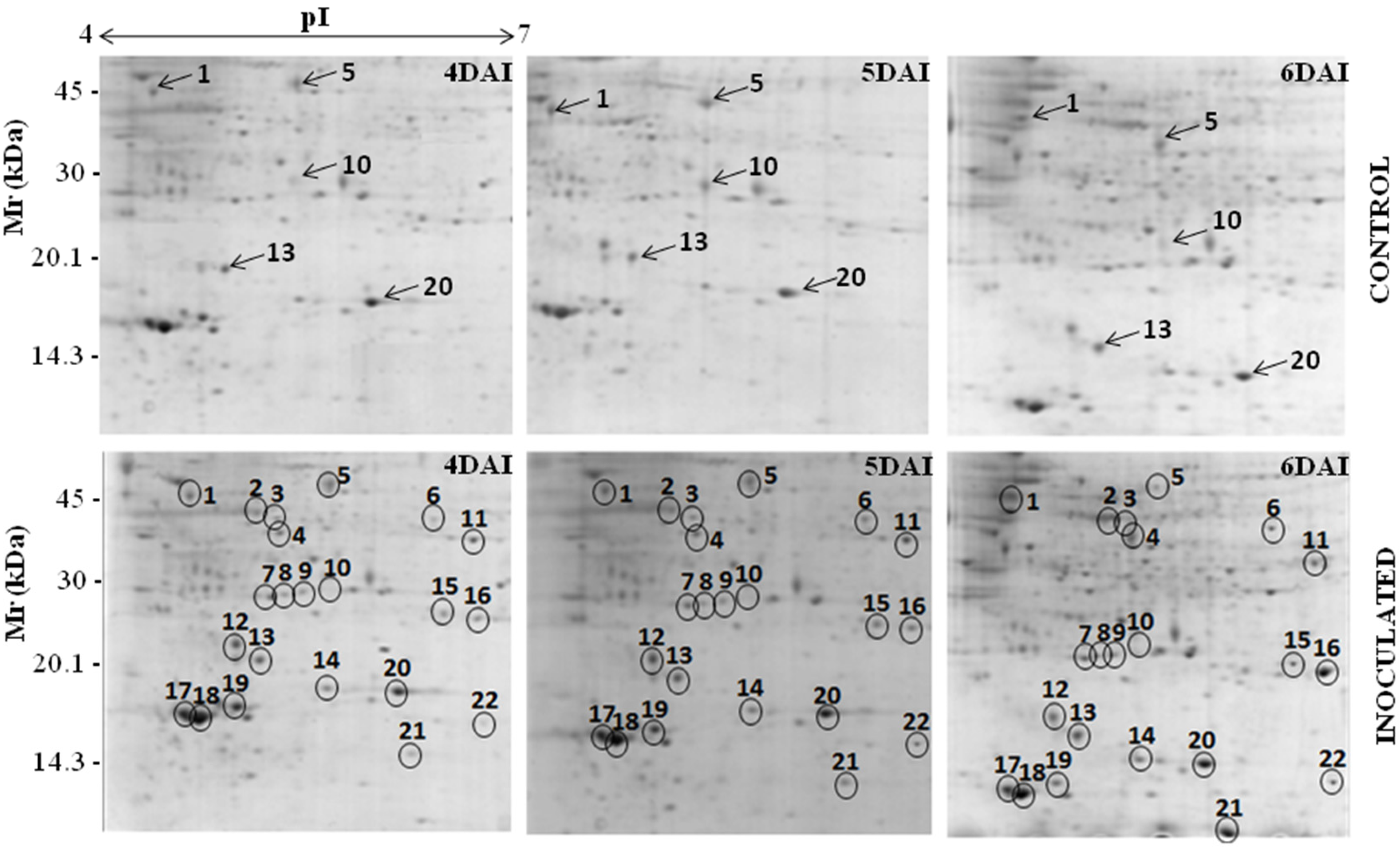
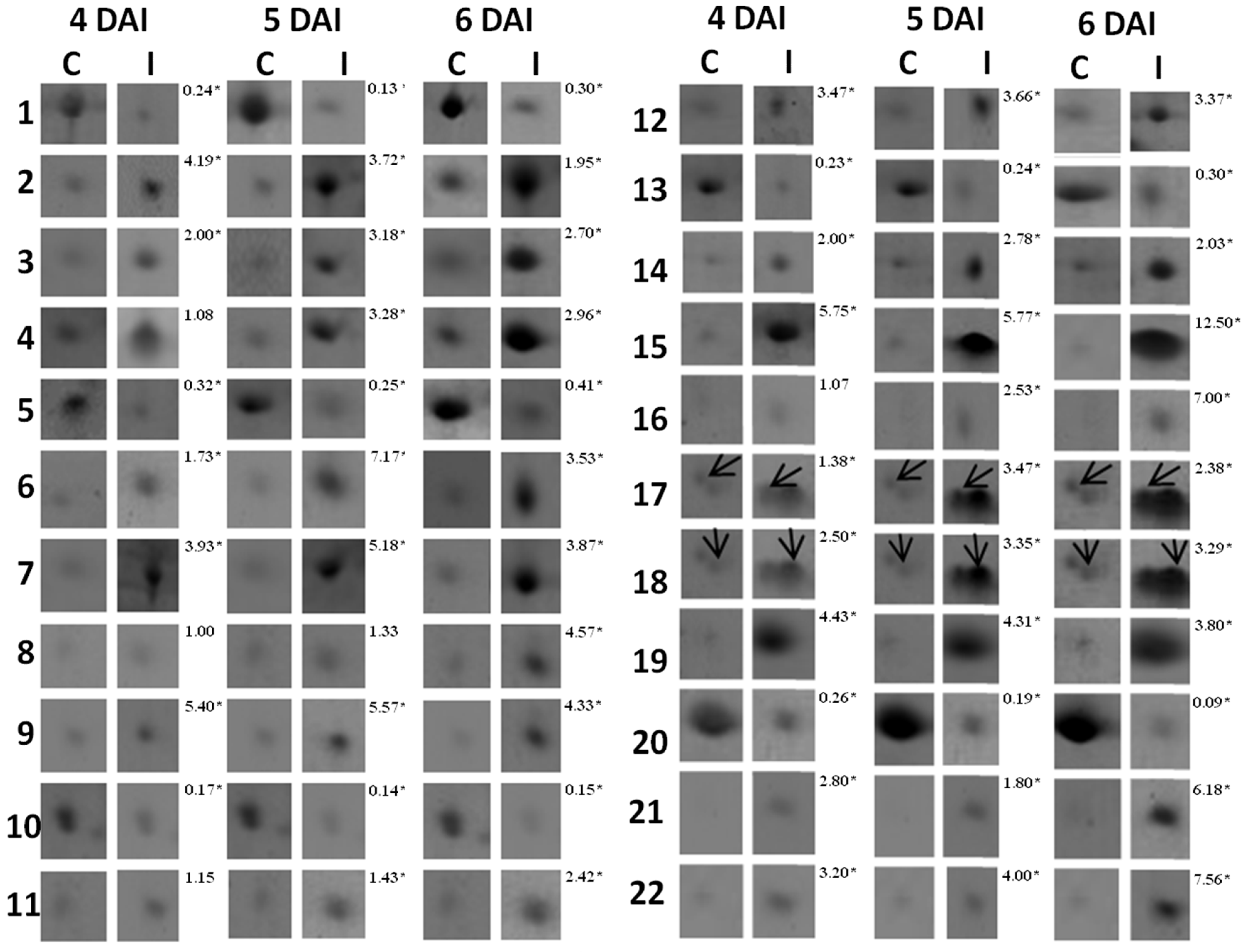
| Spot *No. | Accession No. (NCBI) | Protein Identification | Organism | pI/MW (kDa) | Score | Sequence Covered (%) | Sequences | |
|---|---|---|---|---|---|---|---|---|
| Experimental | Theoretical | |||||||
| Functional category: Disease/defense | ||||||||
| 2↑ | gi/297493 | ACC synthase | Vigna radiata | 4.89/39.678 | 5.40/43.768 | 79 | 7.98 | YFDGWK |
| VHIVYSLSK | ||||||||
| VGTIYSYNDSVVTTAR | ||||||||
| 3↑ | gi/86197901 | ACC oxidase | Vigna radiate | 5.05/38.056 | 5.86/40.236 | 111 | 12.30 | GAAMEMIK |
| EMVANK | ||||||||
| VSNYPPCPTPDLIK | ||||||||
| DDQWIDVPPMR | ||||||||
| 11↑ | gi/42795352 | Ascorbate peroxidase | Vigna unguiculata | 6.62/28238 | 6.67/31.746 | 108 | 11.84 | NCAPLMLR |
| EIVALSGGHTLGR | ||||||||
| SGFDGPWTEDPLK | ||||||||
| 15↑ | gi/13274148 | CuZn-superoxide dismutase | Populus tremula | 6.76/23.114 | 5.87/29.203 | 142 | 14.74 | AVAVLK |
| LTHGAPEDEIR | ||||||||
| GGHELSSTTGNAGGR | ||||||||
| 16↑ | gi/123539 | 17.5 kDa HSP | Glycine max | 5.87/22.268 | 6.12/17.412 | 177 | 18.18 | DFHVPTSSVSAENSAFVSTR |
| VLQISGER | ||||||||
| 17↑ | gi/130829 | PR-1 | Phaseolus vulgaris | 4.44/16.126 | 4.83/16.528 | 168 | 18.58 | ALPDSFK |
| ISFVEDGETK | ||||||||
| LSDGPNGGSLIK | ||||||||
| 18↑ | gi/4850337 | PR-3 (Chitinase) | Vigna unguiculata | 4.55/15.971 | 4.75/16.265 | 122 | 14.28 | ISFLEDGETK |
| LSDGSNGGSVVK | ||||||||
| 19↑ | gi/ 130835 | PR-2 (β-1,3-glucanase) | Phaseolus vulgaris | 5.11/16.352 | 4.85/16.402 | 86 | 10.96 | ISIDSK |
| GDAPPNEDELK | ||||||||
| Functional category: Secondary metabolism | ||||||||
| 6↑ | gi/27530706 | Chalcone-flavone isomerase | Lotus japonicus | 6.41/35666 | 5.94 /36.987 | 97 | 11.11 | SYFLGGAGER |
| STGTYGEAEAAAIGK | ||||||||
| Functional category: Metabolism | ||||||||
| 21↑ | gi/ 20138591 | Leghemoglobin | Vigna unguiculata | 6.00/14.722 | 5.68/15.341 | 225 | 31.03 | ADIPK |
| NLFSFLANGVDATNPK | ||||||||
| ASGGVVADAALGAVHSQK | ||||||||
| EAVGDK | ||||||||
| 22↑ | gi/ 26245403 | Nucleoside diphosphate Kinase | Glycine max | 6.89/15.964 | 6.30/16.254 | 183 | 21.08 | PDGVQSGLIGEIISR |
| IIGATNPAQSEPGTIR | ||||||||
| Functional category: Protein destination and storage | ||||||||
| 1↓ | gi/ 4589396 | Asparaginyl endopeptidase | Vigna mungo | 4.38/49.316 | 5.14/52.982 | 77 | 6.83 | FPIIFVVANLITLVSGGR |
| NSLVPPSK | ||||||||
| APLGSSR | ||||||||
| 4↑ | gi/ 1223922 | Cysteinyl endopeptidase | Vigna radiata | 5.10/35.289 | 5.44/37.332 | 64 | 6.35 | LLWVVLSLSLVLGVANSFDFHEK |
| Functional category: Unknown/Predicted/Uncharacterized | ||||||||
| 7↑ | gi/297849580 | Predicted protein | Arabidopsis lyrata | 5.03/25.370 | 6.22/33.633 | 119 | 12.58 | EVETLPEEAFEEEEDK |
| EILENHGGEER | ||||||||
| IMDEAVNASR | ||||||||
| 9↑ | gi/ 297824991 | Predicted protein | Arabidopsis lyrata | 5.26/25.967 | 7.97/39.819 | 58 | 5.54 | QVVDETEPK |
| VYGSIEEHYYR | ||||||||
| 10↓ | gi/2970051 | ARG 10 | Vigna radiata | 5.33/30.326 | 5.62/25.480 | 133 | 14.34 | DEIFCLFEGALDNLGSLR |
| VVCHLSGSFAFIVFDK | ||||||||
| 12↑ | gi/416640 | Auxin induced protein | Vigna radiata | 4.81/19.354 | 4.65/21.345 | 104 | 11.34 | EGLGLEITELR |
| GYSDLAFALEK | ||||||||
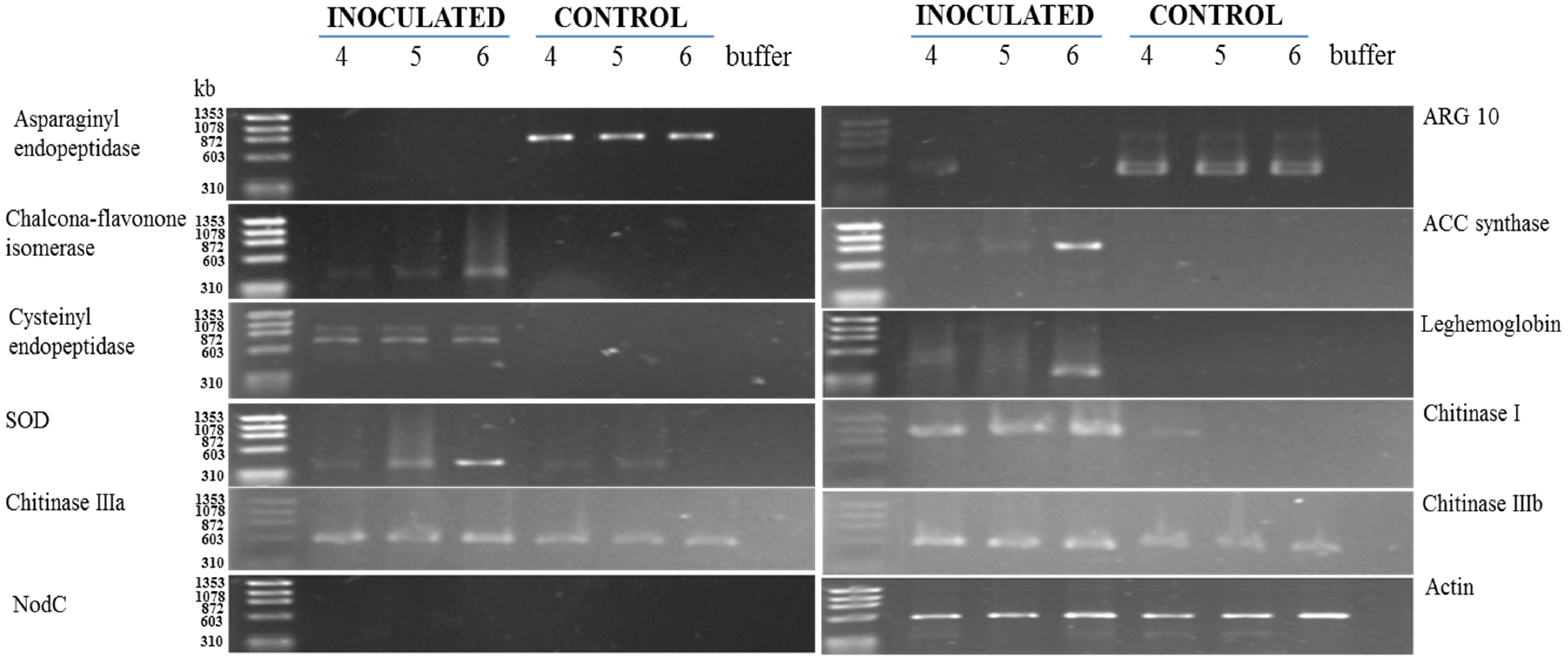
3.2. RT-PCR Analyses of Cowpea Root Gene Expression
4. Discussion
5. Conclusions

Acknowledgments
Author Contributions
Conflicts of Interest
References
- Castagnone-Sereno, P.; Danchin, E.G.J.; Perfus-Barbeoch, L.; Abad, P. Diversity and evolution of root-knot nematodes, genus Meloidogyne: New insights from the genomic era. Annu. Rev. Phytopathol. 2013, 51, 203–220. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Ralph, P. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011, 16, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Quentin, M.; Abad, P.; Favery, B. Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 2013, 4, e53. [Google Scholar] [CrossRef]
- Gheysen, G.; Mitchum, M.G. How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 2011, 14, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Bostock, R.M. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann. Bot. 2002, 89, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Barna, B.; Fodor, J.; Harrach, B.D.; Pogány, M.; Király, Z. The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 2012, 59, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Transcriptional profiling of root-knot nematode induced feeding sites in cowpea (Vigna unguiculata L. Walp.) using a soybean genome array. BMC Genomics 2010, 11, e480. [Google Scholar] [CrossRef]
- Souza e Silva, S.M.; Maia, J.M.; Araújo, Z.B.; Freire-Filho, F.R. Chemical Composition of 45 Cowpea [Vigna unguiculata (L.) Walp.] Genotypes; technical report; The Brazilian Agricultural Research Corporation, Embrapa Meio-Norte: Teresina, Brazil, 2002; Volume 149, pp. 1–2. [Google Scholar]
- Ehlers, J.D.; Matthews, W.C.; Hall, A.E.; Roberts, P.A. Breeding and evaluation of cowpeas with high levels of broad-based resistance to root-knot nematodes. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production, Proceedings of the World Cowpea Conference III, Ibadan, Nigeria, 4–8 September 2000; Fatokun, C.A., Tarawali, S.A., Singh, B.B., Kormawa, P.M., Tamo, M., Eds.; International Institute of Tropical Agriculture (IITA): Ibadan, Nigeria, 2002; pp. 41–51. [Google Scholar]
- Oliveira, J.T.A.; Andrade, N.C.; Martins-Miranda, A.S.; Soares, A.A.; Gondim, D.M.F.; Araújo-Filho, J.H.; Freire-Filho, F.R.; Vasconcelos, I.M. Differential expression of antioxidant enzymes and PR-proteins in compatible and incompatible interactions of cowpea (Vigna unguiculata) and the root-knot nematode Meloidogyne incognita. Plant Physiol. Biochem. 2012, 51, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Silveira, J.A.G.; Costa, R.C.L.; Oliveira, J.T.A. Drought-induced effects and recovery of nitrate assimilation and nodule activity in cowpea plants inoculated with Bradyrhizobium spp. under moderate nitrate level. Braz. J. Microbiol. 2001, 32, 187–194. [Google Scholar] [CrossRef]
- Hussey, R.S.; Davis, E.L.; Baum, T.J. Secrets in secretions: Genes that control nematode parasitism of plants. Braz. J. Plant Physiol. 2002, 14, 183–194. [Google Scholar] [CrossRef]
- Atkinson, H.J. Nematodes. In Molecular Plant Pathology; Gurr, S.J., McPherson, M.J., Bowles, D.J., Eds.; Oxford University Press: Oxford, UK, 1992; Volume 1, pp. 99–107. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 39, 809–821. [Google Scholar]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; Volume 1, pp. 7.1–7.87. [Google Scholar]
- Sarita, S.; Sharma, P.K.; Priefer, U.B.; Prell, J. Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol. Ecol. 2005, 54, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Warner, S. Genomic DNA isolation and lambda library construction. In Plant Gene Isolation: Principles and Practice; Foster, G.D., Twell, D., Eds.; John Wiley & Sons Ltd: Chichester, UK, 1996; pp. 51–53. [Google Scholar]
- Costa, J.H.; Hasenfratz-Sauder, M.P.; Pham-Thi, A.T.; Silva Lima, M.G.; Dizengremel, P.; Jolivet, Y.; Fernandes de Melo, D. Identification in Vigna unguiculata (L.) Walp. of two cDNAs encoding mitochondrial alternative oxidase orthologous to soybean alternative oxidase genes 2a and 2b. Plant Sci. 2004, 167, 233–239. [Google Scholar] [CrossRef]
- Twyman, R.M. Strategies for protein quantitation. In Principles of Proteomics, 2nd ed.; Garland Science, Taylor & Francis Group, LLC: New York, NY, USA, 2014; pp. 69–86. [Google Scholar]
- Xu, W.; Li, L.; Du, L.; Tan, N. Various mechanisms in cyclopeptide production from precursors synthesized independently of non-ribosomal peptide synthetases. Acta Biochem. Biophys. Sin. 2011, 43, 757–762. [Google Scholar] [CrossRef]
- Craik, D.J. Host-defense activities of cyclotides. Toxins 2012, 4, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Bellafiore, S.; Shen, Z.; Rosso, M.-N.; Abad, P.; Shih, P.; Briggs, S.P. Direct identification of the meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog. 2008, 4, e1000192. [Google Scholar] [CrossRef] [PubMed]
- Curtis, R.; Buttle, D.; Behnke, J.; Duce, I.; Shewry, P.; Kurup, S.; Kerry, B.; Kerry, M. Nematicidal effects of cysteine proteinases and methods of use thereof to treat nematode infestation. WO2008087555 A2, 24 July 2008. [Google Scholar]
- Damiani, I.; Baldacci-Cresp, F.; Hopkins, J.; Andrio, E.; Balzergue, S.; Lecomte, P.; Puppo, A.; Abad, P.; Favery, B.; Hérouart, D. Plant genes involved in harbouring symbiotic rhizobia or pathogenic nematodes. New Phytol. 2012, 194, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Glazer, I.; Epstein, E.; Orion, D.; Apelbaum, A. Interactions between auxin and ethylene in root-knot nematode (Meloidogyne javanica) infected tomato roots. Physiol. Mol. Plant Pathol. 1986, 28, 171–179. [Google Scholar] [CrossRef]
- Gutierrez, O.A.; Wubben, M.J.; Howard, M.; Roberts, B.; Hanlon, E.; Wilkinson, J.R. The role of phytohormones ethylene and auxin in plant-nematode interactions. Russ. J. Plant Physiol. 2009, 56, 1–5. [Google Scholar] [CrossRef]
- Neil, S.J.; Desikan, R.; Clarke, A.; Hurst, R.D.; Hancock, J.T. Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 2002, 53, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Vanderspool, M.C.; Kaplan, D.T.; McCollum, T.G.; Wodzinski, R.J. Partial characterization of cytosolic superoxide dismutase activity in the interaction of Meloidogyne incognita with two cultivars of Glycine max. J. Nematol. 1994, 26, 422–429. [Google Scholar] [PubMed]
- Ferrer, J.-L.; Austin, M.B.; Stewart, C., Jr.; Noel, J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Hallmann, J.; Kokalis-Burelle, N.; Waever, D.B.; Rodríguez-Kábana, R.; Tuzun, S. Activity and differential induction of chitinase isozymes in soybean cultivars resistant or susceptible to root-knot nematodes. J. Nematol. 1997, 29, 523–530. [Google Scholar] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Williams, K.L.; Nevalainen, H.K.M. Effects of Paecilomyces lilacinus protease and chitinase on the eggshell structures and hatching of Meloidogyne javanica juveniles. Biol. Control 2004, 31, 346–352. [Google Scholar] [CrossRef]
- Hammargren, J.; Sundström, J.; Johansson, M.; Bergman, P.; Knorpp, C. On the phylogeny, expression and targeting of plant nucleoside diphosphate kinases. Physiol. Plantarum 2007, 129, 79–89. [Google Scholar] [CrossRef]
- Steeg, P.S.; Bevilacqua, G.; Kopper, L.; Thorgeirsson, U.P.; Talmadge, J.E.; Liotta, L.A.; Sobel, M.E. Evidence for a novel gene associated with low tumor metastatic potential. J. Natl. Cancer Inst. 1988, 80, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.; van Dongen, J.T.; Gunther, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvarvi, M.K. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.A.; Husain, S.I. Proline and leghaemoglobin contents and water absorption capability of cowpea roots as influenced by infection with Rotylenchus reniformis, Meloidogyne incognita and Rhizoctonia solani. Nematol. Medit. 1989, 17, 135–137. [Google Scholar]
- Job, D.; Zeba, B.; Puppo, A.; Rigaud, J. Kinetic studies of the reaction of ferric soybean leghemoglobins with hydrogen peroxide, cyanide and nicotinic acid. Eur. J. Biochem. 1980, 107, 491–500. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, J.T.A.; Araujo-Filho, J.H.; Grangeiro, T.B.; Gondim, D.M.F.; Segalin, J.; Pinto, P.M.; Carlini, C.R.R.S.; Silva, F.D.A.; Lobo, M.D.P.; Costa, J.H.; et al. Enhanced Synthesis of Antioxidant Enzymes, Defense Proteins and Leghemoglobin in Rhizobium-Free Cowpea Roots after Challenging with Meloydogine incognita . Proteomes 2014, 2, 527-549. https://doi.org/10.3390/proteomes2040527
Oliveira JTA, Araujo-Filho JH, Grangeiro TB, Gondim DMF, Segalin J, Pinto PM, Carlini CRRS, Silva FDA, Lobo MDP, Costa JH, et al. Enhanced Synthesis of Antioxidant Enzymes, Defense Proteins and Leghemoglobin in Rhizobium-Free Cowpea Roots after Challenging with Meloydogine incognita . Proteomes. 2014; 2(4):527-549. https://doi.org/10.3390/proteomes2040527
Chicago/Turabian StyleOliveira, Jose T. A., Jose H. Araujo-Filho, Thalles B. Grangeiro, Darcy M. F. Gondim, Jeferson Segalin, Paulo M. Pinto, Celia R. R. S. Carlini, Fredy D. A. Silva, Marina D. P. Lobo, Jose H. Costa, and et al. 2014. "Enhanced Synthesis of Antioxidant Enzymes, Defense Proteins and Leghemoglobin in Rhizobium-Free Cowpea Roots after Challenging with Meloydogine incognita " Proteomes 2, no. 4: 527-549. https://doi.org/10.3390/proteomes2040527
APA StyleOliveira, J. T. A., Araujo-Filho, J. H., Grangeiro, T. B., Gondim, D. M. F., Segalin, J., Pinto, P. M., Carlini, C. R. R. S., Silva, F. D. A., Lobo, M. D. P., Costa, J. H., & Vasconcelos, I. M. (2014). Enhanced Synthesis of Antioxidant Enzymes, Defense Proteins and Leghemoglobin in Rhizobium-Free Cowpea Roots after Challenging with Meloydogine incognita . Proteomes, 2(4), 527-549. https://doi.org/10.3390/proteomes2040527