Proteomic Profiling of Primary Human Acute Myeloid Leukemia Cells Does Not Reflect Their Constitutive Release of Soluble Mediators
Abstract
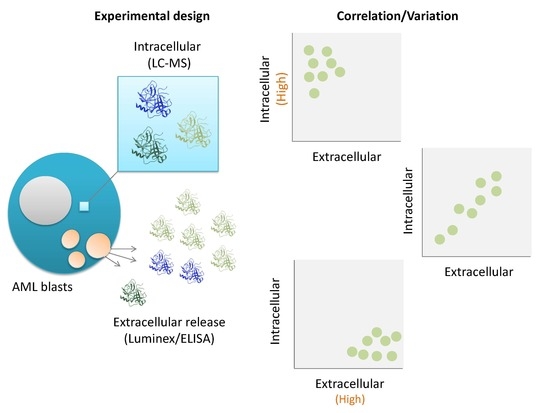
:1. Introduction
2. Materials and Methods
2.1. Collection and Preparation of Primary AML Patient Cells
2.2. Protein Analyses and In Vitro Culture
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Ehninger, A.; Trumpp, A. The bone marrow stem cell niche grows up: Mesenchymal stem cells and macrophages move in. J. Exp. Med. 2011, 208, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Aasebø, E.; Hernandez-Valladares, M.; Tsykunova, G.; Reikvam, H. Therapeutic targeting of leukemic stem cells in acute myeloid leukemia—The biological background for possible strategies. Expert Opin. Drug Discov. 2017, 12, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Brenner, A.K.; Hagen, K.M.; Liseth, K.; Skrede, S.; Hatfield, K.J.; Bruserud, Ø. The cytokine-mediated crosstalk between primary human acute myeloid cells and mesenchymal stem cells alters the local cytokine network and the global gene expression profile of the mesenchymal cells. Stem Cell Res. 2015, 15, 530–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, A.K.; Nepstad, I.; Bruserud, Ø. Mesenchymal Stem Cells Support Survival and Proliferation of Primary Human Acute Myeloid Leukemia Cells through Heterogeneous Molecular Mechanisms. Front. Immunol. 2017, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Barrett, A.J.; Dutra, A.; Pak, E.; Miner, S.; Keyvanfar, K.; Hensel, N.F.; Rezvani, K.; Muranski, P.; Liu, P.; et al. Long term maintenance of myeloid leukemic stem cells cultured with unrelated human mesenchymal stromal cells. Stem Cell Res. 2015, 14, 95–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löwenberg, B.; van Putten, W.L.; Touw, I.P.; Delwel, R.; Santini, V. Autonomous proliferation of leukemic cells in vitro as a determinant of prognosis in adult acute myeloid leukemia. N. Engl. J. Med. 1993, 328, 614–619. [Google Scholar] [CrossRef]
- Aasebø, E.; Mjaavatten, O.; Vaudel, M.; Farag, Y.; Selheim, F.; Berven, F.; Bruserud, Ø.; Hernandez-Valladares, M. Freezing effects on the acute myeloid leukemia cell proteome and phosphoproteome revealed using optimal quantitative workflows. J. Proteom. 2016, 145, 214–225. [Google Scholar] [CrossRef]
- Aasebø, E.; Vaudel, M.; Mjaavatten, O.; Gausdal, G.; Van der Burgh, A.; Gjertsen, B.T.; Døskeland, S.O.; Bruserud, Ø.; Berven, F.S.; Selheim, F. Performance of super-SILAC based quantitative proteomics for comparison of different acute myeloid leukemia (AML) cell lines. Proteomics 2014, 14, 1971–1976. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, E.; Forthun, R.B.; Berven, F.; Selheim, F.; Hernandez-Valladares, M. Global Cell Proteome Profiling, Phospho-signaling and Quantitative Proteomics for Identification of New Biomarkers in Acute Myeloid Leukemia Patients. Curr. Pharm. Biotechnol. 2016, 17, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Valladares, M.; Aasebø, E.; Mjaavatten, O.; Vaudel, M.; Bruserud, Ø.; Berven, F.; Selheim, F. Reliable FASP-based procedures for optimal quantitative proteomic and phosphoproteomic analysis on samples from acute myeloid leukemia patients. Biol. Proced. Online 2016, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Honnemyr, M.; Bruserud, Ø.; Brenner, A.K. The constitutive protease release by primary human acute myeloid leukemia cells. J. Cancer Res. Clin. Oncol. 2017, 143, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.K.; Tvedt, T.H.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Ther. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Hovland, R.; Wergeland, L.; Huang, T.S.; Gjertsen, B.T. Flt3-mediated signaling in human acute myelogenous leukemia (AML) blasts: A functional characterization of Flt3-ligand effects in AML cell populations with and without genetic Flt3 abnormalities. Haematologica 2003, 88, 416–428. [Google Scholar] [PubMed]
- Bruserud, Ø.; Gjertsen, B.T.; Foss, B.; Huang, T.S. New strategies in the treatment of acute myelogenous leukemia (AML): In vitro culture of aml cells—The present use in experimental studies and the possible importance for future therapeutic approaches. Stem Cells 2001, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gjertsen, B.T.; Øyan, A.M.; Marzolf, B.; Hovland, R.; Gausdal, G.; Døskeland, S.O.; Dimitrov, K.; Golden, A.; Kalland, K.H.; Hood, L.; et al. Analysis of acute myelogenous leukemia: Preparation of samples for genomic and proteomic analyses. J. Hematother. Stem Cell Res. 2002, 11, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Ryningen, A.; Olsnes, A.M.; Stordrange, L.; Øyan, A.M.; Kalland, K.H.; Gjertsen, B.T. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 2007, 92, 332–341. [Google Scholar] [CrossRef] [Green Version]
- Brenner, A.K.; Reikvam, H.; Bruserud, Ø. A Subset of Patients with Acute Myeloid Leukemia Has Leukemia Cells Characterized by Chemokine Responsiveness and Altered Expression of Transcriptional as well as Angiogenic Regulators. Front. Immunol. 2016, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Glenjen, N.; Ersvaer, E.; Ryningen, A.; Bruserud, Ø. In vitro effects of native human acute myelogenous leukemia blasts on fibroblasts and osteoblasts. Int. J. Cancer 2004, 111, 858–867. [Google Scholar] [CrossRef]
- Hatfield, K.; Ryningen, A.; Corbascio, M.; Bruserud, Ø. Microvascular endothelial cells increase proliferation and inhibit apoptosis of native human acute myelogenous leukemia blasts. Int. J. Cancer 2006, 119, 2313–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruserud, Ø.; Ryningen, A.; Wergeland, L.; Glenjen, N.I.; Gjertsen, B.T. Osteoblasts increase proliferation and release of pro-angiogenic interleukin 8 by native human acute myelogenous leukemia blasts. Haematologica 2004, 89, 391–402. [Google Scholar] [PubMed]
- Brenner, A.K.; Aasebø, E.; Hernandez-Valladares, M.; Selheim, F.; Berven, F.; Bruserud, Ø. Rethinking the role of osteopontin in human acute myeloid leukemia. Leuk Lymphoma 2017, 58, 1494–1497. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.C.; Dias, S. Internal and external autocrine VEGF/KDR loops regulate survival of subsets of acute leukemia through distinct signaling pathways. Blood 2004, 103, 3883–3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-Duque, P.; Quintanilla, M.; McNeish, I.; Lopes, R.; Romero, J.; Romero, D.; Lemoine, N.R.; Ramón y Cajal, S.; Vassaux, G. Caspase-1 as a radio- and chemo-sensitiser in vitro and in vivo. Int. J. Mol. Med. 2006, 17, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R. Intracellular targets of matrix metalloproteinase-2 in cardiac disease: Rationale and therapeutic approaches. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 211–242. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; Purslow, P.P. The role of matrix metalloproteinases in muscle and adipose tissue development and meat quality: A review. Meat Sci. 2016, 119, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Strongin, A.Y. Proteolytic and non-proteolytic roles of membrane type-1 matrix metalloproteinase in malignancy. Biochim. Biophys. Acta 2010, 1803, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, E.; Dassé, E.; Haye, B.; Petitfrère, E. TIMPs as multifacial proteins. Crit. Rev. Oncol. Hematol. 2004, 49, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zeng, W.; Tang, W.; Long, T.; Zhang, J.; Xie, X.; Kuang, Y.; Chen, M.; Su, J.; Chen, X. CD147 interacts with NDUFS6 in regulating mitochondrial complex I activity and the mitochondrial apoptotic pathway in human malignant melanoma cells. Curr. Mol. Med. 2014, 14, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Han, Q.; Huang, W.; Nan, G.; Xu, B.Q.; Jiang, J.L.; Chen, Z.N. HAb18G/CD147 regulates vinculin-mediated focal adhesion and cytoskeleton organization in cultured human hepatocellular carcinoma cells. PLoS ONE 2014, 9, e102496. [Google Scholar] [CrossRef] [PubMed]
- Soh, H.; Venkatesan, N.; Veena, M.S.; Ravichandran, S.; Zinabadi, A.; Basak, S.K.; Parvatiyar, K.; Srivastava, M.; Liang, L.J.; Gjertson, D.W.; et al. Cystatin E/M Suppresses Tumor Cell Growth through Cytoplasmic Retention of NF-kappaB. Mol. Cell. Biol. 2016, 36, 1776–1792. [Google Scholar] [CrossRef] [PubMed]
- Wallin, H.; Abrahamson, M.; Ekström, U. Cystatin C properties crucial for uptake and inhibition of intracellular target enzymes. J. Biol. Chem. 2013, 288, 17019–17029. [Google Scholar] [CrossRef] [PubMed]
- Riccio, M.; Di Giaimo, R.; Pianetti, S.; Palmieri, P.P.; Melli, M.; Santi, S. Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1). Exp. Cell Res. 2001, 262, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Casado, P.; Hijazi, M.; Britton, D.; Cutillas, P.R. Impact of phosphoproteomics in the translation of kinase-targeted therapies. Proteomics 2017, 17. [Google Scholar] [CrossRef]
- Ryningen, A.; Ersvaer, E.; Øyan, A.M.; Kalland, K.H.; Vintermyr, O.K.; Gjertsen, B.T.; Bruserud, Ø. Stress-induced in vitro apoptosis of native human acute myelogenous leukemia (AML) cells shows a wide variation between patients and is associated with low BCL-2:Bax ratio and low levels of heat shock protein 70 and 90. Leuk. Res. 2006, 30, 1531–1540. [Google Scholar] [CrossRef]
- Wangen, R.; Aasebø, E.; Trentani, A.; Døskeland, S.O.; Bruserud, Ø.; Selheim, F.; Hernandez-Valladares, M. Preservation Method and Phosphate Buffered Saline Washing Affect the Acute Myeloid Leukemia Proteome. Int. J. Mol. Sci. 2018, 19, 296. [Google Scholar] [CrossRef]

| Patient Characteristics | Cell Morphology | Cell Genetics | |||
|---|---|---|---|---|---|
| Age | FAB classification | Cytogenetics | |||
| Median (years) | 52 | M0 | 1 | Favorable | 3 |
| Range (years) | 18–68 | M1 | 5 | Intermediate | 1 |
| M2 | 3 | Normal | 13 | ||
| Gender | M4 | 4 | Adverse | 2 | |
| Females | 7 | M5 | 6 | ||
| Males | 12 | Flt3 mutations | |||
| ITD | 9 1 | ||||
| Disease | CD34 receptor | Wild-type | 10 | ||
| De novo AML | 14 | Negative (≤20%) | 13 | ||
| Secondary AML | 2 | Positive (>20%) | 6 | NPM1 mutations | |
| AML relapse | 3 | Insertion alone | 4 | ||
| Wild-type | 12 | ||||
| Insertion+Flt3-ITD | 3 | ||||
| Mediator | Detectable Constitutive Release During in Vitro Culture | Quantifiable Levels in Proteomic Analysis | Correlation (p-Value) | ||||
|---|---|---|---|---|---|---|---|
| Number | Median | Range | Fold Variation | Number | Fold Variation | ||
| CCL5 | 19 | 56.3 | 17.4–2481 | 143 | 7 | 27 | 0.0480 |
| MMP9 | 12 | 23,957 | 1123–261,935 | 233 | 13 | 2361 | ns |
| TIMP1 (tissue inhibitor of MMPs) | 19 | 2298 | 580–34,789 | 60 | 17 | 95 | ns |
| Caspase 1/CASP1 | 9 | 8.1 | 1.0–28.0 | 28 | 19 | 163 | ns |
| BSG (basigin) | 19 | 401 | 61.2–1876 | 31 | 19 | 4.5 | ns |
| CFD (complement factor D) | 13 | 4106 | 123–18,426 | 150 | 18 | 139 | 0.0553 |
| Cystatin B/CSTB | 18 | 2846 | 165–17,949 | 109 | 18 | 121 | ns |
| Cystatin C/CST3 | 19 | 3075 | 233–52,208 | 224 | 16 | 19 | 0.0078 |
| Neutrophil elastase/ELANE | 13 | 11,224 | 1072–203,665 | 190 | 19 | 835 | ns |
| Serpin E1 | 19 | 1131 | 36.2–23,925 | 661 | 14 | 412 | ns |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aasebø, E.; Hernandez-Valladares, M.; Selheim, F.; Berven, F.S.; Brenner, A.K.; Bruserud, Ø. Proteomic Profiling of Primary Human Acute Myeloid Leukemia Cells Does Not Reflect Their Constitutive Release of Soluble Mediators. Proteomes 2019, 7, 1. https://doi.org/10.3390/proteomes7010001
Aasebø E, Hernandez-Valladares M, Selheim F, Berven FS, Brenner AK, Bruserud Ø. Proteomic Profiling of Primary Human Acute Myeloid Leukemia Cells Does Not Reflect Their Constitutive Release of Soluble Mediators. Proteomes. 2019; 7(1):1. https://doi.org/10.3390/proteomes7010001
Chicago/Turabian StyleAasebø, Elise, Maria Hernandez-Valladares, Frode Selheim, Frode S. Berven, Annette K. Brenner, and Øystein Bruserud. 2019. "Proteomic Profiling of Primary Human Acute Myeloid Leukemia Cells Does Not Reflect Their Constitutive Release of Soluble Mediators" Proteomes 7, no. 1: 1. https://doi.org/10.3390/proteomes7010001