A Paper-Based Multicolor Colorimetric Aptasensor for the Visual Determination of Multiple Sulfonamides Based on Aptamer-Functionalized Magnetic Beads and NADH–Ascorbic Acid-Mediated Gold Nanobipyramids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Apparatus
2.2. Fabrication of the Paper-Based Analytical Device (PAD) and Preparation of Gold Seeds
2.3. Detection of Multiple SAs with PAD-Based Multicolor Colorimetric Aptasensor
2.4. Detection of Fish Muscle Sample
3. Results and Discussion
3.1. The Principle of the PAD-Based Multicolor Colorimetric Aptasensor
3.2. The Feasibility of the Experimental Strategy
3.3. Optimization of PAD-Based Multicolor Colorimetric Aptasensor
3.4. Specificity of the Multicolor Colorimetric Aptasensor
3.5. Analytical Performance of the Multicolor Colorimetric Aptasensor
3.6. Determination of SQ, SMP and SMD in Fish Muscle Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Białk-Bielińska, A.; Stolte, S.; Arning, J.; Uebers, U.; Böschen, A.; Stepnowski, P.; Matzke, M. Ecotoxicity evaluation of selected sulfonamides. Chemosphere 2011, 85, 928–933. [Google Scholar] [CrossRef]
- Apaydin, S.; TöröK, M. Sulfonamide derivatives as multi-target agents for complex disease. Bioorganic Med. Chem. Lett. 2019, 29, 2042–2050. [Google Scholar] [CrossRef]
- Yu, L.; Song, C.; Zhang, C.; Fan, L.; Qiu, L.; Wu, W.; Meng, S.; Hu, G.; Chen, J. Occurrence of sulfonamides in fish in the lower reaches of Yangtze River, China and estimated daily intake for understanding human dietary exposure. Aquaculture 2018, 495, 538–544. [Google Scholar] [CrossRef]
- Tačić, A.; Nikolić, V.; Nikolić, L.; Savić, I. Antimicrobial sulfonamide drugs. Adv. Technol. 2017, 6, 58–71. [Google Scholar] [CrossRef] [Green Version]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Hamscher, G.; Bachour, G. Veterinary drugs in the environment: Current knowledge and challenges for the future. J. Agric. Food Chem. 2018, 66, 751–752. [Google Scholar] [CrossRef] [PubMed]
- Suo, D.C.; Wang, P.L.; Xiao, Z.M.; Zhang, S.; Zhuang, H.T.; Li, Y.; Su, X.O. Multi-residue determination of 27 sulfonamides in poultry feathers and its application to a sulfamethazine pharmacokinetics study on laying hen feathers and sulfonamide residue monitoring on poultry feathers. J. Agric. Food Chem. 2019, 67, 11236–11243. [Google Scholar] [CrossRef]
- Kim, Y.; Jung, J.; Kim, M.; Park, J.; Boxall, A.B.A.; Choi, K. Prioritizing veterinary pharmaceuticals for aquatic environment in Korea. Environ. Toxicol. Pharmacol. 2008, 26, 167–176. [Google Scholar] [CrossRef]
- Baran, W.; Adamek, E.; Ziemiańska, J.; Sobczak, A. Effects of the presence of sulfonamides in the environment and their influence on human health. J. Hazard. Mater. 2011, 196, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhang, J.; Chen, H.; Wang, J.; Sun, W.; Zhang, X.; Yang, Y.; Wang, Q.; Ma, J. Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci. Total Environ. 2017, 607, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Duan, Z.J.; Wang, L.; Zhang, Y.; Wang, S. Hapten synthesis and development of polyclonal antibody-based multi-sulfonamide immunoassays. J. Agric. Food Chem. 2006, 54, 4499–4505. [Google Scholar] [CrossRef]
- Hu, S.P.; Zhao, M.; Xi, Y.Y.; Mao, Q.Q.; Zhou, X.D.; Chen, D.W.; Yan, P.C. Non-targeted screening and determination of sulfonamides: A dispersive micro solid-phase extraction approach to the analysis of milk and honey samples using liquid chromatography high-resolution mass spectrometry. J. Agric. Food Chem. 2017, 65, 1984–1991. [Google Scholar] [CrossRef]
- Li, T.; Wang, C.; Xu, Z.A.; Chakraborty, A. A coupled method of on-line solid phase extraction with the UHPLC-MS/MS for detection of sulfonamides antibiotics residues in aquaculture. Chemosphere 2020, 254, 126765. [Google Scholar] [CrossRef]
- Saxena, S.K.; Rangasamy, R.; Krishnan, A.A.; Singh, D.P.; Uke, S.P.; Malekadi, P.K.; Sengar, A.S.; Mohamed, D.P.; Gupta, A. Simultaneous determination of multi-residue and multi-class antibiotics in aquaculture shrimps by UPLC-MS/MS. Food Chem. 2018, 260, 336–343. [Google Scholar] [CrossRef]
- Zhao, X.L.; Wang, J.Y.; Wang, J.P.; Wang, S. Development of water-compatible molecularly imprinted solid-phase extraction coupled with high performance liquid chromatography-tandem mass spectrometry for the detection of six sulfonamides in animal-derived foods. J. Chromatogr. A 2018, 1574, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, M.Y.; Wu, X.Y.; Dong, S.B.; Zhu, N.F.; Gyimah, E.; Wang, K.; Li, Y. A competitive immunosensor for ultrasensitive detection of sulphonamides from environmental waters using silver nanoparticles decorated single-walled carbon nanohorns as labels. Chemosphere 2019, 225, 282–287. [Google Scholar] [CrossRef]
- Li, Z.B.; Cui, P.L.; Liu, J.; Liu, J.X.; Wang, J.P. Production of generic monoclonal antibody and development of chemiluminescence immunoassay for determination of 32 sulfonamides in chicken muscle. Food Chem. 2020, 311, 125966. [Google Scholar] [CrossRef]
- Wang, Z.X.; Xing, K.Y.; Ding, N.S.; Wang, S.H.; Zhang, G.G.; Lai, W.H. Lateral flow immunoassay based on dual spectral-overlapped fluorescence quenching of polydopamine nanospheres for sensitive detection of sulfamethazine. J. Hazard. Mater. 2022, 423, 127204. [Google Scholar] [CrossRef] [PubMed]
- Galvidis, I.A.; Wang, Z.H.; Nuriev, R.I.; Burkin, M.A. Broadening the detection spectrum of small analytes using a two-antibody-designed hybrid immunoassay. Anal. Chem. 2018, 90, 4901–4908. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Guo, L.L.; Liu, L.Q.; Song, S.S.; Kuang, H.; Xu, C.L. Ultrasensitive immuno-chromatographic strip for fast screening of 27 sulfonamides in honey and pork liver samples based on a monoclonal antibody. J. Agric. Food Chem. 2017, 65, 8248–8255. [Google Scholar] [CrossRef]
- Jiang, W.X.; Wang, Z.H.; Beier, R.C.; Jiang, H.Y.; Wu, Y.N.; Shen, J.Z. Simultaneous determination of 13 fluoroquinolone and 22 sulfonamide residues in milk by a dual-colorimetric enzyme-linked immunosorbent assay. Anal. Chem. 2013, 85, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-X.; Lin, Z.-Z.; Hong, C.-Y.; Yao, Q.-H.; Huang, Z.-Y. A dichromatic label-free aptasensor for sulfadimethoxine detection in fish and water based on AuNPs color and fluorescent dyeing of double-stranded DNA with SYBR Green I. Food Chem. 2020, 309, 125712. [Google Scholar] [CrossRef]
- Okoth, O.K.; Yan, K.; Liu, Y.; Zhang, J.D. Graphene-doped Bi2S3 nanorods as visible-light photoelectrochemical aptasensing platform for sulfadimethoxine detection. Biosens. Bioelectron. 2016, 86, 636–642. [Google Scholar] [CrossRef]
- You, H.; Bai, L.J.; Yuan, Y.H.; Zhou, J.; Bai, Y.; Mu, Z.D. An amperometric aptasensor for ultrasensitive detection of sulfadimethoxine based on exonuclease-assisted target recycling and new signal tracer for amplification. Biosens. Bioelectron. 2018, 117, 706–712. [Google Scholar] [CrossRef]
- Liu, Q.; Shi, T.Y.; Cheng, Y.; Wen, Z.R.; Ding, C.F.; Li, Y.Q.; Wang, K. Amplified photocurrent signal for fabricating photoelectrochemical sulfadimethoxine aptasensor based on carbon nitride photosensitization with visible/near-infrared light responsive zinc phthalocyanine. J. Hazard. Mater. 2021, 406, 124749. [Google Scholar] [CrossRef] [PubMed]
- Mohammad-Razdari, A.; Ghasemi-Varnamkhasti, M.; Izadi, Z.; Rostami, S.; Ensafi, A.A.; Siadat, M.; Losson, E. Detection of sulfadimethoxine in meat samples using a novel electrochemical biosensor as a rapid analysis method. J. Food Compos. Anal. 2019, 82, 103252. [Google Scholar] [CrossRef]
- Yang, Z.H.; Ding, X.F.; Guo, Q.; Wang, Y.; Lu, Z.W.; Ou, H.C.; Luo, Z.F.; Lou, X.H. Second generation of signaling-probe displacement electrochemical aptasensor for detection of picomolar ampicillin and sulfadimethoxine. Sens. Actuators B-Chem. 2017, 253, 1129–1136. [Google Scholar] [CrossRef]
- Yang, L.; Ni, H.J.; Li, C.L.; Zhang, X.Y.; Wen, K.; Ke, Y.B.; Yang, H.J.; Shi, W.M.; Zhang, S.X.; Shen, J.Z.; et al. Development of a highly specific chemiluminescence aptasensor for sulfamethazine detection in milk based on in vitro selected aptamers. Sens. Actuators B-Chem. 2019, 281, 801–811. [Google Scholar] [CrossRef]
- Chen, X.-X.; Lin, Z.-Z.; Yao, Q.-H.; Huang, Z.-Y. A practical aptaprobe for sulfadimethoxine residue detection in water and fish based on the fluorescence quenching of CdTe QDs by poly (dial-lyldimethylammonium chloride). J. Food Compos. Anal. 2020, 91, 103526. [Google Scholar] [CrossRef]
- Wang, Y.R.; Yan, X.-L.; Kou, Q.-M.; Sun, Q.; Wang, Y.-X.; Wu, P.; Yang, L.-L.; Tang, J.-M.; Le, T. An ultrasensitive label-free fluorescent aptasensor platform for detection of sulfamethazine. Int. J. Nanomed. 2021, 16, 2751–2758. [Google Scholar] [CrossRef]
- Song, K.-M.; Jeong, E.; Jeon, W.; Jo, H.; Ban, C. A coordination polymer nanobelt (CPNB)-based aptasensor for sulfadimethoxine. Biosens. Bioelectron. 2012, 33, 113–119. [Google Scholar] [CrossRef]
- Chen, A.L.; Jiang, X.L.; Zhang, W.W.; Chen, G.; Zhao, Y.; Tunio, T.M.; Liu, J.C.; Lv, Z.Z.; Li, C.; Yang, S.M. High sensitive rapid visual detection of sulfadimethoxine by label-free aptasensor. Biosens. Bioelectron. 2013, 42, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.-Y.; Yang, C.; Huang, L.; Lv, W.-C.; Yang, W.-J.; Wu, Y.-N.; Fu, F.-F. Broad-specificity aptamer of sulfonamides: Isolation and its application in simultaneous detection of multiple sulfonamides in fish sample. J. Agric. Food Chem. 2022, 70, 11804–11812. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.S.; Lethies, C.M.; Lin, X.-M.; Schatz, G.C.; Chen, L.-X.; Schaller, R.D. Size-dependent coherent-phonon plasmon modulation and deformation characterization in gold bipyramids and nanojavelins. ACS Photonics 2016, 3, 758–763. [Google Scholar] [CrossRef]
- Lombardi, A.; Loumaigne, M.; Crut, A.I.; Maioli, P.; Del Fatti, N.; Vallée, F.; Spuch-Calvar, M.; Burgin, J.; Majimel, J.R.; Tréguer-Delapierre, M. Surface plasmon resonance properties of single elongated nano-objects: Gold nanobipyramids and nanorods. Langmuir 2012, 28, 9027–9033. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Q.; Rao, H.-H.; Luo, M.-Y.; Xue, X.; Xue, Z.-H.; Lu, X.-Q. Noble metal nanoparticles growth-based colorimetric strategies: From monocolorimetric to multicolorimetric sensors. Coord. Chem. Rev. 2019, 398, 113003. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Wang, H.; Chen, Z.-P.; Wang, X.-Y.; Choo, J.; Chen, L.-X. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: Strategies and applications. Biosens. Bioelectron. 2018, 114, 52–65. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Chen, Q.; Zhong, Y.-Y.; Yu, X.-H.; Wu, Y.-N.; Fu, F.-F. A multicolor immunosensor for sensitive visual detection of breast cancer biomarker based on sensitive NADH-ascorbic-acid-mediated growth of gold nanobipyramids. Anal. Chem. 2020, 92, 1534–1540. [Google Scholar] [CrossRef]
- Ye, X.-Y.; Zhang, F.; Yang, L.; Yang, W.-J.; Zhang, L.-Y.; Wang, Z.-W. Paper-based multicolor sensor for on-site quantitative detection of 2, 4-dichlorophenoxyacetic acid based on alkaline phosphatase-mediated gold nanobipyramids growth and colorimeter-assisted method for quantifying color. Talanta 2022, 245, 123489. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Yang, L.; Ye, X.-Y.; Huang, C.-L.; Yang, W.-J.; Zhang, L.-Y.; Wu, Z.-J.; Fu, F.-F. Multicolor visual screening of total dithiocarbamate pesticides in foods based on sulfydryl-mediated growth of gold nanobipyramids. Anal. Chim. Acta 2020, 1139, 59–67. [Google Scholar] [CrossRef]


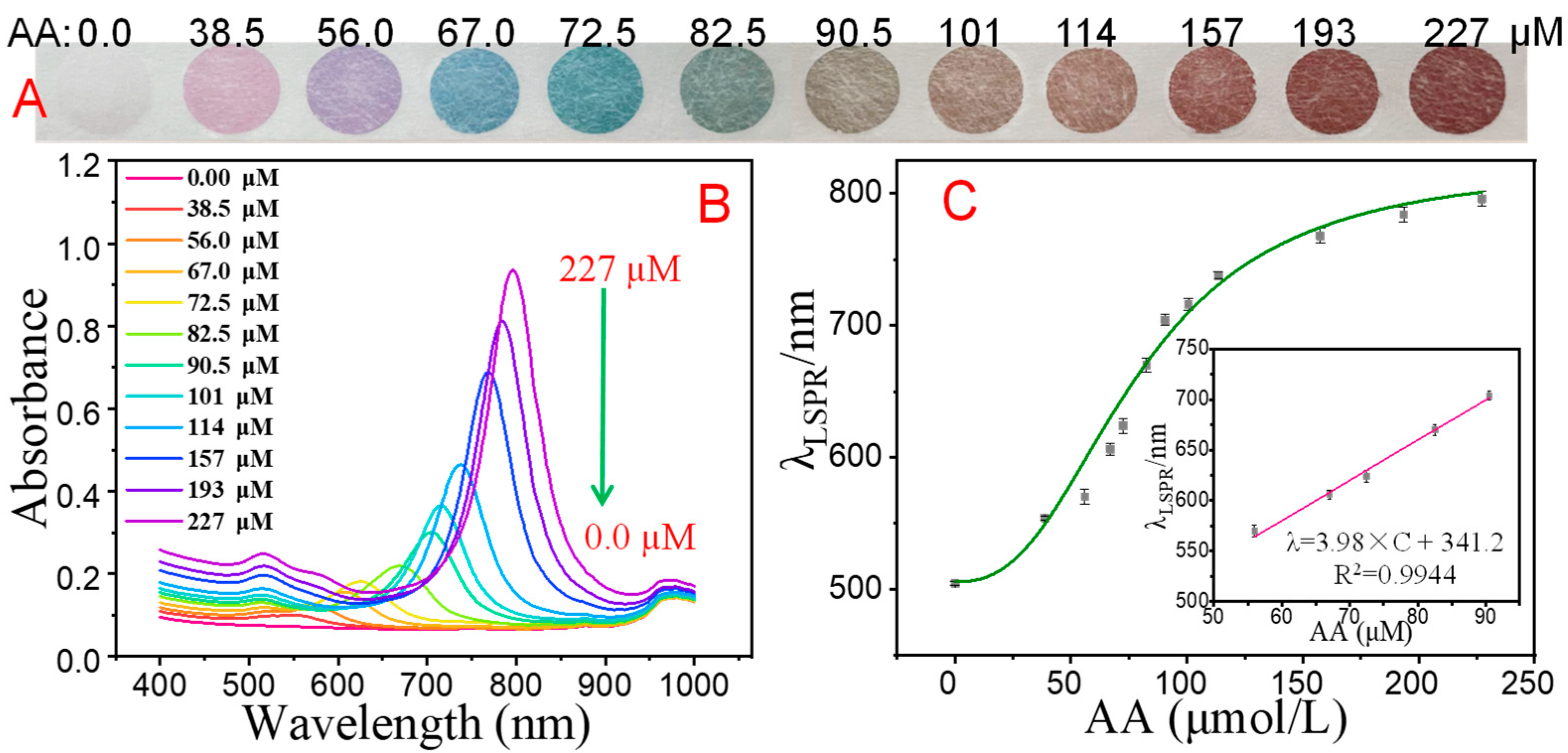
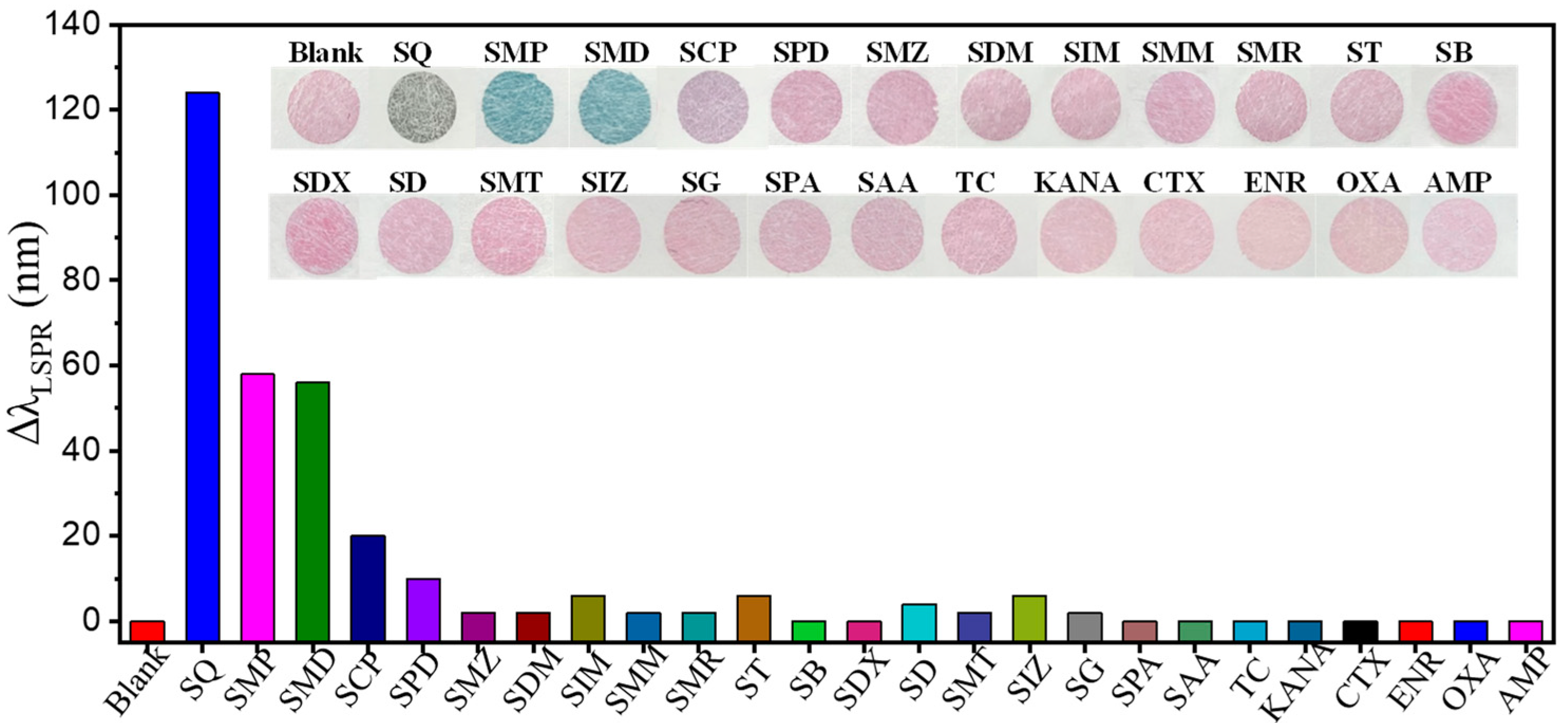
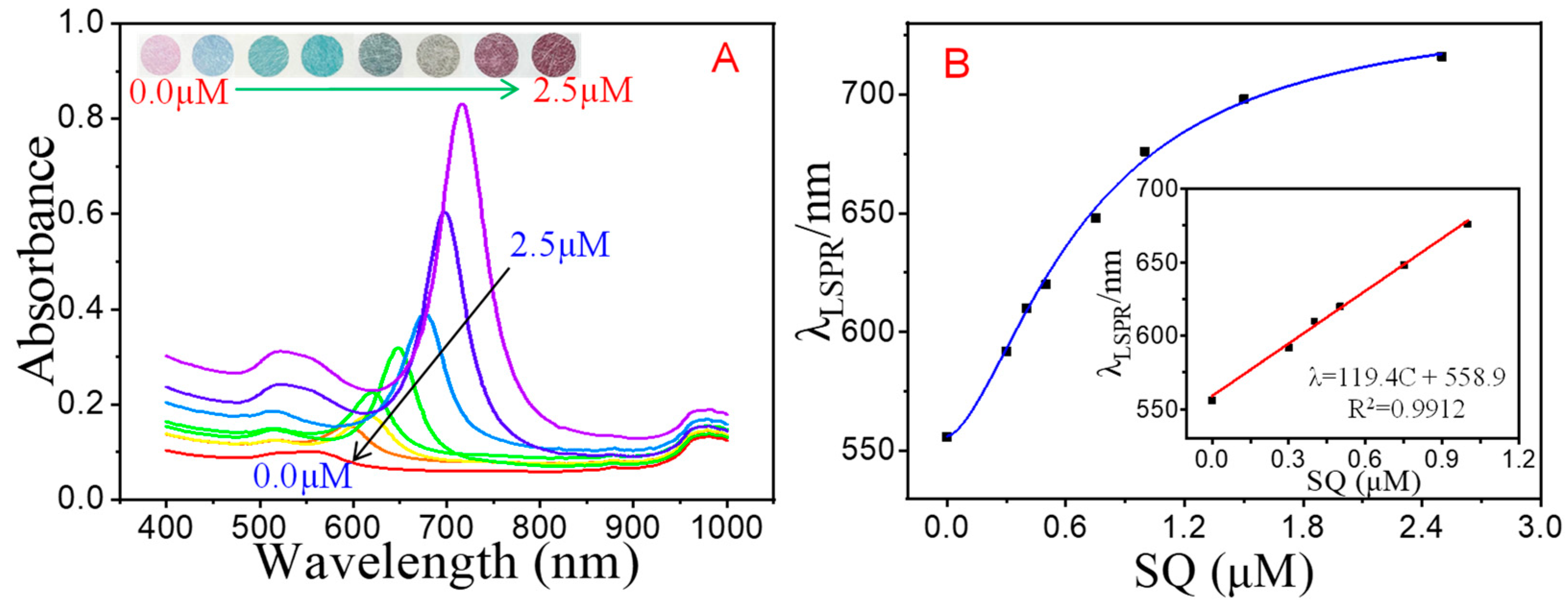
| SAs | Linear Equation | Coefficient (R2) | Linear Range | Visual LOD | Spectrometry | |
|---|---|---|---|---|---|---|
| LOQ | LOD | |||||
| SQ | λ = 119.4 × C + 558.9 | 0.9912 | 0.3–1.0 μM | 0.3 μM | 0.3 μM | 0.09 μM |
| SMP | λ = 56.4 × C + 550.4 | 0.9941 | 0.3–1.0 μM | 0.5 μM | 0.3 μM | 0.09 μM |
| SMD | λ = 20.4 × C + 550.1 | 0.9951 | 0.5–5.0 μM | 1.0 μM | 0.5 μM | 0.15 μM |
| Added SAs | 1 Added Con. (µg/g) | 2 SAs Detected | ||||
|---|---|---|---|---|---|---|
| Our Multicolor Aptasensor | 5 ELISA Kit Con. (µg/g) | |||||
| Color | UV-Vis Spectrometry | |||||
| Con. (µg/g) | RSD (n = 5) | Rec. | ||||
| None | 0.0 |  | - | - | - | - |
| SQ | 3.6 |  | 3.4 | 5% | 94% | 3.7 |
| SMP | 5.7 |  | 5.2 | 6% | 91% | - |
| SMD | 5.6 |  | 5.0 | 8% | 89% | 5.2 |
| 3 SQ + SMD | 5.8 |  | 5.2 | 7% | 90% | 5.4 |
| 4 Mixture | 4.6 |  | 4.1 | 6% | 90% | 3.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ping, M.; Lv, W.; Yang, C.; Chen, Q.; Wang, Z.; Fu, F. A Paper-Based Multicolor Colorimetric Aptasensor for the Visual Determination of Multiple Sulfonamides Based on Aptamer-Functionalized Magnetic Beads and NADH–Ascorbic Acid-Mediated Gold Nanobipyramids. Chemosensors 2023, 11, 386. https://doi.org/10.3390/chemosensors11070386
Ping M, Lv W, Yang C, Chen Q, Wang Z, Fu F. A Paper-Based Multicolor Colorimetric Aptasensor for the Visual Determination of Multiple Sulfonamides Based on Aptamer-Functionalized Magnetic Beads and NADH–Ascorbic Acid-Mediated Gold Nanobipyramids. Chemosensors. 2023; 11(7):386. https://doi.org/10.3390/chemosensors11070386
Chicago/Turabian StylePing, Meiling, Wenchao Lv, Chen Yang, Qian Chen, Zongwen Wang, and Fengfu Fu. 2023. "A Paper-Based Multicolor Colorimetric Aptasensor for the Visual Determination of Multiple Sulfonamides Based on Aptamer-Functionalized Magnetic Beads and NADH–Ascorbic Acid-Mediated Gold Nanobipyramids" Chemosensors 11, no. 7: 386. https://doi.org/10.3390/chemosensors11070386





