Determination of Extra- and Intra-Cellular pH Using Characteristic Absorption of Water by Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Sample Preparation
2.2. Spectral Measurement
2.3. pH Measurement
2.4. Calculation Method
3. Results and Discussion
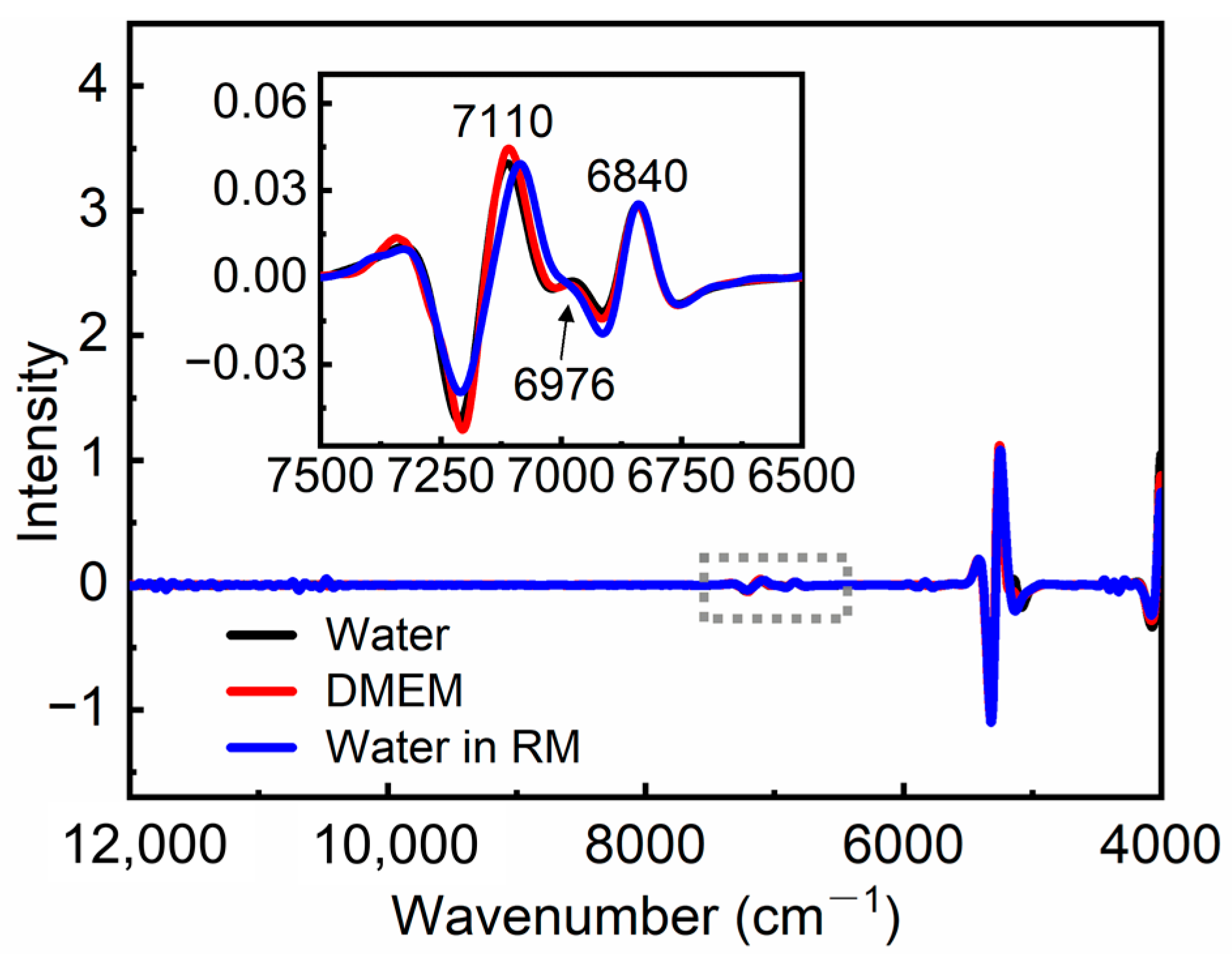
3.1. Spectral Analysis
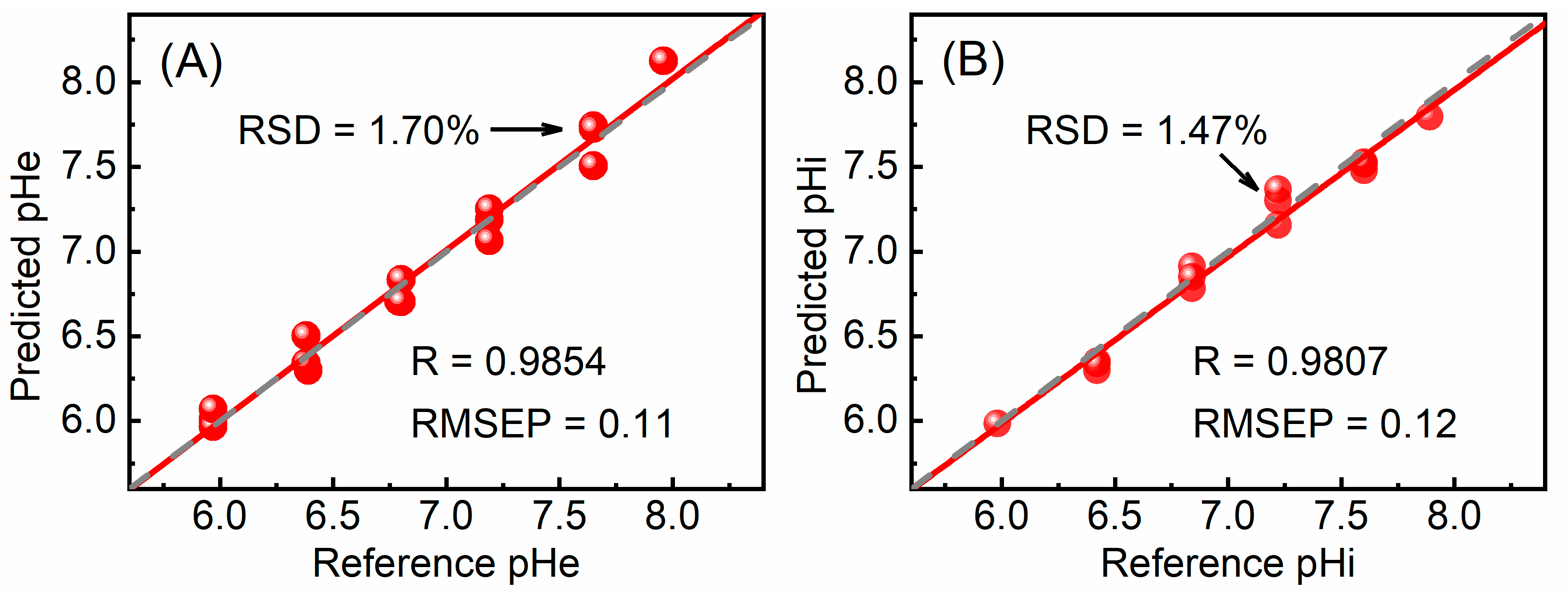
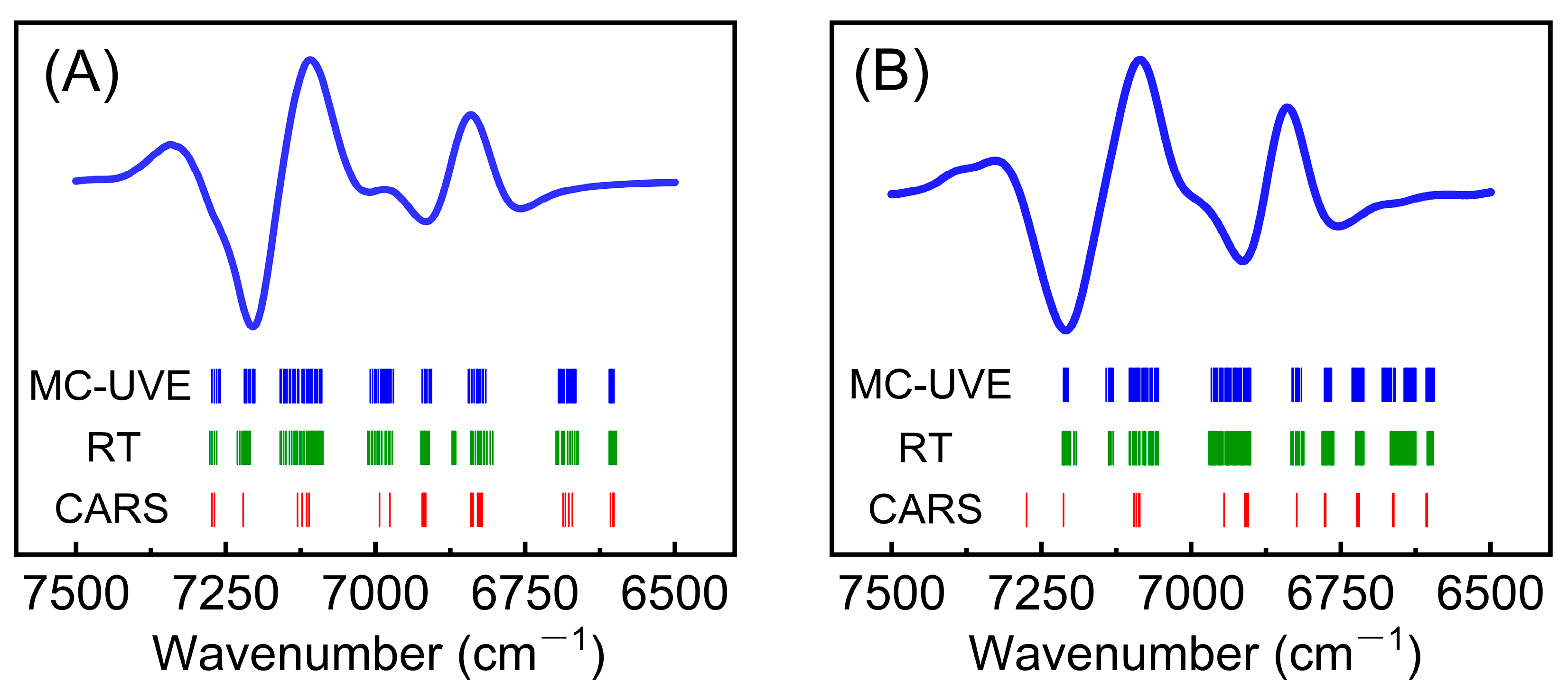
3.2. Quantitative Models and Validation
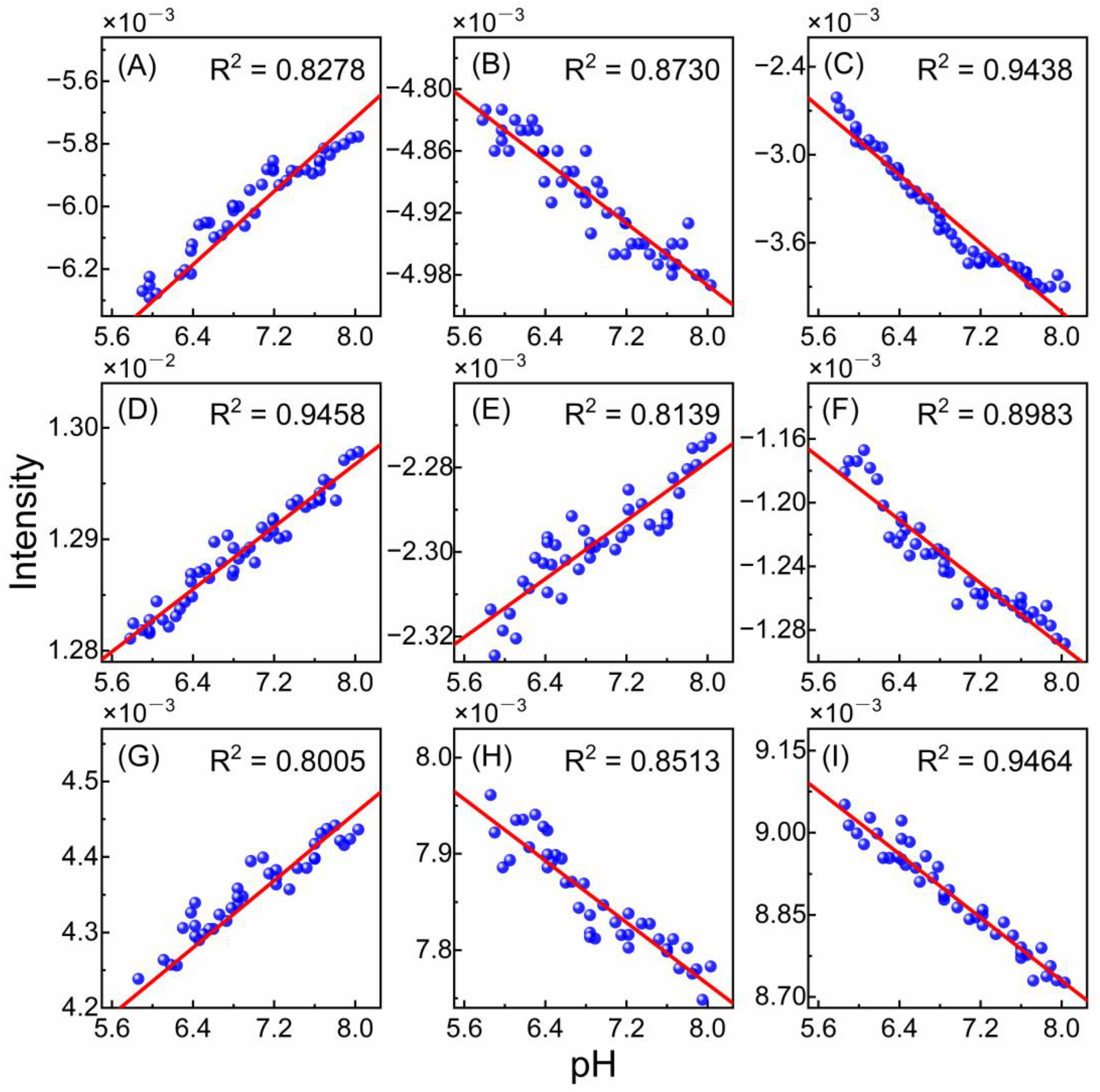
3.3. pH Dependence of Selected Wavenumbers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cialla, M.D.; Zheng, X.S.; Weber, K.; Popp, J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: From cells to clinics. Chem. Soc. Rev. 2017, 46, 3945–3961. [Google Scholar] [CrossRef]
- Kurkdjian, A.; Guern, J. Intracellular pH: Measurement and importance in cell activity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 271–303. [Google Scholar] [CrossRef]
- Sindhu, R.; Binod, P.; Pandey, A. Biological pretreatment of lignocellulosic biomass—An overview. Bioresour. Technol. 2016, 199, 76–82. [Google Scholar] [CrossRef]
- Persi, E.; Duran, F.M.; Damaghi, M.; Roush, W.R.; Aloy, P.; Cleveland, J.L.; Gillies, R.J.; Ruppin, E. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat. Commun. 2018, 9, 2997. [Google Scholar] [CrossRef] [Green Version]
- Stubbs, M.; McSheehy, P.M.; Griffiths, J.R.; Bashford, C.L. Causes and consequences of tumour acidity and implications for treatment. Mol. Med. Today 2000, 6, 15–19. [Google Scholar] [CrossRef]
- Suffrian, K.; Schulz, K.G.; Gutowska, M.A.; Riebesell, U.; Bleich, M. Cellular pH measurements in Emiliania huxleyi reveal pronounced membrane proton permeability. New Phytol. 2011, 190, 595–608. [Google Scholar] [CrossRef] [Green Version]
- Shirmanova, M.V.; Druzhkova, I.N.; Lukina, M.M.; Dudenkova, V.V.; Ignatova, N.I.; Snopova, L.B.; Shcheslavskiy, V.I.; Belousov, V.V.; Zagaynova, E.V. Chemotherapy with cisplatin: Insights into intracellular pH and metabolic landscape of cancer cells in vitro and in vivo. Sci. Rep. 2017, 7, 8911. [Google Scholar] [CrossRef] [Green Version]
- Ball, P. Water—An enduring mystery. Nature 2008, 452, 291–292. [Google Scholar] [CrossRef]
- Ball, P. Water as an active constituent in cell biology. Chem. Rev. 2008, 108, 74–108. [Google Scholar] [CrossRef]
- Czarnik-Matusewicz, B.; Pilorz, S. Study of the temperature-dependent near-infrared spectra of water by two-dimensional correlation spectroscopy and principal components analysis. Vib. Spectrosc. 2006, 40, 235–245. [Google Scholar] [CrossRef]
- Dong, Q.; Guo, X.P.; Li, L.; Yu, C.; Nie, L.; Tian, W.L.; Zhang, H.; Huang, S.; Zang, H.C. Understanding hyaluronic acid induced variation of water structure by near-infrared spectroscopy. Sci. Rep. 2020, 10, 1387. [Google Scholar] [CrossRef] [Green Version]
- Su, C.L.; Han, L.; An, H.L.; Cai, W.S.; Shao, X.G. Structures of water on the surface of anatase TiO2 studied by diffuse reflectance near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 296, 122674. [Google Scholar] [CrossRef]
- Han, G.; Yu, X.Y.; Xia, D.D.; Liu, R.; Liu, J.; Xu, K.X. Preliminary clinical validation of a differential correction method for improving measurement accuracy in noninvasive measurement of blood glucose using near-infrared spectroscopy. Appl. Spectrosc. 2017, 71, 2177–2186. [Google Scholar] [CrossRef]
- Eum, C.; Jang, D.; Kim, J.; Choi, S.; Cha, K.; Chung, H. Improving the accuracy of spectroscopic identification of geographical origins of agricultural samples through cooperative combination of near infrared and laser-induced breakdown spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2018, 149, 281–287. [Google Scholar] [CrossRef]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-line application of near infrared (NIR) spectroscopy in food production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Bec, K.B.; Huck, C.W. Breakthrough potential in near-infrared spectroscopy: Spectra simulation. A review of recent developments. Front. Chem. 2019, 7, 48. [Google Scholar] [CrossRef] [Green Version]
- Bec, K.B.; Grabska, J.; Huck, C.W. Current and future research directions in computer-aided near-infrared spectroscopy: A perspective. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 254, 119625. [Google Scholar] [CrossRef]
- Bec, K.B.; Futami, Y.; Wojcik, M.J.; Nakajima, T.; Ozaki, Y. Spectroscopic and computational study of acetic acid and its cyclic dimer in the near-infrared region. J. Phys. Chem. A 2016, 120, 6170–6183. [Google Scholar] [CrossRef]
- Maeda, H.; Ozaki, Y.; Tanaka, M.; Hayashi, N.; Kojima, T. Near infrared spectroscopy and chemometrics studies of temperature-dependent spectral variations of water: Relationship between spectral changes and hydrogen bonds. J. Near Infrared Spectrosc. 1995, 3, 191–201. [Google Scholar] [CrossRef]
- Tan, J.H.; Sun, Y.; Ma, L.; Feng, H.Y.; Guo, Y.C.; Cai, W.S.; Shao, X.G. Knowledge-based genetic algorithm for resolving the near-infrared spectrum and understanding the water structures in aqueous solution. Chemom. Intell. Lab. Syst. 2020, 206, 104150. [Google Scholar] [CrossRef]
- Wang, Y.; Murayama, K.; Myojo, Y.; Tsenkova, R.; Hayashi, N.; Ozaki, Y. Two-dimensional Fourier transform near-infrared spectroscopy study of heat denaturation of ovalbumin in aqueous solutions. J. Phys. Chem. B 1998, 102, 6655–6662. [Google Scholar] [CrossRef]
- Goto, N.; Bazar, G.; Kovacs, Z.; Kunisada, M.; Morita, H.; Kizaki, S.; Sugiyama, H.; Tsenkova, R.; Nishigori, C. Detection of UV-induced cyclobutane pyrimidine dimers by near-infrared spectroscopy and aquaphotomics. Sci. Rep. 2015, 5, 11808. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.L.; Zhong, L.; Zhang, J.; Zhang, M.Q.; Zeng, Y.Z.; Li, L.; Zang, H.C. Water as a probe to understand the traditional Chinese medicine extraction process with near infrared spectroscopy: A case of Danshen (Salvia miltiorrhiza Bge) extraction process. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 244, 118854. [Google Scholar] [CrossRef]
- Ishigaki, M.; Yasui, Y.; Kajita, M.; Ozaki, Y. Assessment of embryonic bioactivity through changes in the water structure using near-infrared spectroscopy and imaging. Anal. Chem. 2020, 92, 8133–8141. [Google Scholar] [CrossRef]
- Tsenkova, R.N.; Iordanova, I.K.; Toyoda, K.; Brown, D.R. Prion protein fate governed by metal binding. Biochem. Biophys. Res. Commun. 2004, 325, 1005–1012. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L.; Cai, W.S.; Shao, X.G. Interaction between tau and water during the induced aggregation revealed by near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 230, 118046. [Google Scholar] [CrossRef]
- Chatani, E.; Tsuchisaka, Y.; Masuda, Y.; Tsenkova, R. Water molecular system dynamics associated with amyloidogenic nucleation as revealed by real time near infrared spectroscopy and aquaphotomics. PLoS ONE 2014, 9, e101997. [Google Scholar] [CrossRef]
- Jinendra, B.; Tamaki, K.; Kuroki, S.; Vassileva, M.; Yoshida, S.; Tsenkova, R. Near infrared spectroscopy and aquaphotomics: Novel approach for rapid in vivo diagnosis of virus infected soybean. Biochem. Biophys. Res. Commun. 2010, 397, 685–690. [Google Scholar] [CrossRef]
- Alam, M.K.; Rohrscheib, M.R.; Franke, J.E.; Niemczyk, T.M.; Maynard, J.D.; Robinson, M.R. Measurement of pH in whole blood by near-infrared spectroscopy. Appl. Spectrosc. 1999, 53, 316–324. [Google Scholar] [CrossRef]
- Amerov, A.K.; Chen, J.; Small, G.W.; Arnold, M.A. Scattering and absorption effects in the determination of glucose in whole blood by near-infrared spectroscopy. Anal. Chem. 2005, 77, 4587–4594. [Google Scholar] [CrossRef]
- Yadav, J.; Rani, A.; Singh, V.; Murari, B.M. Prospects and limitations of non-invasive blood glucose monitoring using near-infrared spectroscopy. Biomed. Signal Process. Control 2015, 18, 214–227. [Google Scholar] [CrossRef]
- Cui, X.Y.; Liu, X.W.; Yu, X.M.; Cai, W.S.; Shao, X.G. Water can be a probe for sensing glucose in aqueous solutions by temperature dependent near infrared spectra. Anal. Chim. Acta 2017, 957, 47–54. [Google Scholar] [CrossRef]
- Han, L.; Cui, X.Y.; Cai, W.S.; Shao, X.G. Three-level simultaneous component analysis for analyzing the near-infrared spectra of aqueous solutions under multiple perturbations. Talanta 2020, 217, 121036. [Google Scholar] [CrossRef]
- Smith, R.E.; Luisi, P.L. Micellar solubilization of bio-polymers in hydrocarbon solvents III. Empirical definition of an acidity scale in reverse micelles. Helv. Chim. Acta 1980, 63, 2302–2311. [Google Scholar] [CrossRef]
- Fujii, H.; Kawai, T.; Nishikawa, H. Determination of pH in reversed micelles. Bull. Chem. Soc. Jpn. 1979, 52, 2051–2055. [Google Scholar] [CrossRef] [Green Version]
- Shao, X.G.; Leung, A.K.M.; Chau, F.T. Wavelet: A new trend in chemistry. Acc. Chem. Res. 2003, 36, 276–283. [Google Scholar] [CrossRef]
- Shao, X.G.; Ma, C.X. A general approach to derivative calculation using wavelet transform. Chemom. Intell. Lab. Syst. 2003, 69, 157–165. [Google Scholar] [CrossRef]
- Zhu, Z.Y.; Chen, S.B.; Wu, X.Y.; Xing, C.R.; Yuan, J. Determination of soybean routine quality parameters using near-infrared spectroscopy. Food Sci. Nutr. 2018, 6, 1109–1118. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhao, C.; Shao, Q.J.; Yang, Z.D.; Zhang, X.F.; Xu, X.F.; Hassan, M. Determination of water content in corn stover silage using near-infrared spectroscopy. Int. J. Agric. Biol. Eng. 2019, 12, 143–148. [Google Scholar] [CrossRef]
- Tang, G.; Huang, Y.; Tian, K.D.; Song, X.Z.; Yan, H.; Hu, J.; Xiong, Y.M.; Min, S.G. A new spectral variable selection pattern using competitive adaptive reweighted sampling combined with successive projections algorithm. Analyst 2014, 139, 4894–4902. [Google Scholar] [CrossRef]
- Zhang, X.; Li, W.; Yin, B.; Chen, W.Z.; Kelly, D.P.; Wang, X.X.; Zheng, K.Y.; Du, Y.P. Improvement of near infrared spectroscopic (NIRS) analysis of caffeine in roasted Arabica coffee by variable selection method of stability competitive adaptive reweighted sampling (SCARS). Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 114, 350–356. [Google Scholar] [CrossRef]
- Zhang, M.J.; Zhang, S.Z.; Iqbal, J. Key wavelengths selection from near infrared spectra using Monte Carlo sampling–recursive partial least squares. Chemom. Intell. Lab. Syst. 2013, 128, 17–24. [Google Scholar] [CrossRef]
- Qi, L.H.; Cai, W.S.; Shao, X.G. Effect of temperature on near infrared spectra of n-alkanes. Acta Chim Sin. 2016, 74, 172–178. [Google Scholar] [CrossRef]
- Czarnecki, M.A.; Morisawa, Y.; Futami, Y.; Ozaki, Y. Advances in molecular structure and interaction studies using near-infrared spectroscopy. Chem. Rev. 2015, 115, 9707–9744. [Google Scholar] [CrossRef]
- Gowen, A.A.; Amigo, J.M.; Tsenkova, R. Characterisation of hydrogen bond perturbations in aqueous systems using aquaphotomics and multivariate curve resolution-alternating least squares. Anal. Chim. Acta 2013, 759, 8–20. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kajimoto, S.; Nakabayashi, T. Experimental evaluation of the density of water in a cell by Raman microscopy. J. Phys. Chem. Lett. 2017, 8, 5241–5245. [Google Scholar] [CrossRef]
- Tsenkova, R. Aquaphotomics: Dynamic spectroscopy of aqueous and biological systems describes peculiarities of water. J. Near Infrared Spectrosc. 2009, 17, 303–314. [Google Scholar] [CrossRef]
- Muncan, J.; Anantawittayanon, S.; Furuta, T.; Kaneko, T.; Tsenkova, R. Aquaphotomics monitoring of strawberry fruit during cold storage-A comparison of two cooling systems. Front. Nutr. 2022, 9, 1058173. [Google Scholar] [CrossRef]
- Tsenkova, R.; Muncan, J.; Pollner, B.; Kovacs, Z. Essentials of aquaphotomics and its chemometrics approaches. Front. Chem. 2018, 6, 363. [Google Scholar] [CrossRef]

| Methods | Spectral Range (cm−1) | nVar 1 | nLV 2 | Rcv | RMSECV | R | RMSEP |
|---|---|---|---|---|---|---|---|
| pHe | |||||||
| None | Full spectra | 4148 | 7 | 0.9775 | 0.14 | 0.9787 | 0.15 |
| None | 7500–6500 | 520 | 7 | 0.9816 | 0.12 | 0.9903 | 0.10 |
| CWT | Full spectra | 4148 | 7 | 0.9743 | 0.15 | 0.9731 | 0.17 |
| CWT | 7500–6500 | 520 | 7 | 0.9853 | 0.11 | 0.9848 | 0.11 |
| CWT-MC-UVE | 7500–6500 | 95 | 4 | 0.9853 | 0.11 | 0.9910 | 0.09 |
| CWT-RT | 7500–6500 | 101 | 4 | 0.9861 | 0.11 | 0.9906 | 0.09 |
| CWT-CARS | 7500–6500 | 28 | 4 | 0.9890 | 0.08 | 0.9854 | 0.11 |
| pHi | |||||||
| None | Full spectra | 4148 | 4 | 0.9046 | 0.26 | 0.9240 | 0.24 |
| None | 7500–6500 | 520 | 4 | 0.9623 | 0.17 | 0.9611 | 0.15 |
| CWT | Full spectra | 4148 | 4 | 0.9628 | 0.17 | 0.9626 | 0.17 |
| CWT | 7500–6500 | 520 | 5 | 0.9639 | 0.16 | 0.9630 | 0.17 |
| CWT-MC-UVE | 7500–6500 | 119 | 5 | 0.9787 | 0.13 | 0.9795 | 0.12 |
| CWT-RT | 7500–6500 | 127 | 4 | 0.9802 | 0.12 | 0.9828 | 0.12 |
| CWT-CARS | 7500–6500 | 21 | 4 | 0.9834 | 0.11 | 0.9807 | 0.12 |
| Sample Number | Reference | Prediction | Absolute Error | Relative Error (%) | RSD (%) n = 3 |
|---|---|---|---|---|---|
| pHe | |||||
| 1 | 7.96 | 8.12 | 0.16 | 2.07 | - |
| 2 | 7.65 | 7.66 | 0.01 | 0.13 | 1.70 |
| 3 | 7.19 | 7.17 | −0.02 | −0.28 | 1.36 |
| 4 | 6.80 | 6.75 | −0.05 | −0.74 | 1.07 |
| 5 | 6.38 | 6.45 | 0.07 | 1.10 | 1.66 |
| 6 | 5.97 | 6.02 | 0.05 | 0.84 | 0.84 |
| 7 | 6.49 | 6.36 | −0.13 | −2.00 | - |
| 8 | 6.32 | 6.26 | −0.06 | −0.95 | - |
| 9 | 6.22 | 6.41 | 0.19 | 3.05 | - |
| pHi | |||||
| 1 | 7.89 | 7.80 | −0.09 | −1.18 | - |
| 2 | 7.60 | 7.51 | −0.09 | −1.18 | 0.35% |
| 3 | 7.22 | 7.28 | 0.06 | 0.83 | 1.47% |
| 4 | 6.84 | 6.85 | 0.01 | 0.15 | 1.45% |
| 5 | 6.42 | 6.33 | −0.09 | −1.40 | 0.42% |
| 6 | 5.98 | 5.99 | 0.01 | 0.17 | - |
| Wavenumber (cm−1) | Water Structure | |
|---|---|---|
| pHe | pHi | |
| 6671 | 6661 | S3 (A2D1) |
| - | 6719 | S2 (A2D0) |
| 6828 | 6823 | S2 (A1D1) |
| 6916 | 6904 | S1 (A0D1) |
| 7110 | 7095 | S0 (A0D0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liang, F.; Han, L.; Yu, X.; Liu, D.; Cai, W.; Shao, X. Determination of Extra- and Intra-Cellular pH Using Characteristic Absorption of Water by Near-Infrared Spectroscopy. Chemosensors 2023, 11, 425. https://doi.org/10.3390/chemosensors11080425
Li J, Liang F, Han L, Yu X, Liu D, Cai W, Shao X. Determination of Extra- and Intra-Cellular pH Using Characteristic Absorption of Water by Near-Infrared Spectroscopy. Chemosensors. 2023; 11(8):425. https://doi.org/10.3390/chemosensors11080425
Chicago/Turabian StyleLi, Jiani, Fanfan Liang, Li Han, Xiaoxuan Yu, Dingbin Liu, Wensheng Cai, and Xueguang Shao. 2023. "Determination of Extra- and Intra-Cellular pH Using Characteristic Absorption of Water by Near-Infrared Spectroscopy" Chemosensors 11, no. 8: 425. https://doi.org/10.3390/chemosensors11080425





