Abstract
A micro-level technique so-called “microfluidic technology or simply microfluidic” has gained a special place as a powerful tool in bioengineering and biomedical engineering research due to its core advantages in modern science and engineering. Microfluidic technology has played a substantial role in numerous applications with special reference to bioscience, biomedical and biotechnological research. It has facilitated noteworthy development in various sectors of bio-research and upsurges the efficacy of research at the molecular level, in recent years. Microfluidic technology can manipulate sample volumes with precise control outside cellular microenvironment, at micro-level. Thus, enable the reduction of discrepancies between in vivo and in vitro environments and reduce the overall reaction time and cost. In this review, we discuss various integrations of microfluidic technologies into biotechnology and its paradigmatic significance in bio-research, supporting mechanical and chemical in vitro cellular microenvironment. Furthermore, specific innovations related to the application of microfluidics to advance microbial life, solitary and co-cultures along with a multiple-type cell culturing, cellular communications, cellular interactions, and population dynamics are also discussed.
1. Introduction
Eternal aspiration of inquiring biology at a single subsistence level has contrived microfluidic noticeably into the organic (living) space [1]. Invariably, due to its miniaturized length scale, rapidity in the analysis, the selective capacity to minimize the volume of reagent, and reduction in turnaround, small-scale fluidics have facilitated the area of natural science [2,3]. Microfluidic scaffolds are micro-fabricated devices that have gained importance for cellular interactions [4]. Cultured cells are worn in an array of a framework like a cell science, tissue culturing, bacterial biofilms, biomedical designing and additionally pharmacokinetics for medication improvement [5,6,7]. The in vitro model makes it rational to culture cells in the required volume as an alternative to animal-based tests for medical screening. On the other hand, in an in vitro model of cell culture, no polymer casings or structures are available to be held fast instead of cell culture dish. Therefore, cultured cells conventionally change their properties practically corresponding to development frequency, metabolic rate and morphology [8,9,10]. On the premise of the oddity among in vivo and in vitro situations, innovative techniques are obliged to set up in vivo alike microenvironment. The utilization of innovative microfluidic approaches to the transposition of culture media, and oxygen even though the unwanted items by cells are exhausted like a parallel to the human circulatory framework [11]. Moreover, numerous studies have devoted on systematic microstructures brought together into a microfluidic stage that executed specimen blending, cell movement and separation in a microchannel [12,13]. Microfluidic structures can be a response domain for cell assay and the wellspring of an in vivo resembling setting for cell culture. Previous studies illustrated a basic two-dimensional (2-D) microstructure utilized for cells cultivation and mainly employed to fabricate a microfluidic framework with requisite characteristics [14]. Microfluidic tools have attained sophisticated advancement with an end goal to understand in vivo conditions. In this context, tools have been tuned to engineer polymer-based platforms in the form of three dimensional (3-D) microstructures, guaranteeing different layers for co-cultures or 3-D cell development [1]. Herein, the present work covers the advantages and challenges of using microfluidics in cell culture. In addition, the potential developments of microfluidic cells cultivation techniques and their associated bio-applications are presented.
2. Microarrays: Encaging Cell Trapping Concepts
Micro-arrays at large and cell-based microarrays, in particular, integrated with other emerging technologies such as optical, electrical or electrochemical-based detection technologies coupled with a well-designed microfluidic system appeared useful in monitoring changes in the cellular behavior under the influence of the external environment. Moreover, the cell-based micro-arrays can facilitate a proper adherence or growth of a variety of cell types on different substrates [15]. So far, various concepts have been proposed to develop novel arrays such as multiplexed cell-based arrays also called positional arrays, and high-throughput cell-based assays also known as suspension arrays. Figure 1 illustrates a schematic generation of cell-based arrays, i.e., positional and non-positional cellular arrays [16].
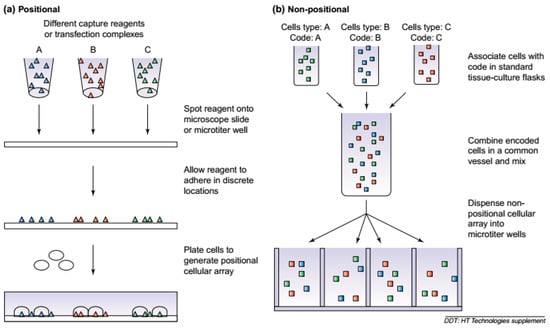
Figure 1.
The generation of cellular arrays. (a) Positional cellular arrays are generated by localizing unique cell types to specific locations. For the Cellomics CellChip™ technology (http://www.cellomics.com), capturing agents are first spotted onto a substrate, and a population of cells is plated onto them. For reverse transfected cellular arrays, transfection reagents are first spotted and the cells plated onto them. (b) When generating non-positional cellular arrays, the different cell types are first associated with a unique code. These encoded cells are then mixed and aliquoted into a microtiter well for subsequent analysis. Reproduced from Beske and Goldbard [16], with permission from Elsevier.
Practice to progression cells online in microfluidic frameworks crop up with the development of propelled microfabrication advancements and configuration. Araci and Brisk [17] elaborated the grasp of aerially controlled low volume elastomer microvalves with the progression of the analogous sample and cells preparing frameworks. A particular cell catching hydro-dynamically at T-intersection in a microfluidic channel favored draft exchanging of required fluids, so that omitted the settled cell. It was approved that time determined to filter ionomycin-intervened calcium flux from Jurkat T-cells [18]. Irima and Toner [19] explained analogies way to pneumatically actuated polydimethylsiloxane (PDMS) valves engaging cells inside. Lee et al. [20] developed a centrifugal power to test microfluidic for trapping and stimulation of single responsive cell. The cell catching spots included smart hollows in the side wall of the molded capillary that exchange cells and confined them in a position within the applied pressure. The cell entrapping efficiency was significant in the region of 50 and 80%. Capillary valves were unified and facilitated the allocation of fluids and samples [21]. The survival rate increased 20–30% higher in comparison to the 96-well plate assay for single cells analysis.
Another design configuration is based on the confinement of cells for seeding in a densely parallel configuration, i.e., micro-well cages. Using a chip surface, individual cells were allowed to settle in each Micro-well [11]. Rettig and Folch [22] reported that PDMS micro-wells with variable diameter and depth reserved ideal to confine rabasophilic leukemia (RBL) and fibroblasts cells. After that, Yamamura et al. [23] described an analogy approach with 3000 wells on each polystyrene chip. Single cell occupancy up to 80% was reported for human blood or mouse spleen lymphocytes. The microarray scanning was practiced checking temporary alteration of Ca2+ level in antigen-specific cells individually when defined anti-mouse IgM antibody, setting off receptor of the B-cell antigen. A course to a swift increase of new monoclonal antibodies is recommended prompting the selection of the antigen specific B-cells from the array, magnifying the DNA recovery by RT-PCR and antibody c-DNA determination [24].
3. Microbial Whole-Cell Arrays
The current development in microfluidic technologies revealed that there is a growing era of biosensors and an array of disciplines providing a base for combined future progress in both fields [25]. Microbial cell array system is distinctive as compared to old cell culture techniques such as multi-well plate methods. The microfluidic system significantly enhances the signals and enrichment of target [26]. These disciplines include immobilization of biological components using different polymers, genetic engineering, advancements of solid surfaces along with cellular viability for cells patterning and deposition [27]. Sensors have been recommended as potent devices to recognize a specific class of toxins, established upon bioavailability and biological activity [11]. Most of the environmentally-related biosensors worked based on the enzyme, antibody or nucleic acid as probe molecules. Thus, such biosensors can only be used to detect a known target which limits their extended and multi-detection applications. This is also because of their highly specific nature of detections which is dependent on the specific interaction between probe molecules and the target [15]. Therefore, to tackle this limitation, based on microbial whole-cell arrays principle, a wide spectrum of cell-based biosensors with an added value of living cells as a sensing element have been developed for various applications in different bio- and non-bio sectors of the modern world. Figure 2 shows a generalized working principle of cellular sensors [28]. Rider et al. [29] developed a genetically engineered cell lines-based sensor for a rapid identification of pathogens using membrane incorporated pathogen-specific antibodies. Such cell-based sensors are usually coupled with an appropriate recognition element which is often a membrane receptor that responds to respective toxins which result in cytotoxicity (Figure 3) [28].
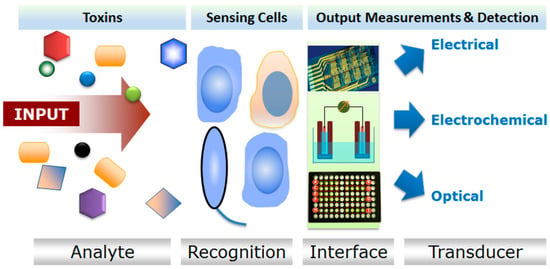
Figure 2.
A generalized working principle of cellular sensors. Reproduced from Banerjee et al. [28], an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
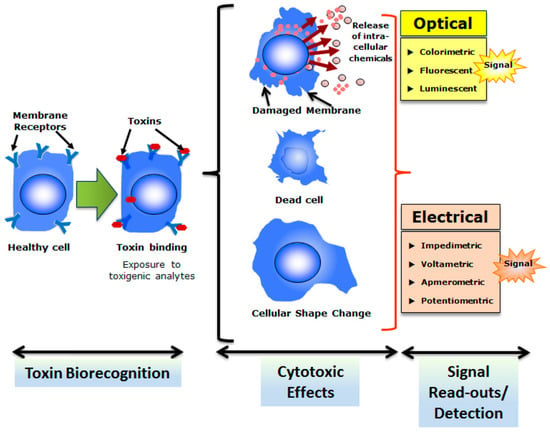
Figure 3.
The overall strategy of cell-based biosensing exploiting cytotoxicity of select toxins. Cells with appropriate recognition elements (often membrane receptors) respond to respective toxins which result in cytotoxicity. Reproduced from Banerjee et al. [28], an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
4. Cell Array Biochips
4.1. Prokaryotic Array Biochips
Eukaryotic arrays, mainly engineered on mammalian frameworks, have the selective advantage of clearly simulating the human cell reactions. Whereas, prokaryotic cell frameworks have different balancing advantages. The accessibility of cells for growth and maintenance promptly available from huge homogenous populations and appropriate techniques are used for cell immobilization and cell preservation. Prokaryotic cells are likewise most powerful and additionally have lower sensitivity to the biochemical and physical environment. However, they are vulnerable to biochemical infection [30,31]. Moreover, microscopic species are also susceptible to biological and physiochemical stimuli mandatory during the array formatting. Precisely, bacterial species can transmit their signals either chemically or physically in the established environment upon the availability of the target compounds or any change sensed in the surrounding environment. In general, this condition is attained by the combination of detecting components and particular promoter to a described molecular system [32,33,34,35]. A couple of expressed independent reporter system has been reported in the single organism [36,37,38]. In addition, reported sensors could identify particular analyte in distinctive media including soil, gas as well as water [39,40,41].
4.2. Eukaryotic Array Biochips
Eukaryotic cell array biochips have enormous potentialities to provide an adequate pattern to engineer high-throughput screening in the region of the whole cell array. It has been recommended that eukaryotic cell arrays can be utilized in different bio-applications such as micro-psychometry, gene and single cell analysis, bio-sensing and bio-identification of therapeutic agents [42,43,44,45,46]. The techniques utilized for developing cell arrays include various cells positioning that is different from lithographic approaches to inkjet technology [47,48,49]. Peculiar patterns involve utilizing microcontact printing for designing of self-assembled monolayers and cell adhesion [50], photo- as well as electro-patterning of hydrogel-encapsulated fibroblasts arrays [51] and inkjet printing of sustainable mammalian cells [52]. Koh et al. [53] fabricated a 3-D array by introducing polyethylene glycol (PEG) hydrogel to test multi-phenotype of encapsulated cells (hepatocytes, fibroblasts, and macrophages). The equal distribution of cells in each PEG hydrogel microstructure controlled via changing the cell density [53].
5. Microfluidics Approaches in Bio-Applications
The microfluidic framework consists of the manipulation as well as control of fluid near to the micron level (10−9 to 10−18). Along microchannels near to sub-millimeter extension, fluid flow is relatively different from a macro-scale system with characteristics of laminar flow, subject to the Reynolds number. The Reynolds number is an important dimensionless quantity in fluid mechanics used to predict flow patterns in different fluid flow situations. For instance, turbulent flow only exists at macroscales, and laminar flow is dominant at microscale. This property can also influence shear forces as well as mixing near the surface. Shear stress of fluid that designates the degree of applied force practiced to a surface parallel or tangentially is an essential characteristic of biofilm adhesion mechanism [54]. The simple microfluidic system holds straight channel that exhibits perfect laminar flow with no active or molecular based mixing determined by Stoke’s drag as well as general effects surface tension. With an extensive series of daily life applications, the fabrication of devices is considered an emerging technology, stabilizing various micro-machining techniques, soft and polymer-based lithography such as PDMS devices. Moreover, the assimilation of multi-functional Micro Electro Mechanical Systems (MEMS) devices: Micropumps and valves as well as other dynamic mixers [55,56]. The active and passive mixing flow the integration of mini-actuators can easily achieve velocity and control over the reagents into microchannels. The development of chemical composition and spatiotemporal gradients of velocity in the microenvironment of laminar flow provides exceptional control over the flow. The different concentration gradients in the passive devices with no actuator can be developed in the microchannel by using dilution or mixing media in various logarithmic or linear levels [57,58].
The advantages of the diffusion-based microfluidic system to create steady and random chemical gradients outweighs the disadvantages while freely moving single cell bacteria analysis. The bacterial chemotaxis quantification, efficient analysis of dynamics of single or multicellular colonies in laminar flow as well as extraordinary throughput makes microfluidics an innovative application. As developments of in vivo bacterial biofilms intensify the protection as well as persistence of infections, the fabrication of microenvironment and utilization for in vitro study should be preferred over traditional approaches to assess antimicrobial agents. Minimum biofilm eradication concentration (MBEC) of yeast embedded biofilms can be 10 to 10,000 than minimum inhibitory concentration (MIC) of their planktonic counterparts [59]. However, antimicrobial susceptibility analyses of phenotypes of biofilm isolates are yet regularly assessed with classical methods covered by standard protocols. The dilution titer method is generally used for MIC calculation which helps to determine the antimicrobial agent’s precise concentration required for growth inhibition or killing of planktonic microbial cells [54].
5.1. Microfluidic Study of Microbial Biofilm
The biofilm characterization has been carried out through various systems since the determination of “animalcules” by Van Leeuwenhoek in his teeth plaque in the 17th century. That notwithstanding, until 1978, not even a single common biofilm theory was published. According to the concept, the maximum bacterial growth took place in the matrix embedded cells which are adherent to nutrient-rich surfaces of aquatic ecosystems. The sessile nature of these matrix embedded bacterial cells distinguished them from their planktonic cells [60]. Aquatic bacteria, in turn, stick to solid surfaces and construct bacterial community [61]. These bacterial communities are supported by extracellular polymeric substances (EPS). Microbes possess a protective shield provided by the biofilm matrix and apply mainly to various clinical trials, comprising (1) Symptomatic inflammation, (2) antibiotic resistance as well as (3) the proliferation of dangerous emboli [61,62,63,64,65]. Cells can exist within critical conditions, for example at elevated temperatures or antibiotics availability in biofilms. The biofilm matrix boasts a microbe’s chance to spread within the structure. The dental hygiene and nosocomial diseases can be elaborated by such applications [66,67].
Various studies on biofilm evolution have observed genetic and ecological factors, comprising shearing stress [68], the topography of surface [69], quorum sensing [70,71], thermodynamics and nutritional level [72,73]. Nevertheless, precise insights of the mechanism associated with an evaluation of biofilm and consequences of all factors have not been properly determined. Microfluidics provides specific control over real time-flow, online observation accessibility, high-throughput trials as well as in vivo explicitly biological environments. An understanding to the protocol of development of bacterial biofilm and along with that, the utilization of microfluidic assay might help to determine keys therapeutic approaches for biofilm infections or biofilm-concerned issues. Biofilm formation by Pseudomonas aeruginosa has been explained by various researchers via a sequence of levels as seen in Figure 4 [74]. The inceptive two stages are distinguished by loose planktonic bacterial cells adhesiveness to a surface top as well as the development of EPS. Planktonic cells motility under the surface helps to determine the physical forces, such as van der Wall and gravitational forces, hydrophobic coordination as well as Brownian motion and electrostatic surface charge respectively. When cells of bacteria reach another cell or come near to the surface, explicit coordination among the respective bodies turns noteworthy [75]. The planktonic cells accumulation and biofilm maturation during biofilm formation process carried out at third and fourth steps. Biofilm structure is dependent on the nutritional levels and can be figured as a slab or mushroom fit as a fiddle [76]. At fifth level, detachment and dispersion of planktonic cells from the matrix of biofilm takes place [77,78]. However, many investigations are carried out to examine the preliminary steps involved in biofilm evolution. Particularly the studies about detachment reveal that it is the largest source of biofilms associated diseases which would be helpful to develop better therapeutic strategies [79].
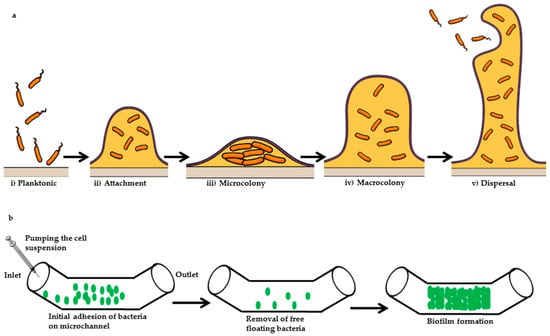
Figure 4.
(a) Illustration of a general biofilm formation; (b) microfluidic approach of biofilm formation. Initially, the bacterial cell suspension is injected into the inlet of the microchannel of a microfluidic device. Cells are allowed to adhere on the surface of the microchannel, and nonadherent cells are removed from the microchannel via using a continuous flow of cells grown media. In the subsequent final stage, cells are allowed to form biofilm inside the microchannel.
Conveniently, devices are fabricated of wafers, glass, elastomers, plastics, and papers. Various studies enforced microfluidic technology because of its significant compatibility, low volume liquid control, cells confining along with particles in a structural geometry, control of temperature and along with that specific gradient development, allowing moderate cost, fast and additionally accurate analysis. Bacterial biofilm studies are a promising stage given by microfluidic devices with an illustration presented in Figure 5. In a closed system, bacterial biofilms could coordinate with hydrodynamic settings. It permits formulating mathematical configuration that explain the effects of such coordination and disclosing the consequences of the hydrodynamic environment (e.g., shear stress) on biofilms formation [80]. In such devices, the fluid flow is relatively steady. It generates rapid response times because of the less Reynolds number and develops a gradient of chemical attractant as well as monitors bacterial chemotaxis. Concentrations, as well as translucency, allow online observation of the biofilm development by utilizing high-throughput arrays. The limited diffusion time at limited scale supports planktonic cells to develop biofilm because dynamically favorable conditions are arranged. In microfluidic devices, the conditions can be utilized to develop structures of the in vivo conditions in 3-D developmental stage [81]. Such characterizations are valuable not only for analyzing the benefactions of all moving elements in the development of a biofilm, yet for reporting the allowed responses. An approach based on microfluidic can probably disclose the procedure in the biofilm development to fix various problems related to biofilm.
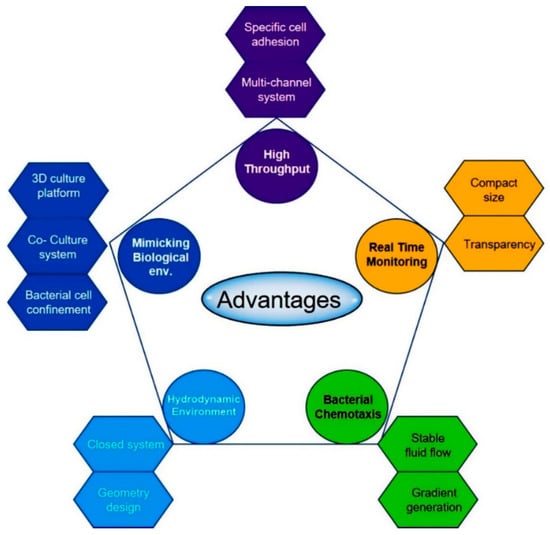
Figure 5.
Advantages of a microfluidic approach for biofilms formation and eradication. Microfluidics provides unprecedented control over the flow condition, accessibility to real-time observation, high throughput testing and in vivo like environment. The microfluidic approach helps to understand the mechanism underlying biofilm formation and to identify the problem.
Micro-channels have been fabricated using PDMS. They allow online checking on the hydrodynamic environment where planktonic cells are cultivated. According to Bahar et al. [82], chambers of the microfluidic array are fabricated to determine the adhesiveness of Acidovorax citrulli. Lee et al. [83] determined the spatial alterations under shear stress inflicted in a PDMS device exhibited by biofilms. During the dispersion stages or biofilm development in mature biofilms shear stress impacts negatively; although, throughout the initial moreover adhesion periods, shear stress facilitates biofilm development by proposing more nutrients as well as a chance for dispersion [84]. Lack of stress facilitates the growth of a bacterial cell; biofilms development gradually correlates to the growth of cells under stressful situations. The geometry of the flow channel can be fabricated to regulate the distribution of shear stress on biofilm [85]. Near various sites inside a channel, cells of bacteria can encounter different levels of shear stress. It alters the coverage of biofilm, thickness, and viability. Assembled microfluidic channels explicate the alloyed impact of various effectors on the development of biofilm. The impact on biofilm eradication or detachment of Dispersin B hydrolyzing enzyme or rifampicin antibiotic is configured by utilizing poly-channel devices. Dispersin B, as well as rifampicin medication, activate most of the biofilms removal. Although at the intersection, because of inadequate shear flow the biofilm remained unmarked. The alloyed impacts of the hydraulics and medicines provide a constructive tool for the eradication of biofilms [83]. A strategy based on microfluidic could be priceless for constructing arithmetical models with these experimental works. Arithmetical model formulated by Janakiraman et al. [80] imparted on the development of biofilm within a closed system, where the growth of biofilm, as well as hydraulics conditions, are interconnected.
In microfluidic devices, steady flow environment enhances flow-free development along with a balanced gradient. In the growth of biofilm, the chemotaxis of uncontrolled swimming or bacterial cells adhered to surface exhibit an essential role. In microfluidic channels, different cases of gradient development for the schematic study of bacterial chemotaxis are dependent on the flow. Devices having analogous or collateral movement entitled T-sensors control relayed on the convergence of trio stream that attaches into a distinct micro-channel [86,87,88]. At the end of the channel, the bacterial dispersion is figured, generating a collective reaction to the emerging on streamlined gradient accomplished alongside the micro-channel [89]. Generators having flow-free chemotaxis design a degree of a slope relayed only on molecular dissemination and no flow is occurring. In this example, to build a preliminary gradient a flow structure is fabricated then flow is switched off by permitting the gradient to emerge by diffusion alone [90]. On a brief countdown when operated, the gradient in these devices can be proximate as being in steady-state. Whenever the countdown of the gradient relaxation is extended than the times of the sensing as well as behavioral mechanisms, these gradients are beneficial for chemotaxis quantification [89]. Integration of perforate substances like hydrogels or membranes allows the assimilation of stable chemical gradients in a condition within microfluidic devices relinquish of flow or shear [90]. P. aeruginosa adhesion on a surface altered the concentration of oxygen, which was fabricated utilizing a gas permeable membrane and the introduction of dyes to the microstructures as described by Skolimowski et al. [72].
5.2. Microfluidic Approach to Antimicrobial Synergism
The utilization of unified microfluidics is an attractive option to test antibiotic susceptibility. This is because of its associated benefits such as low sample volume requirements, high-throughput, single cell sensitivity, the ability to obtain accurate bacterial cells information, and the propensity to quantify bacterial interactions in polymicrobial cultures. Various microfluidic platforms have supported these advantages for mono-microbial antibiotic susceptibility [5]. Moreover, microfluidic platforms have been used for studying co-cultures in addition to cell interaction in 2-D and 3-D environment for several applications such as tissue engineering [91]. The auto-inducer conciliated cell–cell signaling method is Quorum sensing (QS) among bacteria. It supports the development of a biofilm, resentment as well as various phenotypes of other multicellular. As antimicrobials, QS inhibitors are being examined due to their compatibility to lessen the manifestations of infectious disease and in the meantime minimizing the development of resistant strains. Inhibition of genotypic QS responses has been explained by auto-inducer-2 (AI-2) analogs between various bacteria. Firstly, Roy et al. [92] explained the potential of C1-alkyl AI-2 analogs and isobutyl-DPD, particularly inhibition to the maturation of in vitro cultivated Escherichia coli biofilms. Utilizing a peculiar microfluidic system that integrates versatile, real-time computations, density and an alloyed method of biofilms is identified. According to this method, the combination of gentamicin and isobutyl-DPD is extremely implicated in yielding near total eradication of already available E. coli biofilms. Eradication of already available biofilms has lasted a serious issue of health care. These computations approve meditations of a new protocol established on the alloying of “quenching” QS signal transduction methods accompanied by classy antibiotic therapy.
Using microfluidic platforms, the efficiency of an antimicrobial candidate has been investigated against wound model methicillin-resistant Staphylococcus pseudintermedius (MRSP) biofilms. Dispersin B, a glycoside hydrolase enzyme, has been shown to constrain biofilm development in Staphylococcus species effectively, particularly S. epidermidis [83,93]. Dispersin B can be an effective entity for analyzing the wound infection prototypes viability; obtained results can be confirmed by utilizing well-recognized methods of micro-titer plate assay [94,95]. The efficacy of Dispersin B was previously analyzed utilizing classical approaches to investigate wound associated bacterial species viz Klebsiella pneumoniae and Staphylococcus epidermidis [96]. Nevertheless, the associated impact has not been determined on an alternative wound infection triggering MRSP bacterium. Multidrug-resistant pathogens may across at wound location, leading to huge indisposition and mortality rate as well as healthcare expenses for humans and animal respectively. The development of biofilm has been assumed the presence of exceedingly resistant as well as MRSP pathogenic clones [97]. Terry and Neethirajan [98], assembled a microfluidic array to investigate antimicrobial agents’ effect on wound biofilms associated microorganisms including MRSP.
Traditional approaches to analyze antibiotic resistance are comprised of disk diffusion and broth dilution methods [99]. Microfluidic devices can develop infectious diseases management at point-of-care as well as the implementation of antimicrobial susceptibility test (AST) at clinics [55,100]. An essential condition for the prompt growth of bacterial species is the sufficient availability of oxygen in the microenvironment [101]. In traditional orbital shaking bacterial cultivation approaches, bacterial growth sustained via the supply of oxygen in the media through vigorous shaking. Sufficient oxygen supply to cell and tissue cultures carried out by oxygenator usually comprised of bioreactors and perfusion circuits [102]. On the contrary, microfluidic systems have a larger surface to volume ratio due to the small length scale. This gives an easy, still effective, approach for oxygenation within a microfluidic cell culture system [103,104]. Leibovitz et al. [105] analyzed that P. aeruginosa colonized in the oropharynx of those elderly patients which are disposed to feed with nasogastric tubes (NGT). Antibiotic susceptibility comparison was carried out between isolates of oropharyngeal derived P. aeruginosa and mucus cultures acquired from their hospital’s bacteriologic research centre. Isolates derived from oropharyngeal P. aeruginosa were found high resistance; noteworthy differences were found for amikacin antibiotic (p < 0.03). The profile of pulsed-field gel electrophoresis for these microorganisms was analogues to oropharyngeal isolates of P. aeruginosa. Patients fed with (NGT) might behave as a carrier of P. aeruginosa resistant strains.
5.3. Microfluidic Approaches for In Vitro Fertilisation
Notably, most research has centered on the upgrading progression of an embryo in vitro relying on the chemical composition of culture media. Although such studies have been verified to be highly fruitful and have doubtlessly participated mainly to enhance progress level subsequently supported reproduction. The sequential pair, as well as systems comprising monoculture media, has been improved. The growth of better-quality blastocysts in vitro is currently an interested area [106,107]. Although the chemical demands of the growing embryo do not only require being valued, yet potential physical demands can also be major characteristics in the pursuing of upgraded in vitro environment. It is imperative that movement of the embryo via the reproductive tract of female additionally results in disclosure of the embryo to an alternating liquid, yet it gives steady mechanical stimulation, that can affect the progression of the embryo [108,109]. Moreover, physical characteristics of the culture platform have an impact on the chemical composition of media through synchronizing the chemical gradients that create about the progressive embryo. Understanding of the pre-implantation embryo enhances as well as recent methods of analysis and technologies evolve, testing of different unique platforms of culture to identify the consequence of physical along with motorized refinements resting on the embryo can assist in the in vitro sequence of supplementary adaptations [110,111]. The method used to restrict embryos to a limited area is micro-drops, to acquire the assistance of the probable gain of factors: trophic autocrine or else paracrine. Conventional magnitudes of such drops primarily dimensioned in the range of 10 to 50 microliters [112]. Efficient micro drop plates are newly accessible to exaggerate this feature: drop/embryo disarticulation. They may be substantial in only some laboratories for a succession of the embryo by supporting revelation with the supervision of the embryos [112]. Additional to these, some other scaffold-free 3D cell culture methods are illustrated in Figure 6 [113]. Some of these such as the conventional hanging drop culture method has been used for initiating the development of embryonic bodies.
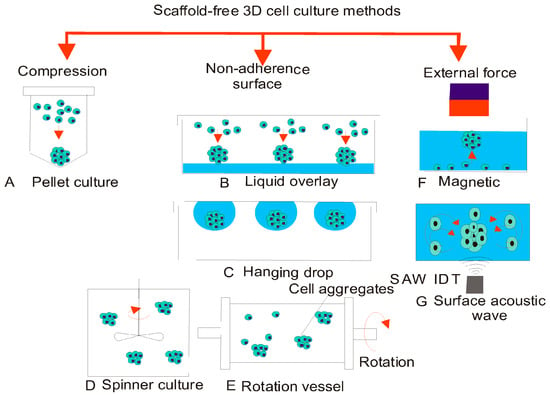
Figure 6.
Conventional methods for spheroid generation: (A) pellet culture; (B) liquid overlay; (C) hanging drop; (D) spinner culture; (E) rotating vessel; (F) magnetic force; and (G) surface acoustic wave. (Reproduced from Ref. [113], an open-access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Moreover, the progression of the embryo, although much tempting is the ability of versatile systems of culture, major perfusional system and unifies different protocols of laboratory over a unit device. It was recently demonstrated by unifying IVF and the approaching culture of embryo promisingly over the correspondent microfluidic device retaining embryos of mouse [114,115]. Importantly perfusion culture platforms lead clearly by different media in the progression embryos, therefore by minimizing interlinked pressure from the persisted organization of the embryos. Among dish on the way to the dish, they are shifted and minimizing stress from sudden alterations of varying media contents and concentration. According to this method, embryos would no longer be constrained to the demonstration to the single isolated unit or else progressive integer of culture media through 1 or 2 refilling in the culture duration. Such a notion of manifesting gametes in addition to embryos gradually altering mediums can be beneficial for other ideas of cryopreservation, either steady rate freezing or verification. In two cryopreservation procedures, manual bit-by-bit alterations in the cryopreservation media indices activate shrinkage of the cell as well as re-expansion which may be lethal to later gamete or embryo cryosurgical plus/or else application. The potential to automatise deliberate cryo-solution alteration in the cell, even if pondering the cell in an inactive stage, could have numerous bio-technical returns [110,111,112,116]. Low efficacy of Assisted Reproductive Technology (ART) stimulated researchers and technologists to explore new possible methods. The possible transformation of IVF primary mechanisms has been recognized in various studies. The superiority of the microfluidic system during sperm isolation and collection illustrated through various advantages (short time analysis, high efficacy, online observation, natural microenvironment and automation of devices) over traditional assays. Although, it is considered that the evolution of advanced technology can compile entire experimental steps on a single device [117].
The non-invasive consequence of metabolism next to the single cell level will have numerous applications in organizing cellular physiology. The solitary clinical application would be to compute the metabolic performance of embryos developed by way of assisted reproduction. There is validation that embryos with better developmental capability have distinct metabolic profiles [118]. Solitary of the criterion means pro evaluating embryonic metabolism has been to estimate consumption along with the production of various input active substrates (glucose, pyruvate, in addition to lactate) exploiting microfluorometric enzymatic assays. These assays are showed by hand exploiting pipes, which significantly confines the efficacy of this scheme. In the course of multilayer soft lithography, Urbanski et al. [117] have premeditated a microfluidic device that can disclose these assays in an automated manner. This scheme shows a sample with enzyme cocktail aliquotting, integration of reagents, and records acquisition in addition to facts investigation exclusive of operator intervention. Among the improved throughput along with flexibility of this scheme, numerous obstacles to the organizing metabolism of embryos along with single cells are expelled. As a testimony of standard, metabolic actions of individual murine embryos were arranged via exploiting this device.
5.4. Microfluidic Approaches for DNA Analysis
DNA sequencing is an important step in genomics. Conversely, the Human Genome Project [108] was investigated many years back, DNA sequencing considers as a core course in studies designed at perceptive a range of diseases alongside by identifying prospective drug targets [119]. Diverse microdevices for DNA sequencing recognized ones that exploit microfluidic methodologies for Sanger sequencing, to facilitate the most uncomplicated conventional approach for de novo genomic sequencing [120,121,122,123]. Three stages including thermal cycling, sample purification, and capillary electrophoresis assimilated and practiced on the unique microdevice by Blazej et al. [124]. On the other hand, the sequencing length has got to be improved to accomplish the current sequencing stage; these microdevices lead to the theory that assimilated microfluidic devices can be recognized for sequencing for additional uses such as low-cost proper sequencing or else for single-cell genome scrutiny [125,126].
Nucleic acid intensification techniques: polymerase chain reaction (PCR) plus the current thermal amplification is obligatory in every field of biology [127,128]. Pro-nucleic acid escalation, microfluidics leads to various recompenses while compared to conventional approaches such as reduced reagent expenditure, fewer escalation times, upgraded diagnostic throughput, with minimized risk of contamination, amplified sensitivity with assimilation. In the direction of plan an optimal taster in gene analysis scheme, micro-PCR has been recognized for utilising continuous-low, droplet-based microreactors with valve-actuated PCR micro-chambers [129,130]. The fabricated chip comprises 2.20 cm in length with 90 µm and 40 µm width and depth of channels respectively. The individual channel frequently crossed via three precise temperature zones (95, 77, 60 °C) utilizing thermal settled copper blocks [129].
The exploitation of droplet-based microfluidics restrains the walls of channels from consequent by way of the polymerase with template DNA also apart from the bound DNA or enzyme that may lead to imitating the results, enhances reaction yield, also reduce contamination of samples. Hindson et al. [131] designed a digital droplet PCR (ddPCR) device for quantification of DNA. The structure was facilitated to process, in sync, 20,000 PCR reactions as of just about 20 µL of reagent mixture. Ottesen et al. [132] fabricated a micro-PCR for single cell detection. Hindson et al. [131] fabricated a single step real-time PCR system to incorporate cell lysis with PCR on a chip to identify bacterial seeds. The assimilation of various cell lysis techniques in micro-PCR is exploiting thermal, chemical, physical and electrical methods [133]. PCR microfluidic chip exploits small volumes; appropriate consideration must be halted to restrain reagent evaporation while the thermocycling action [134]. Isothermal intensification techniques are economical and not labor intensive to restrain their exploitation in schedule investigations; first isothermal escalation approaches are an exceptional alternative to identify nucleic acids in microfluidic systems. Recently, different DNA-based microfluidic probes have been exploited along with dynamic dimension approaches such as surface plasmon resonance, imaging and fluorescence [135]. Hong et al. [136] designed a nanofluidic system for DNA recovery exhibiting three parallel processors. To get some useful forensic information, several steps can be distinguished as shown in Figure 7 [137], in which the conventional techniques, as well as their existing microfluidic counterparts, are listed.
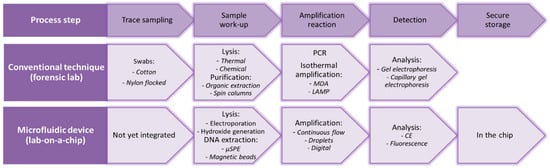
Figure 7.
In the top row, the consecutive steps in the process of forensic DNA analysis are listed; within the middle row, the conventional technique with some examples. In the bottom row, microfluidic analogies can be found with some examples. (Reproduced from Ref. [137], an open-access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
The potential to create synthetic genes leads to a potent tool for genetic investigative fields: genomics plus transcriptomic also for small-molecule fabrication. Even if, high expenditure in recent times hinders de novo incorporation of DNA theories. Microfluidic oligonucleotide incorporation identifies a financial reserve to conventional approaches, need lesser amounts of reagents [138]. Kong et al. [139] have identified multi-chamber fabricated microfluidic coordination for synthesising a 1 kb elongated gene to exploits small amount of oligonucleotides (1025 nM) and reagents than expected techniques. Presently, a microdevice fabricated for 16 oligonucleotides that can gather into a 200 bp elongated DNA [140]. The device is a valve-fabricated microfluidic device, coordinate to allow entity trial management and product assortment. Kosuri et al. [141] integrated a 35 kb of DNA encoding 47 genes containing 13,000 oligonucleotides.
In vitro genotoxicity, investigations categorise carcinogens that are measured to proceed mainly in the course of a method consisting of straight genetic damage. Ahn and Gu [142] fabricated a Geno-Tox cell array chip for genotoxicity investigation; eight recombinant biolumi nascent bacteria were exploited to formulate a Geno-Tox cell array chip productively. Four well-characterized DNA damage chemicals were designated to set up the competence of this Geno-Tox array chip as well as all strain was typically alert according to the precise form of the genotoxic act. Even though, this Geno-Tox cell array chip could be exploited to categorise and value the genotoxic modes of impact; accordingly, it could be exploited to grant the genotoxic means of action for novel drugs or anonymous or recently formulated chemicals in cooking also the atmosphere.
In previous years, many gene expression characterising processes have established and also progressively applied to research. They comprise discrepancy show, subsequent investigation of gene expression and microarrays [143,144]. Sixteen prostate cancer specimens were differentiated from nine Benign prostatic hyperplasia (BPH) samples by Luo et al. [145] that was consisted of gene expression modifications as tested on 6500 cDNA microarrays. Different groups utilizing subtractive hybridization as well as microarray analysis have characterized many prospective immunomodulatory candidates for prostate cancer treatment: Protein, the six-transmembrane epithelial antigen of the prostate (STEAP), and prostate cancer antigen 3 (PCA3) [52,146]. Prostate cancer cell line is recruited by Virolle et al. [147] which exhibited a higher constitutive level of Egr1 protein, a transcription factor overexpressed mostly in advanced tumorigenic prostate cancer cells. They occupied a pair of consequent subtractive DNA as well as testing of the microarray to compute novel genes principally for squamous cell sarcoma of the head and also neck (HNSCC) as potential markers of tumor and antiserum entities. Nine recognized genes were acknowledged to be correctly overexpressed in HNSCC relatively to healthy tissue. Likewise, in sarcomas subset, four unique genes were overexpressed.
The transcriptome in oral cavity squamous cell sarcoma was acknowledged by Alevizos et al. [148]. The identifications of six hundred participant genes that comprised transcription factors, malignant genes, differentiation markers, tumor suppressors, metastatic proteins also xenobiotic enzymes which were vigorously tinted in oral cancer, certifying merely a trio of these genes via PCR. The breast sarcoma technical heterogeneity, microarray system may be an excellent tool for accurate auxiliary presentation. Primal experiments exploiting microarray-based expression profiling discussed its capacity to organise estrogen receptor accurately-negative with breast sarcomas consisting estrogen receptor-positive and to categorise BRCA1-equivalence sarcoma commencing BRCA2-analogy also sporadic tumors [149,150,151].
Alizadeh et al. [152] studied eight hundred and sixty-four DNA constituents screened in resistant to ten ovarian cancer cells lines as well as 5 in the pink epithelial cell lines exploiting limited cell seeding to enlarge the epithelium surface of ovarian before the exclusion of RNA. They documented the 2 types of diffuse large B-cell lymphoma (DLBCL) comprised on gene expression protocols which are indicators of different levels of B-cell isolation. This categorization of molecules has characteristic value independent of stratification by the uncomplicated scientific grading. In lymphoid malignancies to elucidate gene expression, a strong coordinating community recognized a proficient microarray: Lymphochip. It is supplemented in genes which are correctly exhibited in lymphocytes auxiliary in adaptable lymphocyte function genes. Marcy et al. [153] have developed a conventional microfluidic device that permits the isolation as well as genome amplification of entity microbial cells, thus, permitting organism genomic investigation of complicated microbial environments’ devoid of the need for culture.
5.5. Microfluidic Outlook for Protein Analysis
A precise detection, quantification, and purification of cellular proteins have central importance in various life sciences related fields including drug discovery or pathogen detection [154]. Within the cell, however, proteins are mostly found in intensively limited copy numbers that develop identification of the proteins in the adventurous single-cell analysis [155,156]. Protein analysis among multiple or single cells microfluidic reactors has been carried out [157]. Taniguchi et al. [158] have designed an automated image rotated microfluidic platform to compute the proteome of E. coli as well as in single cells transcriptome along with its single-molecule sensitivity. A chip of PDMS was encompassed of 96 separate channels allocated to clench 96 discrete samples in collateral channels [158]. All channel quantifications: (w) × (l) × (h) = 150 µm × 10 mm × 25 µm and to immobilise bacteria within the micro-channels, these channels were pre-coated using poly-l-lysine [159]. An additional method is flow cytometry for organizing protein expression with the purpose to count microscopic details [160]. At this time, microdevices scrutinized antibody staining as well as transfection competency of the green fluorescent protein (GFP) [161,162]. For instance, a commercial lab on a chip device (Bio-analyzer, Technologies GmbH) exploited to ensure protein expression in cells [163].
Enzyme-linked immunosorbent assay (ELISA) plates all along concurrent with bacterial cells solid phase immunoassays immobilized at the same time as antigens have been with altered bacteria exploiting antigen–antibody dexterity on cell facade. Principally, whole cell assays are favorable when the antigens that respond all along with the antibodies in opposition to a definite pathogen are unidentified [164]. Likewise, pure antigens segregation is a bit complex and labor exhaustive [165]. Joint reimbursement of a cell-based immunoassay with the compensation of a restrain array configuration. A bacterial array biochip was pre-printed based on nitrocellulose-coated glass substrates to reveal antibody related interactions and to detect Gram-negative and Gram-positive strains [166]. In tissues, a protein expression acquired from squamous cell carcinomas of the oral cavity via an antibody microarray technique has been ascertained by Hanash and Schliekelman [167]. Micro-dissection ambushed by exploiting laser to prompt complete proteins driving out of microscopic cellular communities. They inspected potent expression of diversified proteins in stromal cells encompassing as well as in vicinity suburbs of diseased epithelium which are merely equated along with tumor progression of the epithelium. Paweletz et al. exploited a reverse emphasising arrangement of proteomics microarray that provoked proteins deriving out of patient specimen was pre-printed onto a facial and also in prostate cancer permitting disguise of biochemical signaling arrays [168]. Rapid investigations perceived autoantibodies amid peculiar intracellular as well as facial antigens which were attributable in the serum driving out of patients along with peculiar cancer categories [169].
6. Conclusions
Over the years, many researchers have proposed and developed unique technological tools, both at the micro- and nano-levels, for fabricating microfluidic-based network devices, cell-based analyses, cell-based manipulation systems such as optical, electrical, mechanical and magnetic manipulation methods. Despite such advancement, several standard procedures still need to be evolved comprehensively as sometimes the entire fabricating process is reliant on various critical aspects that are hinged on: (i) application, (ii) device channel geometry (dimensions), (iii) integration type (single or 3D), (iv) network integration with electronics or sensors and (v) material system. As discussed above, microfluidic technology offers a wider spectrum of potential applications with opportunities for enhanced performance through automation, controlled flow speed, portability, cost-effective ratio, minimal energy consumption and little or no waste products. Microfluidic-based techniques for the study of cell bioprocessing, e.g., cell separation and cell lysis are well developed and documented in the literature. However, many new developments such as organs-on-a-chip and lab-on-a-chip are taking place to revolutionize the current microfluidic technologies at an advanced level. In this context, organs-on-a-chip and lab-on-a-chip programs are likely to remain the subject of intensive investigations in the different sectors of research and material-based industries. In summary, the integration of modern technology in the fabrication of microfluidic devices will not only enhance the productive output as well as it will make an attractive platform for different bio-applications.
Acknowledgments
All authors are grateful to their representative institutes for providing literature facilities.
Conflicts of Interest
All authors declare no conflict of interest in any capacity.
References
- Li, R.; Lv, X.; Zhang, X.; Saeed, O.; Deng, Y. Microfluidics for cell-cell interactions: A review. Front. Chem. Sci. Eng. 2016, 10, 90–98. [Google Scholar] [CrossRef]
- Das, T.; Chakraborty, S. Biomicrofluidics: Recent trends and future challenges. Sadhana 2009, 34, 573. [Google Scholar] [CrossRef]
- Chan, A.S.; Danquah, M.K.; Agyei, D.; Hartley, P.G.; Zhu, Y. A simple microfluidic chip design for fundamental bioseparation. J. Anal. Methods Chem. 2014, 2014, 175457. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Datta, P.; Ayan, B.; Ozbolat, V.; Sosnoski, D.; Ozbolat, I.T. 3d bioprinting for drug discovery and development in pharmaceutics. Acta Biomater. 2017, 57, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Ahmed, I.; Hwang, J.; Seo, Y.; Lee, E.; Choi, J.; Moon, S.; Hong, J.W. A microfluidic approach to investigating a synergistic effect of tobramycin and sodium dodecyl sulfate on Pseudomonas aeruginosa biofilms. Anal. Sci. 2016, 32, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Fan, S.; Xing, W.; Liu, C. Microfluidic cell culture system studies and computational fluid dynamics. Math. Comput. Model. 2010, 52, 2036–2042. [Google Scholar] [CrossRef]
- Yi, C.; Li, C.-W.; Ji, S.; Yang, M. Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar] [CrossRef]
- Weibel, D.B.; Garstecki, P.; Whitesides, G.M. Combining microscience and neurobiology. Curr. Opin. Neurobiol. 2005, 15, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Beyond the petri dish. Nat. Biotechnol. 2004, 22, 151. [Google Scholar] [CrossRef] [PubMed]
- Saltzman, W.M.; Olbricht, W.L. Building drug delivery into tissue engineering design. Nat. Rev. Drug Discov. 2002, 1, 177. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Du, Y.; Lin, J.; Qi, M.; Shu, B.; Wen, X.; Liang, G.; Chen, B.; Liu, D. Simultaneous identification and antimicrobial susceptibility testing of multiple uropathogens on a microfluidic chip with paper-supported cell culture arrays. Anal. Chem. 2016, 88, 11593–11600. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.M.; Aizenberg, J.; Analoui, M.; Andrews, A.M.; Bisker, G.; Boyden, E.S.; Kamm, R.D.; Karp, J.M.; Mooney, D.J.; Oklu, R. Emerging trends in micro-and nanoscale technologies in medicine: From basic discoveries to translation. ACS Nano 2017, 11, 5195–5214. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Song, J.W.; Gu, W.; Futai, N.; Warner, K.A.; Nor, J.E.; Takayama, S. Computer-controlled microcirculatory support system for endothelial cell culture and shearing. Anal. Chem. 2005, 77, 3993–3999. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.J.; Koom, W.S.; Koh, W.G. Cell microarray technologies for high-throughput cell-based biosensors. Sensors 2017, 17, 1293. [Google Scholar] [CrossRef] [PubMed]
- Beske, O.E.; Goldbard, S. High-throughput cell analysis using multiplexed array technologies. Drug Discov. Today 2002, 7, S131–S135. [Google Scholar] [CrossRef]
- Araci, I.E.; Brisk, P. Recent developments in microfluidic large scale integration. Curr. Opin. Biotechnol. 2014, 25, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, A.R.; Throndset, W.R.; Whelan, R.J.; Leach, A.M.; Zare, R.N.; Liao, Y.H.; Farrell, K.; Manger, I.D.; Daridon, A. Microfluidic device for single-cell analysis. Anal. Chem. 2003, 75, 3581–3586. [Google Scholar] [CrossRef] [PubMed]
- Irimia, D.; Toner, M. Cell handling using microstructured membranes. Lab Chip 2006, 6, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Lee, I.-H.; Ryu, S.-S.; Kwak, S.-M.; Shin, K.-S.; Kang, J.-Y.; Jung, H.-I.; Kim, T.-S. Single cell assay on cd-like lab chip using centrifugal single cell trap, Micro Electro Mechanical Systems, 2007. In Proceedings of the IEEE 20th International Conference on MEMS, Hyogo, Japan, 21–25 January 2007; pp. 557–560. [Google Scholar]
- Li, B.; Yu, L.; Qi, J.; Fu, L.; Zhang, P.; Chen, L. Controlling capillary-driven fluid transport in paper-based microfluidic devices using a movable valve. Anal. Chem. 2017, 89, 5707–5712. [Google Scholar] [CrossRef] [PubMed]
- Rettig, J.R.; Folch, A. Large-scale single-cell trapping and imaging using microwell arrays. Anal. Chem. 2005, 77, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Kishi, H.; Tokimitsu, Y.; Kondo, S.; Honda, R.; Rao, S.R.; Omori, M.; Tamiya, E.; Muraguchi, A. Single-cell microarray for analysing cellular response. Anal. Chem. 2005, 77, 8050–8056. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Evander, M.; Hammarström, B.; Laurell, T. Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta 2009, 649, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Strommenger, B.; Schmidt, C.; Werner, G.; Roessle-Lorch, B.; Bachmann, T.T.; Witte, W. DNA microarray for the detection of therapeutically relevant antibiotic resistance determinants in clinical isolates of Staphylococcus aureus. Mol. Cell. Probes 2007, 21, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, I.; Bendali, A.; Tabnaoui, S.; Alexandre, L.; Srbova, J.; Bilkova, Z.; Deegan, S.; Joshi, L.; Viovy, J.-L.; Malaquin, L. A new microfluidic approach for the one-step capture, amplification and label-free quantification of bacteria from raw samples. Chem. Sci. 2017, 8, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.-G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.-C.; Lee, J.C. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Kintzios, S.; Prabhakarpandian, B. Biotoxin detection using cell-based sensors. Toxins 2013, 5, 2366–2383. [Google Scholar] [CrossRef] [PubMed]
- Rider, T.H.; Petrovick, M.S.; Nargi, F.E.; Harper, J.D.; Schwoebel, E.D.; Mathews, R.H.; Hollis, M.A. AB cell-based sensor for rapid identification of pathogens. Science 2003, 301, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Elad, T.; Lee, J.H.; Belkin, S.; Gu, M.B. Microbial whole-cell arrays. Microb. Biotechnol. 2008, 1, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-F.; Tsai, Y.-C.; Yagur-Kroll, S.; Palevsky, N.; Belkin, S.; Cheng, J.-Y. Water pollutant monitoring by a whole cell array through lens-free detection on ccd. Lab Chip 2015, 15, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Belkin, S.; Smulski, D.R.; Dadon, S.; Vollmer, A.C.; Van Dyk, T.K.; Larossa, R.A. A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Res. 1997, 31, 3009–3016. [Google Scholar] [CrossRef]
- Vollmer, A.C.; Belkin, S.; Smulski, D.R.; Van Dyk, T.K.; LaRossa, R.A. Detection of DNA damage by use of Escherichia coli carrying reca’: Lux, uvra’: Lux, or alka’: Lux reporter plasmids. Appl. Environ. Microbiol. 1997, 63, 2566–2571. [Google Scholar] [PubMed]
- Lee, H.; Gu, M. Construction of a soda: Luxcdabe fusion Escherichia coli: Comparison with a katg fusion strain through their responses to oxidative stresses. Appl. Microbiol. Biotechnol. 2003, 60, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.J.; Janzen, W.P.; Kireev, D.; Singleton, S.F. High-throughput screening for reca inhibitors using a transcreener adenosine 5′-o-diphosphate assay. Assay Drug Dev. Technol. 2012, 10, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.J.; Gu, M.B. Construction and characterisation of novel dual stress-responsive bacterial biosensors. Biosens. Bioelectron. 2004, 19, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.J.; Gu, M.B. An Escherichia coli biosensor capable of detecting both genotoxic and oxidative damage. Appl. Microbiol. Biotechnol. 2004, 64, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Hever, N.; Belkin, S. A dual-color bacterial reporter strain for the detection of toxic and genotoxic effects. Eng. Life Sci. 2006, 6, 319–323. [Google Scholar] [CrossRef]
- Auchtung, J.M.; Robinson, C.D.; Britton, R.A. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (mbras). Microbiome 2015, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Gil, G.C.; Mitchell, R.J.; Chang, S.T.; Gu, M.B. A biosensor for the detection of gas toxicity using a recombinant bioluminescent bacterium. Biosens. Bioelectron. 2000, 15, 23–30. [Google Scholar] [PubMed]
- Gu, M.B.; Chang, S.T. Soil biosensor for the detection of pah toxicity using an immobilised recombinant bacterium and a biosurfactant. Biosens. Bioelectron. 2001, 16, 667–674. [Google Scholar] [CrossRef]
- Webster, D.P.; TerAvest, M.A.; Doud, D.F.; Chakravorty, A.; Holmes, E.C.; Radens, C.M.; Sureka, S.; Gralnick, J.A.; Angenent, L.T. An arsenic-specific biosensor with genetically engineered shewanella oneidensis in a bioelectrochemical system. Biosens. Bioelectron. 2014, 62, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Ziauddin, J.; Sabatini, D.M. Microarrays of cells expressing defined cdnas. Nature 2001, 411, 107. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Váchová, L.; Valer, M.; Preckel, T. Single-cell analysis of yeast, mammalian cells, and fungal spores with a microfluidic pressure-driven chip-based system. Cytom. Part A J. Int. Soc. Anal. Cytol. 2004, 59, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.L.; Miller, A.D.; Freeman, T.C. Identification and characterisation of human apoptosis inducing proteins using cell-based transfection microarrays and expression analysis. BMC Genom. 2006, 7, 145. [Google Scholar]
- Hung, P.J.; Lee, P.J.; Sabounchi, P.; Lin, R.; Lee, L.P. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol. Bioeng. 2005, 89, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yeon, J.H.; Ryu, H.R.; Chung, M.; Hu, Q.P.; Jeon, N.L. In vitro formation and characterisation of a perfusable three-dimensional tubular capillary network in microfluidic devices. Lab Chip 2012, 12, 2815–2822. [Google Scholar] [CrossRef] [PubMed]
- Park, T.H.; Shuler, M.L. Integration of cell culture and microfabrication technology. Biotechnol. Prog. 2003, 19, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Basaran, O.A.; Gao, H.; Bhat, P.P. Nonstandard inkjets. Annu. Rev. Fluid Mech. 2013, 45, 85–113. [Google Scholar] [CrossRef]
- Wendeln, C.; Ravoo, B.J. Surface patterning by microcontact chemistry. Langmuir 2012, 28, 5527–5538. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, D.R.; Tsang, V.L.; Sah, R.L.; Bhatia, S.N. Photo-and electropatterning of hydrogel-encapsulated living cell arrays. Lab Chip 2005, 5, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kalos, M.; Stolk, J.A.; Zasloff, E.J.; Zhang, X.; Houghton, R.L.; Maltez Filho, A.; Nolasco, M.; Badaró, R.; Reed, S.G. Identification and characterisation of prostein, a novel prostate-specific protein. Cancer Res. 2001, 61, 1563–1568. [Google Scholar] [PubMed]
- Koh, W.-G.; Itle, L.J.; Pishko, M.V. Molding of hydrogel microstructures to create multiphenotype cell microarrays. Anal. Chem. 2003, 75, 5783–5789. [Google Scholar] [CrossRef] [PubMed]
- DiCicco, M. Assessment of Novel Antimicrobial Therapy against Methicillin-resistant Staphylococcus pseudintermedius Biofilm with Conventional Assays and a Microfluidic Platform; University of Guelph: Guelph, ON, Canada, 2013. [Google Scholar]
- Ahmed, I.; Iqbal, H.M.; Akram, Z. Microfluidics engineering: Recent trends, valorization, and applications. Arab. J. Sci. Eng. 2018, 43, 23–32. [Google Scholar] [CrossRef]
- Ochs, C.J.; Kasuya, J.; Pavesi, A.; Kamm, R.D. Oxygen levels in thermoplastic microfluidic devices during cell culture. Lab Chip 2014, 14, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, Y.V.; Jiang, L.; Tu, Y.; Wu, M. Logarithmic sensing in Escherichia coli bacterial chemotaxis. Biophys. J. 2009, 96, 2439–2448. [Google Scholar] [CrossRef] [PubMed]
- Toetsch, S.; Olwell, P.; Prina-Mello, A.; Volkov, Y. The evolution of chemotaxis assays from static models to physiologically relevant platforms. Integr. Boil. 2009, 1, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Almshawit, H.; Macreadie, I.; Grando, D. A simple and inexpensive device for biofilm analysis. J. Microbiol. Methods 2014, 98, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Belas, R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Prince, A.A.; Steiger, J.D.; Khalid, A.N.; Dogrhamji, L.; Reger, C.; Claire, S.E.; Chiu, A.G.; Kennedy, D.W.; Palmer, J.N.; Cohen, N.A. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am. J. Rhinol. 2008, 22, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative role of β-1, 3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Deligianni, E.; Pattison, S.; Berrar, D.; Ternan, N.G.; Haylock, R.W.; Moore, J.E.; Elborn, S.J.; Dooley, J.S. Pseudomonas aeruginosa cystic fibrosis isolates of similar rapd genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Sawasdidoln, C.; Taweechaisupapong, S.; Sermswan, R.W.; Tattawasart, U.; Tungpradabkul, S.; Wongratanacheewin, S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS ONE 2010, 5, e9196. [Google Scholar] [CrossRef] [PubMed]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: Relationship among Candida spp. Front. Microbiol. 2015, 6, 205. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Nakaishi, L.A. Managing the complexity of a dynamic biofilm. J. Am. Dent. Assoc. 2006, 137, S10–S15. [Google Scholar] [CrossRef]
- Vergidis, P.; Patel, R. Novel approaches to the diagnosis, prevention, and treatment of medical device-associated infections. Infect. Dis. Clin. 2012, 26, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Rochex, A.; Godon, J.-J.; Bernet, N.; Escudié, R. Role of shear stress on composition, diversity and dynamics of biofilm bacterial communities. Water Res. 2008, 42, 4915–4922. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.A.; Demir, K.; Hartanto, T.G.; Schmitt, E.; Levkin, P.A. Droplet-microarray on superhydrophobic–superhydrophilic patterns for high-throughput live cell screenings. RSC Adv. 2016, 6, 38263–38276. [Google Scholar] [CrossRef]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Skolimowski, M.; Nielsen, M.W.; Emnéus, J.; Molin, S.; Taboryski, R.; Sternberg, C.; Dufva, M.; Geschke, O. Microfluidic dissolved oxygen gradient generator biochip as a useful tool in bacterial biofilm studies. Lab Chip 2010, 10, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, L.K.; Hermanowicz, S.W. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci. Technol. 1998, 37, 211–214. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Gottenbos, B.; van der Mei, H.C.; Klatter, F.; Nieuwenhuis, P.; Busscher, H.J. In vitro and in vivo antimicrobial activity of covalently coupled quaternary ammonium silane coatings on silicone rubber. Biomaterials 2002, 23, 1417–1423. [Google Scholar] [CrossRef]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A spider web strategy of type iv pili-mediated migration to build a fibre-like psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2013, 15, 2238–2253. [Google Scholar] [CrossRef] [PubMed]
- Lecuyer, S.; Rusconi, R.; Shen, Y.; Forsyth, A.; Vlamakis, H.; Kolter, R.; Stone, H.A. Shear stress increases the residence time of adhesion of Pseudomonas Aeruginosa. Biophys. J. 2011, 100, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, J.-C.; Coufort, C.; Escudié, R.; Liné, A.; Paul, E. Influence of non-uniform distribution of shear stress on aerobic biofilms. Chem. Eng. Sci. 2007, 62, 3672–3684. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Gibbs, K.A.; Hager, P.W.; Phibbs, P.V.; Kolter, R. The global carbon metabolism regulator crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Janakiraman, V.; Englert, D.; Jayaraman, A.; Baskaran, H. Modeling growth and quorum sensing in biofilms grown in microfluidic chambers. Ann. Biomed. Eng. 2009, 37, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.-D.; Chung, S. Microfluidic approaches to bacterial biofilm formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef] [PubMed]
- Bahar, O.; De La Fuente, L.; Burdman, S. Assessing adhesion, biofilm formation and motility of acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol. Lett. 2010, 312, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kaplan, J.B.; Lee, W.Y. Microfluidic devices for studying growth and detachment of staphylococcus epidermidis biofilms. Biomed. Microdevices 2008, 10, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Jeong, H.-H.; Lee, J.; Kim, K.P.; Lee, C.-S. Effect of shear stress on the formation of bacterial biofilm in a microfluidic channel. BioChip J. 2011, 5, 236. [Google Scholar] [CrossRef]
- Salek, M.M.; Jones, S.M.; Martinuzzi, R.J. The influence of flow cell geometry related shear stresses on the distribution, structure and susceptibility of Pseudomonas aeruginosa 01 biofilms. Biofouling 2009, 25, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Groisman, A.; Lobo, C.; Cho, H.; Campbell, J.K.; Dufour, Y.S.; Stevens, A.M.; Levchenko, A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2005, 2, 685. [Google Scholar] [CrossRef] [PubMed]
- Keenan, T.M.; Folch, A. Biomolecular gradients in cell culture systems. Lab Chip 2008, 8, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Ford, R.M. Enhanced transverse migration of bacteria by chemotaxis in a porous t-sensor. Environ. Sci. Technol. 2009, 43, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Shimizu, T.S.; Stocker, R. Microfluidics for bacterial chemotaxis. Integr. Boil. 2010, 2, 604–629. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Young, L.; Kim, S.; Fogarty, E.A.; Heilman, S.M.; Zhou, P.; Shuler, M.L.; Wu, M.; DeLisa, M.P. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip 2006, 6, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, M.; Kim, S. Microfluidic cell coculture methods for understanding cell biology, analyzing bio/pharmaceuticals, and developing tissue constructs. Anal. Biochem. 2011, 2, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Roy, V.; Meyer, M.T.; Smith, J.A.; Gamby, S.; Sintim, H.O.; Ghodssi, R.; Bentley, W.E. Ai-2 analogs and antibiotics: A synergistic approach to reduce bacterial biofilms. Appl. Microbiol. Biotechnol. 2013, 97, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Brindle, E.R.; Miller, D.A.; Stewart, P.S. Hydrodynamic deformation and removal of Staphylococcus epidermidis biofilms treated with urea, chlorhexidine, iron chloride, or dispersinb. Biotechnol. Bioeng. 2011, 108, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- DiCicco, M.; Neethirajan, S.; Singh, A.; Weese, J.S. Efficacy of clarithromycin on biofilm formation of methicillin-resistant Staphylococcus pseudintermedius. BMC Vet. Res. 2012, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Gawande, P.V.; Leung, K.P.; Madhyastha, S. Antibiofilm and antimicrobial efficacy of dispersinb®-ksl-w peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 2014, 68, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Osland, A.M.; Vestby, L.K.; Fanuelsen, H.; Slettemeås, J.S.; Sunde, M. Clonal diversity and biofilm-forming ability of methicillin-resistant Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 2012, 67, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Terry, J.; Neethirajan, S. A novel microfluidic wound model for testing antimicrobial agents against Staphylococcus pseudintermedius biofilms. J. Nanobiotechnol. 2014, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (mic) of antimicrobial substances. Nat. Protoc. 2008, 3, 163. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Moy, C.K.; Danquah, M.K.; Ongkudon, C.M. Development and characteristics of polymer monoliths for advanced lc bioscreening applications: A review. J. Chromatogr. B 2016, 1015, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.C.; Mastali, M.; Li, Y.; Gau, V.; Suchard, M.A.; Babbitt, J.; Gornbein, J.; Landaw, E.M.; McCabe, E.R.; Churchill, B.M. Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection. J. Mol. Diagn. 2007, 9, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Paul, A.; Vrana, N.E.; Zhao, X.; Memic, A.; Hwang, Y.-S.; Dokmeci, M.R.; Khademhosseini, A. Microfluidic techniques for development of 3d vascularized tissue. Biomaterials 2014, 35, 7308–7325. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Wong, P.K.; Park, J.; Levchenko, A.; Sun, Y. Microengineered platforms for cell mechanobiology. Annu. Rev. Biomed. Eng. 2009, 11, 203–233. [Google Scholar] [CrossRef] [PubMed]
- Maharbiz, M.M.; Holtz, W.J.; Sharifzadeh, S.; Keasling, J.D.; Howe, R.T. A microfabricated electrochemical oxygen generator for high-density cell culture arrays. J. Microelectromech. Syst. 2003, 12, 590–599. [Google Scholar] [CrossRef]
- Leibovitz, A.; Dan, M.; Zinger, J.; Carmeli, Y.; Habot, B.; Segal, R. Pseudomonas aeruginosa and the oropharyngeal ecosystem of tube-fed patients. Emerg. Infect. Dis. 2003, 9, 956. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.; Gardner, D.K. Embryo culture medium: Which is the best? Best Prat. Res. Clin. Obstet. Gynaecol. 2007, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Casanova, E.A.; Okoniewski, M.J.; Cinelli, P. Cross-species genome wide expression analysis during pluripotent cell determination in mouse and rat preimplantation embryos. PLoS ONE 2012, 7, e47107. [Google Scholar] [CrossRef] [PubMed]
- Ezzati, M.; Djahanbakhch, O.; Arian, S.; Carr, B.R. Tubal transport of gametes and embryos: A review of physiology and pathophysiology. J. Assist. Reprod. Genet. 2014, 31, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Fauci, L.J.; Dillon, R. Biofluidmechanics of reproduction. Annu. Rev. Fluid Mech. 2006, 38, 371–394. [Google Scholar] [CrossRef]
- Smith, G.; Swain, J.; Bormann, C. Microfluidics for Gametes, Embryos, and Embryonic Stem Cells; Seminars in Reproductive Medicine; Thieme Medical Publishers: Stuart, Germany, 2011; pp. 5–14. [Google Scholar]
- Swain, J.; Smith, G. Advances in embryo culture platforms: Novel approaches to improve preimplantation embryo development through modifications of the microenvironment. Hum. Reprod. Update 2011, 17, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Zhang, Q.; Ma, R.; Xie, L.; Qiu, T.; Wang, L.; Mitchelson, K.; Wang, J.; Huang, G.; Qiao, J. Integration of single oocyte trapping, in vitro fertilization and embryo culture in a microwell-structured microfluidic device. Lab Chip 2010, 10, 2848–2854. [Google Scholar] [CrossRef] [PubMed]
- Vadivelu, R.K.; Kamble, H.; Shiddiky, M.J.; Nguyen, N.T. Microfluidic technology for the generation of cell spheroids and their applications. Micromachines 2017, 8, 94. [Google Scholar] [CrossRef]
- Ma, R.; Xie, L.; Han, C.; Su, K.; Qiu, T.; Wang, L.; Huang, G.; Xing, W.; Qiao, J.; Wang, J. In vitro fertilization on a single-oocyte positioning system integrated with motile sperm selection and early embryo development. Anal. Chem. 2011, 83, 2964–2970. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, S.M.; Sadasivam, M.; Tasoglu, S. Microfluidics for sperm research. Trends Biotechnol. 2015, 33, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Urbanski, J.P.; Johnson, M.T.; Craig, D.D.; Potter, D.L.; Gardner, D.K.; Thorsen, T. Noninvasive metabolic profiling using microfluidics for analysis of single preimplantation embryos. Anal. Chem. 2008, 80, 6500–6507. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, M.; Kruhne, U.; Laursen, S. Full in vitro fertilization laboratory mechanization: Toward robotic assisted reproduction? Fertil. Steril. 2012, 97, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Kubinyi, H. Drug research: Myths, hype and reality. Nat. Rev. Drug Discov. 2003, 2, 665. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Gibson, J.F.; Nikbakht, H.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Next-generation DNA barcoding: Using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Resour. 2014, 14, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Blazej, R.G.; Kumaresan, P.; Cronier, S.A.; Mathies, R.A. Inline injection microdevice for attomole-scale sanger DNA sequencing. Anal. Chem. 2007, 79, 4499–4506. [Google Scholar] [CrossRef] [PubMed]
- Kartalov, E.P.; Quake, S.R. Microfluidic device reads up to four consecutive base pairs in DNA sequencing-by-synthesis. Nucleic Acids Res. 2004, 32, 2873–2879. [Google Scholar] [CrossRef] [PubMed]
- Aborn, J.H.; El-Difrawy, S.A.; Novotny, M.; Gismondi, E.A.; Lam, R.; Matsudaira, P.; Mckenna, B.K.; O’Neil, T.; Streechon, P.; Ehrlich, D.J. A 768-lane microfabricated system for high-throughput DNA sequencing. Lab Chip 2005, 5, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Blazej, R.G.; Kumaresan, P.; Mathies, R.A. Microfabricated bioprocessor for integrated nanoliter-scale sanger DNA sequencing. Proc. Natl. Acad. Sci. USA 2006, 103, 7240–7245. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, H.; Utsunomiya, S.; Nakamura, S.; Yamamoto, R.; Harada, A.; Kaji, T.; Hazama, M.; Ohashi, T.; Inami, A.; Ikegami, T. Large-scale microfabricated channel plates for high-throughput, fully automated DNA sequencing. Electrophoresis 2008, 29, 4723–4732. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Mathies, R.A. Integrated microfluidic systems for high-performance genetic analysis. Trends Biotechnol. 2009, 27, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Sanchez, S.; Hanning, I.; Pendleton, S.; D’Souza, D. Next-generation sequencing: The future of molecular genetics in poultry production and food safety. Poult. Sci. 2013, 92, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Craw, P.; Balachandran, W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip 2012, 12, 2469–2486. [Google Scholar] [CrossRef] [PubMed]
- Stals, A.; Mathijs, E.; Baert, L.; Botteldoorn, N.; Denayer, S.; Mauroy, A.; Scipioni, A.; Daube, G.; Dierick, K.; Herman, L. Molecular detection and genotyping of noroviruses. Food Environ. Virol. 2012, 4, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xing, D.; Zhang, C. Rapid detection of genetically modified organisms on a continuous-flow polymerase chain reaction microfluidics. Anal. Biochem. 2009, 385, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C. High-throughput droplet digital pcr system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, E.A.; Hong, J.W.; Quake, S.R.; Leadbetter, J.R. Microfluidic digital pcr enables multigene analysis of individual environmental bacteria. Science 2006, 314, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Privorotskaya, N.; Liu, Y.-S.; Lee, J.; Zeng, H.; Carlisle, J.A.; Radadia, A.; Millet, L.; Bashir, R.; King, W.P. Rapid thermal lysis of cells using silicon–diamond microcantilever heaters. Lab Chip 2010, 10, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Santra, T.S.; Tseng, F.G. Recent trends on micro/nanofluidic single cell electroporation. Micromachines 2013, 4, 333–356. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.W.; Studer, V.; Hang, G.; Anderson, W.F.; Quake, S.R. A nanoliter-scale nucleic acid processor with parallel architecture. Nat. Biotechnol. 2004, 22, 435. [Google Scholar] [CrossRef] [PubMed]
- Bruijns, B.; van Asten, A.; Tiggelaar, R.; Gardeniers, H. Microfluidic devices for forensic DNA analysis: A review. Biosensors 2016, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Javanmard, M.; Davis, R. A microfluidic platform for electrical detection of DNA hybridisation. Sens. Actuators B Chem. 2011, 154, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.S.; Carr, P.A.; Chen, L.; Zhang, S.; Jacobson, J.M. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007, 35, e61. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-C.; Snyder, T.M.; Quake, S.R. A microfluidic oligonucleotide synthesiser. Nucleic Acids Res. 2010, 38, 2514–2521. [Google Scholar] [CrossRef] [PubMed]
- Kosuri, S.; Eroshenko, N.; LeProust, E.M.; Super, M.; Way, J.; Li, J.B.; Church, G.M. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010, 28, 1295. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-M.; Gu, M.B. Geno-tox: Cell array biochip for genotoxicity monitoring and classification. Appl. Biochem. Biotechnol. 2012, 168, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Granjeaud, S.; Bertucci, F.; Jordan, B.R. Expression profiling: DNA arrays in many guises. Bioessays 1999, 21, 781–790. [Google Scholar] [CrossRef]
- Luo, J.; Duggan, D.J.; Chen, Y.; Sauvageot, J.; Ewing, C.M.; Bittner, M.L.; Trent, J.M.; Isaacs, W.B. Human prostate cancer and benign prostatic hyperplasia: Molecular dissection by gene expression profiling. Cancer Res. 2001, 61, 4683–4688. [Google Scholar] [PubMed]
- Day, J.R.; Jost, M.; Reynolds, M.A.; Groskopf, J.; Rittenhouse, H. Pca3: From basic molecular science to the clinical lab. Cancer Lett. 2011, 301, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Virolle, T.; Krones-Herzig, A.; Baron, V.; De Gregorio, G.; Adamson, E.D.; Mercola, D. Egr1 promotes growth and survival of prostate cancer cells identification of novel egr1 target genes. J. Boil. Chem. 2003, 278, 11802–11810. [Google Scholar] [CrossRef] [PubMed]
- Alevizos, I.; Mahadevappa, M.; Zhang, X.; Ohyama, H.; Kohno, Y.; Posner, M.; Gallagher, G.T.; Varvares, M.; Cohen, D.; Kim, D. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 2001, 20, 6196. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A. Molecular portraits of human breast tumours. Nature 2000, 406, 747. [Google Scholar] [CrossRef] [PubMed]
- West, M.; Blanchette, C.; Dressman, H.; Huang, E.; Ishida, S.; Spang, R.; Zuzan, H.; Olson, J.A.; Marks, J.R.; Nevins, J.R. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. USA 2001, 98, 11462–11467. [Google Scholar] [CrossRef] [PubMed]
- Ismail, R.S.; Baldwin, R.L.; Fang, J.; Browning, D.; Karlan, B.Y.; Gasson, J.C.; Chang, D.D. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 2000, 60, 6744–6749. [Google Scholar] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X. Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503. [Google Scholar] [CrossRef] [PubMed]
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Lösekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated tm7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.L. Systems biology in drug discovery and development. Drug Discov. Today 2014, 19, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Friedman, N.; Xie, X.S. Stochastic protein expression in individual cells at the single molecule level. Nature 2006, 440, 358. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Young, J.W.; Alon, U.; Swain, P.S.; Elowitz, M.B. Gene regulation at the single-cell level. Science 2005, 307, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Hellmich, W.; Pelargus, C.; Leffhalm, K.; Ros, A.; Anselmetti, D. Single cell manipulation, analytics, and label-free protein detection in microfluidic devices for systems nanobiology. Electrophoresis 2005, 26, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Choi, P.J.; Li, G.-W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yamaji, H.; Fukuda, H. Immobilisation of Escherichia coli cells using porous support particles coated with cationic polymers. J. Biosci. Bioeng. 2007, 104, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Freire, J.M.; Gaspar, D.; de la Torre, B.G.; Veiga, A.S.; Andreu, D.; Castanho, M.A. Monitoring antibacterial permeabilisation in real time using time-resolved flow cytometry. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Schrum, D.P.; Culbertson, C.T.; Jacobson, S.C.; Ramsey, J.M. Microchip flow cytometry using electrokinetic focusing. Anal. Chem. 1999, 71, 4173–4177. [Google Scholar] [CrossRef] [PubMed]
- Preckel, T.; Luedke, G.; Chan, S.D.; Wang, B.N.; Dubrow, R.; Buhlmann, C. Detection of cellular parameters using a microfluidic chip-based system. JALA J. Assoc. Lab. Autom. 2002, 7, 85–89. [Google Scholar] [CrossRef]
- Watanabe, K.; Joh, T.; Seno, K.; Sasaki, M.; Todoroki, I.; Miyashita, M.; Tochikubo, K.; Itoh, M. Development and clinical application of an immunoassay using intact helicobacter pylori attached to a solid phase as an antigen. Clin. Biochem. 2001, 34, 291–295. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme immunoassay (eia)/enzyme-linked immunosorbent assay (elisa). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-H.; Lee, S.-H.; Kenrick, S.A.; Daugherty, P.S.; Soh, H.T. Microfluidic protein detection through genetically engineered bacterial cells. J. Proteome Res. 2006, 5, 3433–3437. [Google Scholar] [CrossRef] [PubMed]
- Thirumalapura, N.; Ramachandran, A.; Morton, R.; Malayer, J. Bacterial cell microarrays for the detection and characterisation of antibodies against surface antigens. J. Immunol. Methods 2006, 309, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.; Schliekelman, M. Proteomic profiling of the tumor microenvironment: Recent insights and the search for biomarkers. Genome Med. 2014, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Paweletz, C.P.; Charboneau, L.; Bichsel, V.E.; Simone, N.L.; Chen, T.; Gillespie, J.W.; Emmert-Buck, M.R.; Roth, M.J.; Petricoin, E.F., III; Liotta, L.A. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 2001, 20, 1981. [Google Scholar] [CrossRef] [PubMed]
- Mintz, P.J.; Kim, J.; Do, K.-A.; Wang, X.; Zinner, R.G.; Cristofanilli, M.; Arap, M.A.; Hong, W.K.; Troncoso, P.; Logothetis, C.J. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat. Biotechnol. 2003, 21, 57. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).