SOX9 Protein in Pancreatic Cancer Regulates Multiple Cellular Networks in a Cell-Specific Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Cell Cultures
2.2. Knockdown of SOX9 via RNA Interference
2.3. Plasmid DNA Transfection
2.4. Western Blot Analysis
2.5. Immunofluorescence
2.6. RNA Isolation, Reverse Transcription, and Quantitative PCR Analysis
2.7. RNA-Sequencing (RNA-Seq)
2.8. Cell Proliferation Assays and Cell Cycle Analysis
2.9. Flow Cytometry Analysis of Apoptosis
2.10. Senescence-Associated β-Galactosidase (SA-β-Gal) Activity Assay
2.11. Caspase Activity Assay
2.12. Xenotransplantation of Cancer Cells into Danio rerio Embryos
2.13. Statistical Analysis
3. Results
3.1. Expression Levels of the SOX9 Protein and RNA in Pancreatic Cancer Cell Lines with Different Levels of Differentiation
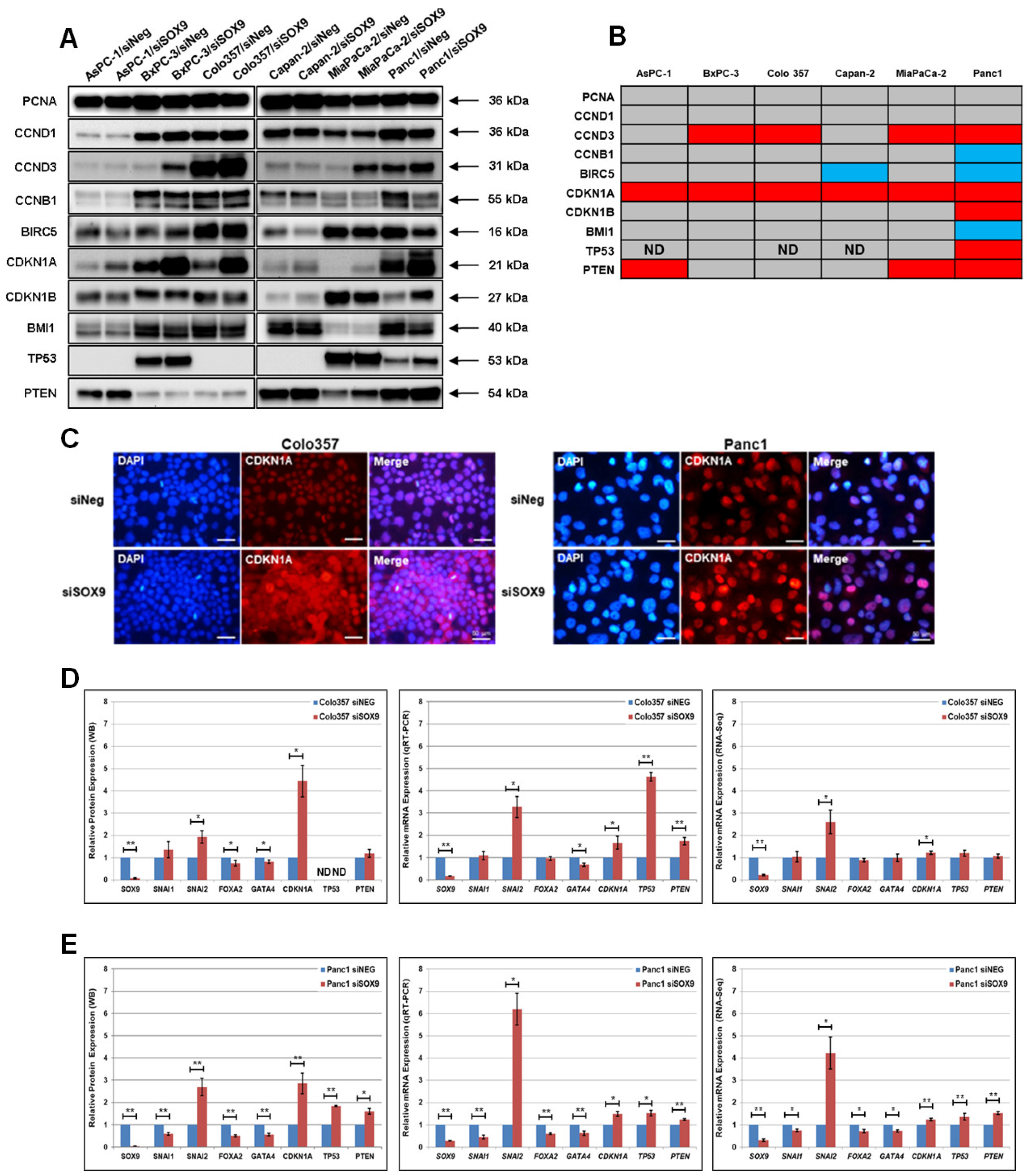
3.2. The Cell-Specific Effect of SOX9 on the Expression Levels of Protein Markers of Pancreatic Cancer Differentiation
3.3. SOX9 and the Expression of Developmental Transcription Factors in Pancreatic Cancer Cells
3.4. The Downregulation of SOX9 Affects the Expression of Cell Cycle Proteins
3.5. The Downregulation of the SOX9 Protein Affects the Transcriptional Activity of SNAI2, GATA4, CDK1A, TP53, and PTEN Genes in Pancreatic Cancer Cells
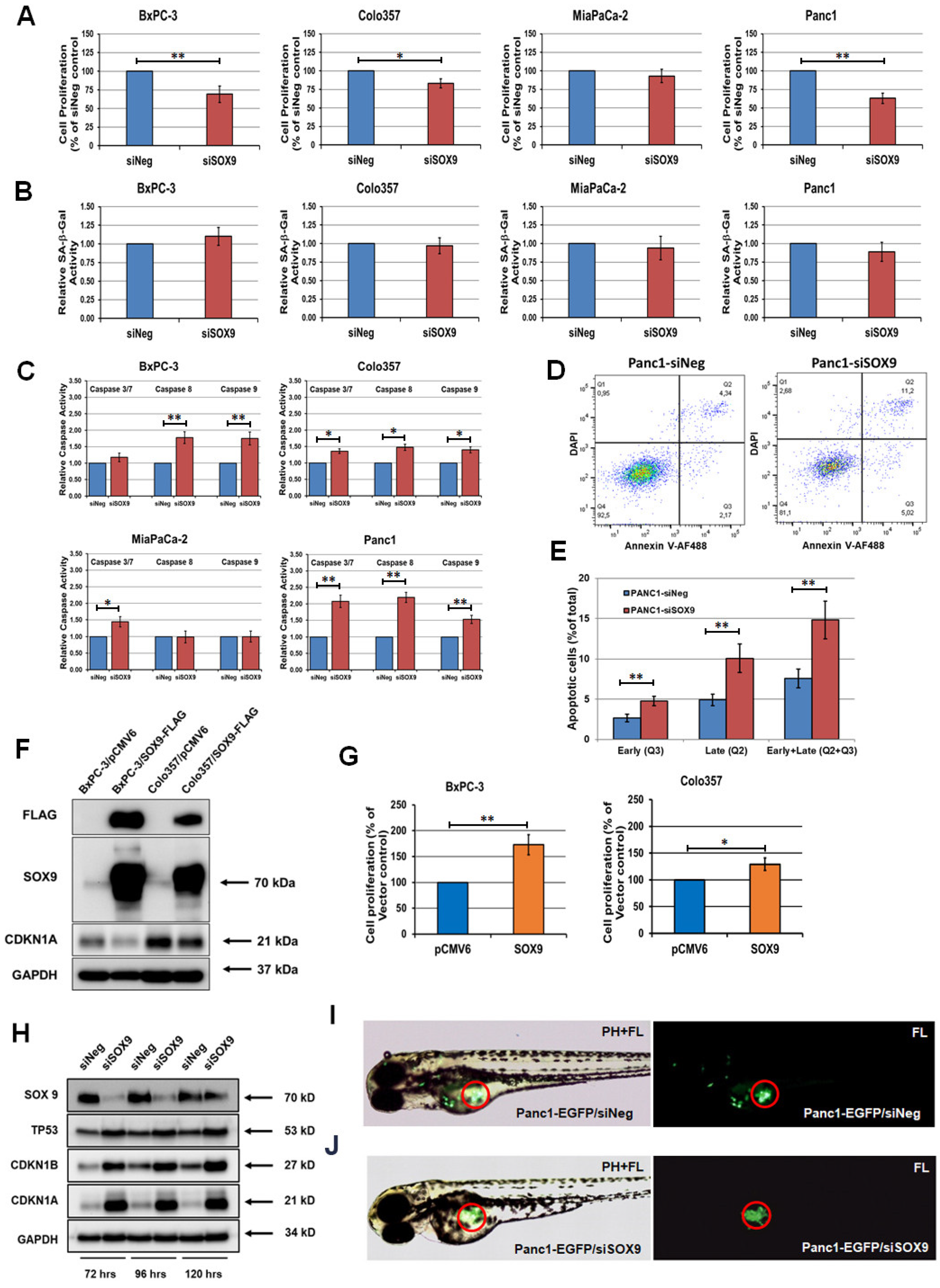
3.6. SOX9 Regulates Apoptosis and Affects the Migratory Potential of Pancreatic Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef]
- Hong, S.-M.; Park, J.Y.; Hruban, R.H.; Goggins, M. Molecular signatures of pancreatic cancer. Arch. Pathol. Lab. Med. 2011, 135, 716–727. [Google Scholar] [CrossRef]
- Makohon-Moore, A.; Iacobuzio-Donahue, C.A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 2016, 16, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Weissmueller, S.; Manchado, E.; Saborowski, M.; Morris, J.P., IV; Wagenblast, E.; Davis, C.A.; Moon, S.-H.; Pfister, N.T.; Tschaharganeh, D.F.; Kitzing, T.; et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 2014, 157, 382–394. [Google Scholar] [CrossRef] [Green Version]
- Mann, K.M.; Ying, H.; Juan, J.; Jenkins, N.A.; Copeland, N.G. KRAS-related proteins in pancreatic cancer. Pharmacol. Ther. 2016, 168, 29–42. [Google Scholar] [CrossRef]
- Kopp, J.L.; von Figura, G.; Mayes, E.; Liu, F.-F.; Dubois, C.L.; Morris, J.P., IV; Pan, F.C.; Akiyama, H.; Wright, C.V.; Jensen, K.; et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 22, 737–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Qin, Y.; Ji, S.; Ling, J.; Fu, J.; Zhuang, Z.; Fan, X.; Song, L.; Yu, X.; Chiao, P.J. SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene 2018, 37, 912–923. [Google Scholar] [CrossRef]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.-J.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014, 1, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Larsimont, J.-C.; Youssef, K.K.; Sanchez-Danes, A.; Sukumaran, V.; Defrance, M.; Delatte, B.; Liagre, M.; Baatsen, P.; Marine, J.-C.; Lippens, S.; et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 2015, 17, 60–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomsen, M.K.; Ambroisine, L.; Wynn, S.; Cheah, K.S.; Foster, C.S.; Fisher, G.; Berney, D.M.; Møller, H.; Reuter, V.E.; Scardino, P.; et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010, 70, 979–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.S.; Fang, W.-T.; Hou, Y.-H.; Huang, S.-F.; Yen, B.L.; Chang, J.-L.; Li, S.-M.; Liu, H.-P.; Liu, Y.-L.; Huang, C.-T.; et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin. Cancer Res. 2010, 16, 4363–4373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Shepherd, J.; Zhao, D.; Bollu, L.R.; Tahaney, W.M.; Hill, J.; Zhang, Y.; Mazumdar, A.; Brown, P.H. SOX9 Is Essential for Triple-Negative Breast Cancer Cell Survival and Metastasis. Mol. Cancer Res. 2020, 18, 1825–1838. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Lian, P.; Zheng, P.-S. SOX9, a potential tumor suppressor in cervical cancer, transactivates p21WAF1/CIP1 and suppresses cervical tumor growth. Oncotarget 2015, 6, 20711–20722. [Google Scholar] [CrossRef] [Green Version]
- Saegusa, M.; Hashimura, M.; Suzuki, E.; Yoshida, T.; Kuwata, T. Transcriptional up-regulation of Sox9 by NF-κB in endometrial carcinoma cells, modulating cell proliferation through alteration in the p14(ARF)/p53/p21(WAF1) pathway. Am. J. Pathol. 2012, 181, 684–692. [Google Scholar] [CrossRef]
- Shakhova, O.; Cheng, P.; Mishra, P.J.; Zingg, D.; Schaefer, S.M.; Debbache, J.; Häusel, J.; Matter, C.; Guo, T.; Davis, S.; et al. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet. 2015, 11, e1004877. [Google Scholar] [CrossRef] [Green Version]
- Prévostel, C.; Rammah-Bouazza, C.; Trauchessec, H.; Canterel-Thouennon, L.; Busson, M.; Ychou, M.; Blache, P. SOX9 is an atypical intestinal tumor suppressor controlling the oncogenic Wnt/ß-catenin signaling. Oncotarget 2016, 7, 82228–82243. [Google Scholar] [CrossRef] [Green Version]
- Seymour, P.A. Sox9: A master regulator of the pancreatic program. Rev. Diabet. Stud. 2014, 11, 51–83. [Google Scholar] [CrossRef] [Green Version]
- Cros, J.; Raffenne, J.; Couvelard, A.; Poté, N. Tumor heterogeneity in pancreatic adenocarcinoma. Pathobiology 2018, 85, 64–71. [Google Scholar] [CrossRef]
- Morgan, R.T.; Woods, L.K.; Moore, G.E.; Quinn, L.A.; McGavran, L.; Gordon, S.G. Human cell line (COLO 357) of metastatic pancreatic adenocarcinoma. Int. J. Cancer 1980, 25, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Kopantzev, E.P.; Kopantseva, M.R.; Grankina, E.V.; Mikaelyan, A.; Egorov, V.I.; Sverdlov, E.D. Activation of IGF/IGF-IR signaling pathway fails to induce epithelial-mesenchymal transition in pancreatic cancer cells. Pancreatology 2019, 19, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Ganger, M.T.; Dietz, G.D.; Headley, P.; Ewing, S.J. Application of the common base method to regression and analysis of covariance (ANCOVA) in qPCR experiments and subsequent relative expression calculation. BMC Bioinform. 2020, 21, 423. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Batut, B.; Hiltemann, S.; Bagnacani, A.; Baker, D.; Bhardwaj, V.; Blank, C.; Bretaudeau, A.; Brillet-Guéguen, L.; Cech, M.; Chilton, J.; et al. Community-driven data analysis training for biology. Cell Syst. 2018, 6, 752–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gary, R.K.; Kindell, S.M. Quantitative assay of senescence-associated beta-galactosidase activity in mammalian cell extracts. Anal. Biochem. 2005, 343, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Kondratyeva, L.G.; Safina, D.R.; Chernov, I.P.; Kopantzev, E.P.; Kostrov, S.V.; Sverdlov, E.D. PDX1, a key factor in pancreatic embryogenesis, can exhibit antimetastatic activity in pancreatic ductal adenocarcinoma. Cancer Manag. Res. 2019, 11, 7077–7087. [Google Scholar] [CrossRef] [Green Version]
- Sipos, B.; Möser, S.; Kalthoff, H.; Török, V.; Löhr, M.; Klöppel, G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: Towards the establishment of an in vitro research platform. Virchows Arch. 2003, 442, 444–452. [Google Scholar] [CrossRef]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Miyauchi, Y.; Kojima, H.; Yamaoka, R.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; et al. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci. Rep. 2016, 6, 30489. [Google Scholar] [CrossRef] [Green Version]
- Francis, J.C.; Capper, A.; Ning, J.; Knight, E.; de Bono, J.; Swain, A. SOX9 is a driver of aggressive prostate cancer by promoting invasion, cell fate and cytoskeleton alterations and epithelial to mesenchymal transition. Oncotarget 2018, 9, 7604–7615. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.-Q.; Wei, F.-K.; Xu, X.-L.; Ye, S.-X.; Song, J.-W.; Ding, P.-K.; Zhu, J.; Li, H.-F.; Luo, X.-P.; Gong, H.; et al. SOX9 drives the epithelial–mesenchymal transition in non-small-cell lung cancer through the Wnt/β-catenin pathway. J. Transl. Med. 2019, 17, 143. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Ye, H.; He, H.H.; Gerrin, S.J.; Chen, S.; Tanenbaum, B.A.; Cai, C.; Sowalsky, A.G.; He, L.; Wang, H.; et al. SOX9 drives WNT pathway activation in prostate cancer. J. Clin. Investig. 2016, 126, 1745–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasco-Garcia, E.; Lopez, L.; Moncho-Amor, V.; Carazo, F.; Aldaz, P.; Collado, M.; Bell, D.; Gaafar, A.; Karamitopoulou, E.; Tzankov, A.; et al. SOX9 triggers different epithelial to mesenchymal transition states to promote pancreatic cancer progression. Cancers 2022, 14, 916. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Medel, J.; Flores, K.; Perry, C.; Galvez, A.; Sandoval, M.; Rivera, S.; Pedroza, D.A.; Penner, E.; Chitti, M.; et al. Hepatocyte nuclear factor 1 alpha influences pancreatic cancer growth and metastasis. Sci. Rep. 2020, 10, 20225. [Google Scholar] [CrossRef]
- Karafin, M.S.; Cummings, C.T.; Fu, B.; Iacobuzio-Donahue, C.A. The developmental transcription factor Gata4 is overexpressed in pancreatic ductal adenocarcinoma. Int. J. Clin. Exp. Pathol. 2009, 3, 47–55. [Google Scholar]
- Martinelli, P.; Carrillo-de Santa Pau, E.; Cox, T.; Sainz, B., Jr.; Dusetti, N.; Greenhalf, W.; Rinaldi, L.; Costello, E.; Ghaneh, P.; Malats, N.; et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2017, 66, 1665–1676. [Google Scholar] [CrossRef] [Green Version]
- Milan, M.; Balestrieri, C.; Alfarano, G.; Polletti, S.; Prosperini, E.; Spaggiari, P.; Zerbi, A.; Diaferia, G.R.; Natoli, G. FOXA2 controls the cis-regulatory networks of pancreatic cancer cells in a differentiation grade-specific manner. EMBO J. 2019, 38, e102161. [Google Scholar] [CrossRef]
- Matheu, A.; Collado, M.; Wise, C.; Manterola, L.; Cekaite, L.; Tye, A.J.; Canamero, M.; Bujanda, L.; Schedl, A.; Cheah, K.S.E.; et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012, 72, 1301–1315. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Chen, W.; Du, X.; Ning, H.; Chen, H.; Shi, R.; Lin, S.; Xu, R.; Zhu, J.; Wu, S.; et al. Upregulation of sex-determining region Y-box 9 (SOX9) promotes cell proliferation and tumorigenicity in esophageal squamous cell carcinoma. Oncotarget 2015, 6, 31241–31254. [Google Scholar] [CrossRef] [Green Version]
- Huilgol, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. Transcription factors that govern development and disease: An Achilles heel in cancer. Genes 2019, 10, 794. [Google Scholar] [CrossRef] [Green Version]
- Yachida, S.; Iacobuzio-Donahue, A. Evolution and dynamics of pancreatic cancer progression. Oncogene 2013, 32, 5253–5260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Kondratyeva, L.G.; Sveshnikova, A.A.; Grankina, E.V.; Chernov, I.; Kopantseva, M.R.; Kopantzev, E.P.; Sverdlov, E.D. Downregulation of expression of mater genes SOX9, FOXA2, and GATA4 in pancreatic cancer cells stimulated with TGFβ1 epithelial–mesenchymal transition. Dokl. Biochem. Biophys. 2016, 469, 257–259. [Google Scholar] [CrossRef]
- Agnihotri, S.; Wolf, A.; Munoz, D.M.; Smith, C.J.; Gajadhar, A.; Restrepo, A.; Clarke, I.D.; Fuller, G.N.; Kesari, S.; Dirks, P.B.; et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J. Exp. Med. 2011, 208, 689–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldaz, P.; Otaegi-Ugartemendia, M.; Saenz-Antoñanzas, A.; Garcia-Puga, M.; Moreno-Valladares, M.; Flores, J.M.; Gerovska, D.; Arauzo-Bravo, M.J.; Samprón, N.; Matheu, A.; et al. SOX9 promotes tumor progression through the axis BMI1-p21 CIP. Sci. Rep. 2020, 10, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.-B.; Li, J.; Liao, W.-T.; Feng, Y.; Yu, C.-P.; Hu, L.-J.; Kong, Q.-L.; Xu, L.-H.; Zhang, X.; Liu, W.-L.; et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J. Clin. Investig. 2009, 119, 3626–3636. [Google Scholar] [CrossRef] [Green Version]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Sirivatanauksorn, V.; Sirivatanauksorn, Y.; Gorman, P.A.; Davidson, J.M.; Sheer, D.; Moore, P.S.; Scarpa, A.; Edwards, P.A.; Lemoine, N.R. Non-random chromosomal rearrangements in pancreatic cancer cell lines identified by spectral karyotyping. Int. J. Cancer 2001, 91, 350–358. [Google Scholar] [CrossRef]
- Deer, E.L.; González-Hernández, J.; Coursen, J.D.; Shea, J.E.; Ngatia, J.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010, 39, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, W.H.; Biade, S.; Zilfou, J.T.; Chen, J.; Murphy, M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002, 277, 3247–3257. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopantzev, E.; Kondratyeva, L.; Kopantseva, M.; Kashkin, K.; Gnatenko, D.; Grigorieva, E.; Alekseenko, I.; Safina, D.; Chernov, I. SOX9 Protein in Pancreatic Cancer Regulates Multiple Cellular Networks in a Cell-Specific Manner. Biomedicines 2022, 10, 1466. https://doi.org/10.3390/biomedicines10071466
Kopantzev E, Kondratyeva L, Kopantseva M, Kashkin K, Gnatenko D, Grigorieva E, Alekseenko I, Safina D, Chernov I. SOX9 Protein in Pancreatic Cancer Regulates Multiple Cellular Networks in a Cell-Specific Manner. Biomedicines. 2022; 10(7):1466. https://doi.org/10.3390/biomedicines10071466
Chicago/Turabian StyleKopantzev, Eugene, Liya Kondratyeva, Marina Kopantseva, Kirill Kashkin, Dmitry Gnatenko, Elizaveta Grigorieva, Irina Alekseenko, Dina Safina, and Igor Chernov. 2022. "SOX9 Protein in Pancreatic Cancer Regulates Multiple Cellular Networks in a Cell-Specific Manner" Biomedicines 10, no. 7: 1466. https://doi.org/10.3390/biomedicines10071466





