Green Synthesized Silver Nanoparticle-Loaded Liposome-Based Nanoarchitectonics for Cancer Management: In Vitro Drug Release Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Tabebuia pallida Leaf Extract
2.3. Preparation of Silver Nanoparticles
2.4. Preparation of Silver Nanoparticle-Loaded Liposomes
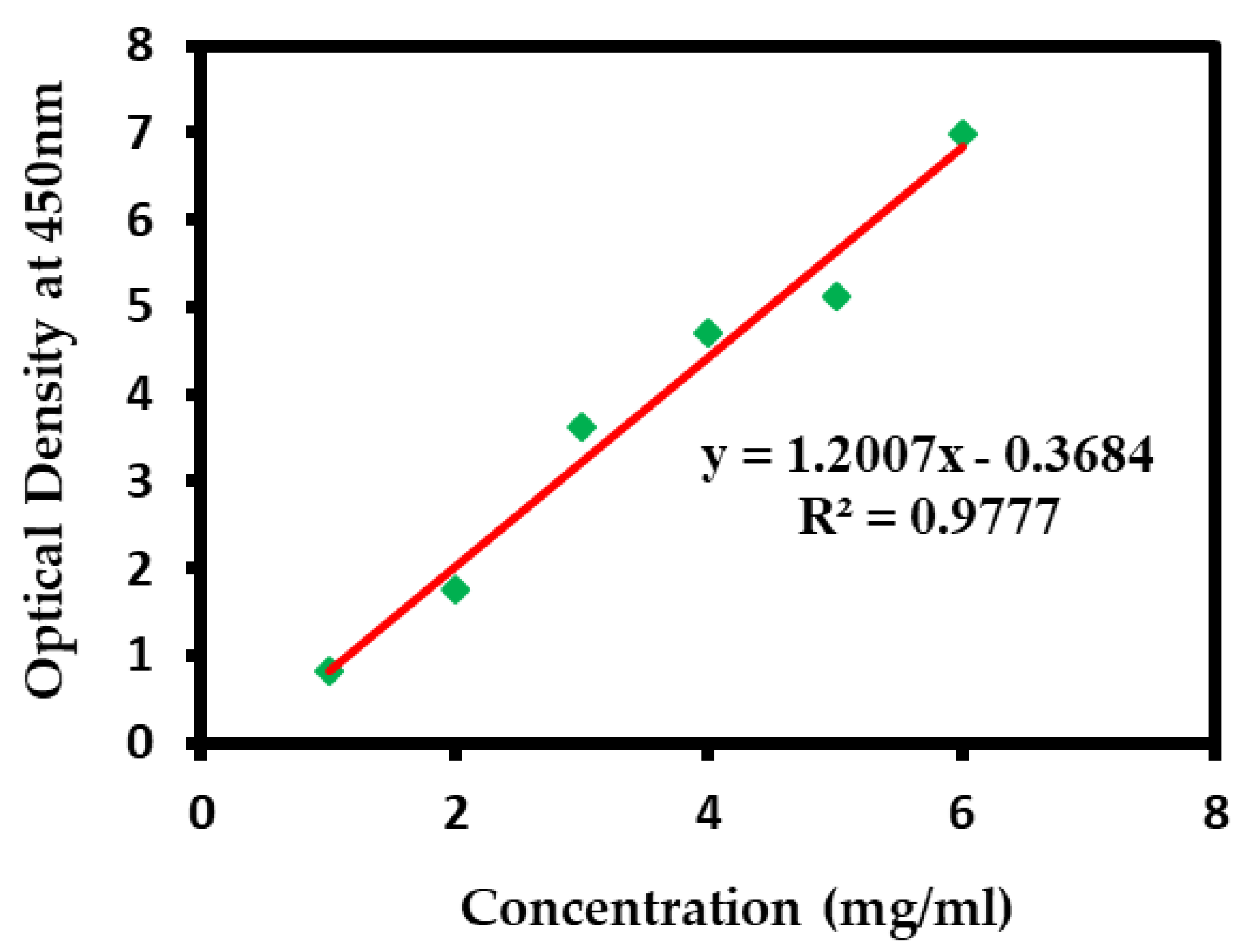
2.5. Encapsulation Efficiency of Silver Nanoparticle-Loaded Liposomes
2.6. Characterization of Silver Nanoparticle-Loaded Liposomes
2.7. In Vitro Drug Release Study
2.8. Application of Mathematical Models in Release Kinetics of Silver Nanoparticle-Loaded Liposomes
3. Results and Discussion
3.1. Encapsulation Efficiency of Silver Nanoparticle-Loaded Liposomes
3.2. Characterization of Silver Nanoparticle-Loaded Liposomes
3.2.1. Dynamic Light Scattering
3.2.2. Field Emission Scanning Electron Microscope
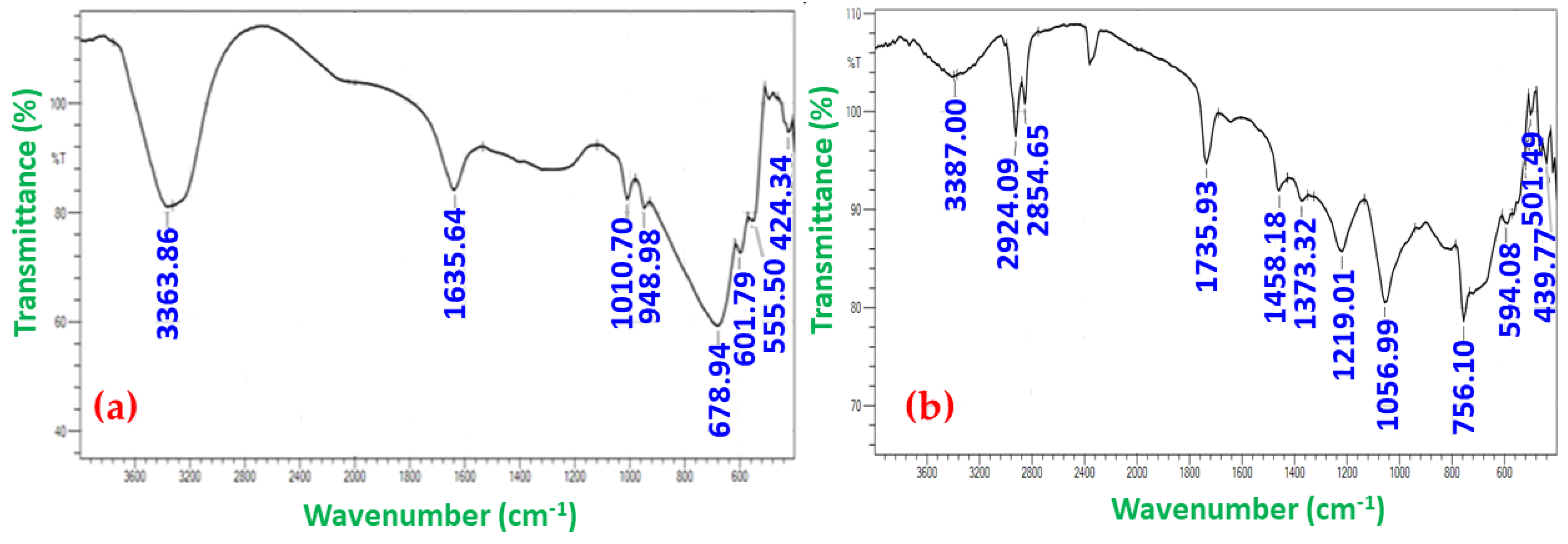
3.2.3. Fourier Transform Infrared Spectroscopy
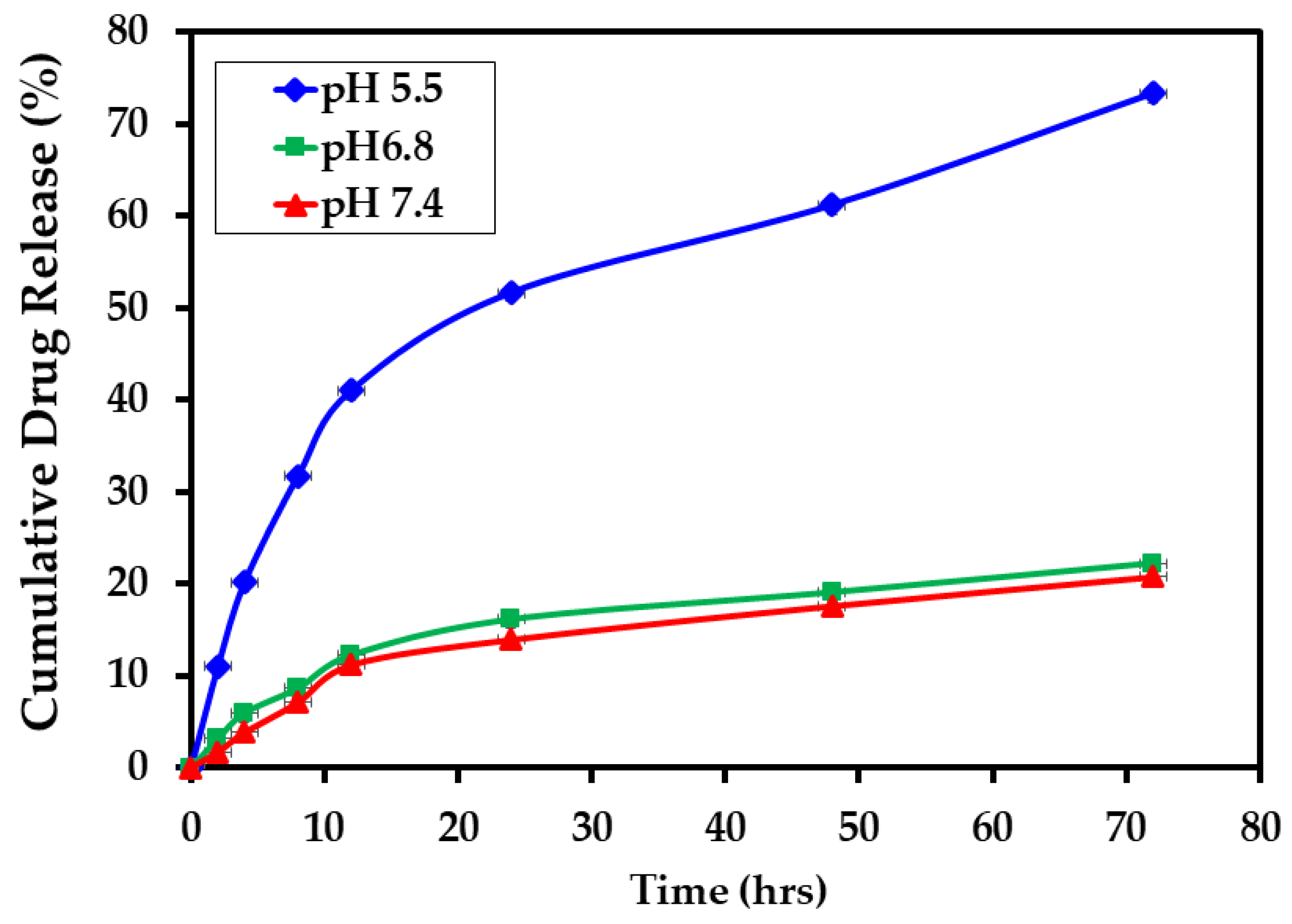
3.3. In Vitro Drug Release Study
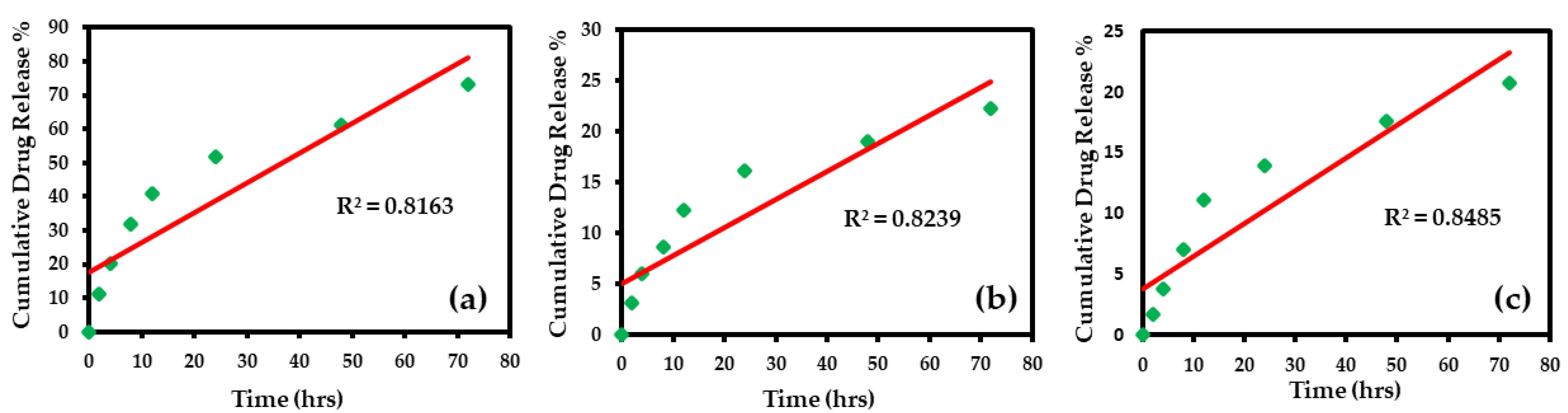
3.4. Application of Mathematical Models in Release Kinetics of Silver Nanoparticle-Loaded Liposomes
3.4.1. Zero-Order Model
3.4.2. First-Order Model
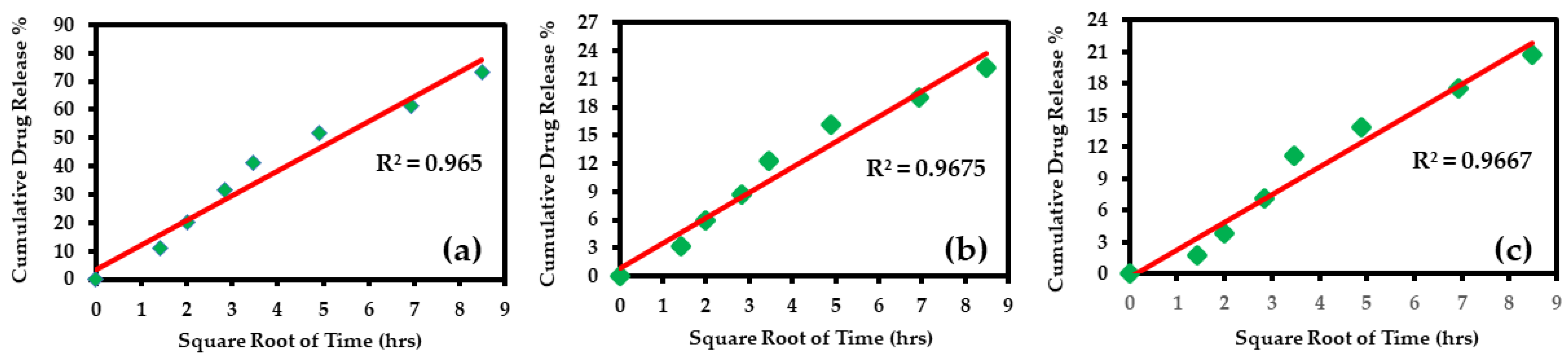
3.4.3. Higuchi Model
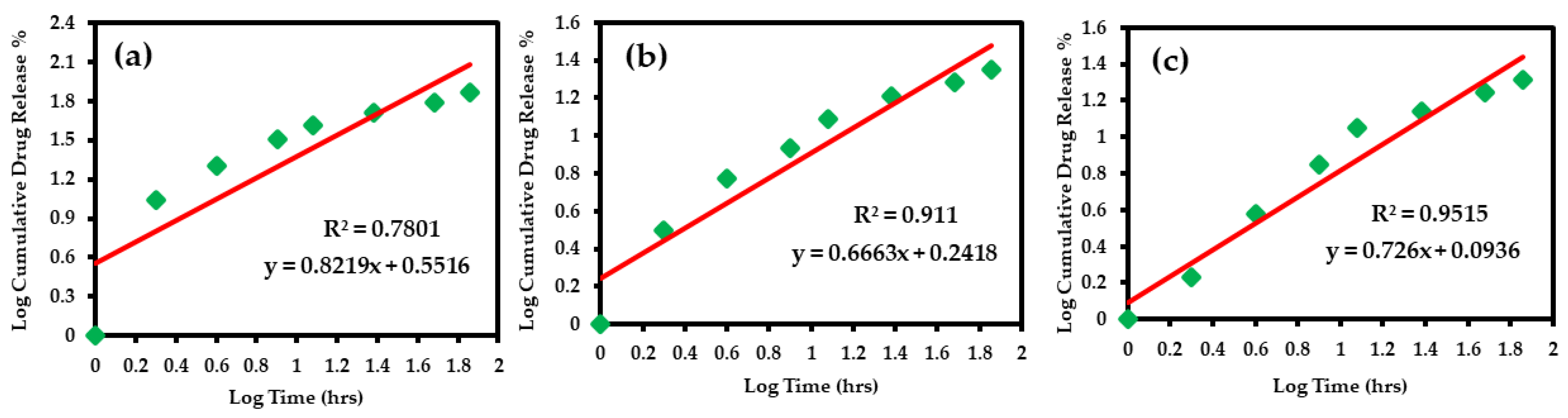
3.4.4. Korsmeyer–Peppas Model
3.4.5. Hixson–Crowell Model
3.5. Results of Various Mathematical Models
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hassanpour, S.H.; Dehghani, M. Review of cancer from perspective of molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Khan, M.; Khan, M.; Al-Marri, A.H.; Al-Warthan, A.; Alkhathlan, H.Z.; Siddiqui, M.R.H.; Nayak, V.L.; Kamal, A.; Adil, S.F. Apoptosis inducing ability of silver decorated highly reduced graphene oxide nanocomposites in A549 lung cancer. Int. J. Nanomed. 2016, 11, 873. [Google Scholar]
- Dotto, G.P.; Rustgi, A.K. Squamous cell cancers: A unified perspective on biology and genetics. Cancer Cell 2016, 29, 622–637. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute, USA. Available online: https://www.cancer.gov/about-cancer/understanding/what-is-cancer (accessed on 5 May 2021).
- Crafton, S.M.; Salani, R. Beyond chemotherapy: An overview and review of targeted therapy in cervical cancer. Clin. Ther. 2016, 38, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment. Int. J. Oncol. 2019, 54, 407–419. [Google Scholar] [PubMed]
- Yao, Y.; Ji, C.; He, Y.; Pan, Y. Relationship between Helicobacter pylori infection and vomiting induced by gastrointestinal cancer chemotherapy. Intern. Med. J. 2017, 47, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Altun, I.; Sonkaya, A. The most common side effects experienced by patients were receiving first cycle of chemotherapy. Iran. J. Public Health 2018, 47, 1218–1219. [Google Scholar]
- Asimuddin, M.; Shaik, M.R.; Adil, S.F.; Siddiqui, M.R.H.; Alwarthan, A.; Jamil, K.; Khan, M. Azadirachta indica based biosynthesis of silver nanoparticles and evaluation of their antibacterial and cytotoxic effects. J. King Saud Univ. Sci. 2020, 32, 648–656. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M.; Mudila, H.; Gupta, G.; Sharma, A.K.; Kumar, D.; Bakshi, H.A.; Negi, P.; Kapoor, D.N.; Chellappan, D.K. Emerging trends in clinical implications of bio-conjugated silver nanoparticles in drug delivery. Colloid Interface Sci. Commun. 2020, 35, 100244. [Google Scholar] [CrossRef]
- Khan, M.; Al-Hamoud, K.; Liaqat, Z.; Shaik, M.R.; Adil, S.F.; Kuniyil, M.; Alkhathlan, H.Z.; Al-Warthan, A.; Siddiqui, M.R.H.; Mondeshki, M. Synthesis of Au, Ag, and Au–Ag Bimetallic Nanoparticles Using Pulicaria undulata Extract and Their Catalytic Activity for the Reduction of 4-Nitrophenol. Nanomaterials 2020, 10, 1885. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Bulut, O.; Some, S.; Mandal, A.K.; Yilmaz, M.D. Green synthesis of silver nanoparticles: Biomolecule-nanoparticle organizations targeting antimicrobial activity. RSC Adv. 2019, 9, 2673–2702. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Munir, S.; Zeb, N.; Ullah, A.; Khan, B.; Ali, J.; Bilal, M.; Omer, M.; Alamzeb, M.; Salman, S.M. Green nanotechnology: A review on green synthesis of silver nanoparticles—An ecofriendly approach. Int. J. Nanomed. 2019, 14, 5087. [Google Scholar] [CrossRef] [Green Version]
- Moradi, F.; Sedaghat, S.; Moradi, O.; Arab Salmanabadi, S. Review on green nano-biosynthesis of silver nanoparticles and their biological activities: With an emphasis on medicinal plants. Inorg. Nano-Met. Chem. 2021, 51, 133–142. [Google Scholar] [CrossRef]
- Garg, D.; Sarkar, A.; Chand, P.; Bansal, P.; Gola, D.; Sharma, S.; Khantwal, S.; Mehrotra, R.; Chauhan, N.; Bharti, R.K. Synthesis of silver nanoparticles utilizing various biological systems: Mechanisms and applications—A review. Prog. Biomater. 2020, 9, 81–95. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Haidere, M.F.; Nurunnabi, M.; Shahriar, S.M.; Ahammad, A.S.; Shim, Y.Y.; Reaney, M.J.; Cho, J.Y. Green chemistry synthesis of silver nanoparticles and their potential anticancer effects. Cancers 2020, 12, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Israel, L.L.; Galstyan, A.; Holler, E.; Ljubimova, J.Y. Magnetic iron oxide nanoparticles for imaging, targeting and treatment of primary and metastatic tumors of the brain. J. Control. Release 2020, 320, 45–62. [Google Scholar] [CrossRef]
- Ali, M.A.; Ahmed, T.; Wu, W.; Hossain, A.; Hafeez, R.; Islam Masum, M.M.; Wang, Y.; An, Q.; Sun, G.; Li, B. Advancements in plant and microbe-based synthesis of metallic nanoparticles and their antimicrobial activity against plant pathogens. Nanomaterials 2020, 10, 1146. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Shaik, M.R.; Adil, S.F.; Khan, S.T.; Al-Warthan, A.; Siddiqui, M.R.H.; Tahir, M.N.; Tremel, W. Plant extracts as green reductants for the synthesis of silver nanoparticles: Lessons from chemical synthesis. Dalton Trans. 2018, 47, 11988–12010. [Google Scholar] [CrossRef]
- Rahman, M.; Hossain, A.; Mostofa, M.; Khan, M.A.; Ali, R.; Mosaddik, A.; Sadik, M.; Alam, A. Evaluation of anti-ROS and anticancer properties of Tabebuia pallida L. Leaves. Clin. Phytoscience 2019, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- El-Hawary, S.S.; Mohammed, R.; Tawfike, A.F.; AbouZid, S.F.; Taher, M.A.; Abdelmohsen, U.R.; Amin, E. Metabolic profiling of cytotoxic metabolites from five Tabebuia species supported by molecular correlation analysis. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Ferraz-Filha, Z.S.; Ferrari, F.C.; Araújo, M.C.d.P.M.; Bernardes, A.C.F.P. Effects of the aqueous extract from Tabebuia roseoalba and phenolic acids on hyperuricemia and inflammation. Evid. -Based Complement. Altern. Med. 2017, 2017, 2712108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regalado, A.I.; Mancebo, B.; Paixão, A.; López, Y.; Merino, N.; Sánchez, L.M. Antinociceptive activity of methanol extract of Tabebuia hypoleuca (C. Wright ex Sauvalle) Urb. stems. Med. Princ. Pract. 2017, 26, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayachandran, P.; Ilango, S.; Nirmaladevi, R. Evaluation of antioxidant potential of green synthesized tabebuia pallida silver nanoparticles. J. Adv. Sci. Res. 2021, 12, 76–85. [Google Scholar] [CrossRef]
- Mbanga, O.; Cukrowska, E.; Gulumian, M. Dissolution kinetics of silver nanoparticles: Behaviour in simulated biological fluids and synthetic environmental media. Toxicol. Rep. 2022, 9, 788–796. [Google Scholar] [CrossRef]
- Prasher, P.; Singh, M.; Mudila, H. Oligodynamic effect of silver nanoparticles: A review. BioNanoScience 2018, 8, 951–962. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M. Antimicrobial properties of surface-functionalized silver nanoparticles. In Silver Nanomaterials for Agri-Food Applications; Elsevier: Amsterdam, The Netherlands, 2021; pp. 39–66. [Google Scholar]
- Ravindran, A.; Chandran, P.; Khan, S.S. Biofunctionalized silver nanoparticles: Advances and prospects. Colloids Surf. B Biointerfaces 2013, 105, 342–352. [Google Scholar] [CrossRef]
- Sarkar, K.; Banerjee, S.L.; Kundu, P.P.; Madras, G.; Chatterjee, K. Biofunctionalized surface-modified silver nanoparticles for gene delivery. J. Mater. Chem. B 2015, 3, 5266–5276. [Google Scholar] [CrossRef] [Green Version]
- Rezazadeh, N.H.; Buazar, F.; Matroodi, S. Synergistic effects of combinatorial chitosan and polyphenol biomolecules on enhanced antibacterial activity of biofunctionalized silver nanoparticles. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, S.; Huang, Y.; Zhang, T.; Liu, X.; Hu, Y.; Zhang, Z.; Tang, M. Acute toxic effects and gender-related biokinetics of silver nanoparticles following an intravenous injection in mice. J. Appl. Toxicol. 2012, 32, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Barbinta-Patrascu, M.E.; Ungureanu, C.; Iordache, S.M.; Bunghez, I.R.; Badea, N.; Rau, I. Green silver nanobioarchitectures with amplified antioxidant and antimicrobial properties. J. Mater. Chem. B 2014, 2, 3221–3231. [Google Scholar] [CrossRef]
- Yusuf, A.; Brophy, A.; Gorey, B.; Casey, A. Liposomal encapsulation of silver nanoparticles enhances cytotoxicity and causes induction of reactive oxygen species-independent apoptosis. J. Appl. Toxicol. 2018, 38, 616–627. [Google Scholar] [CrossRef]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 601, 120571. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.K.; Riaz, M.A.; Zhang, X.; Lin, C.; Wong, K.H.; Chen, X.; Zhang, G.; Lu, A.; Yang, Z. Surface functionalization and targeting strategies of liposomes in solid tumor therapy: A review. Int. J. Mol. Sci. 2018, 19, 195. [Google Scholar] [CrossRef] [Green Version]
- Shivanna, A.T.; Dash, B.S.; Chen, J.-P. Functionalized Magnetic Nanoparticles for Alternating Magnetic Field-or Near Infrared Light-Induced Cancer Therapies. Micromachines 2022, 13, 1279. [Google Scholar] [CrossRef] [PubMed]
- Large, D.E.; Abdelmessih, R.G.; Fink, E.A.; Auguste, D.T. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021, 176, 113851. [Google Scholar] [CrossRef]
- TS, A.; Shalumon, K.; Chen, J.-P. Applications of magnetic liposomes in cancer therapies. Curr. Pharm. Des. 2019, 25, 1490–1504. [Google Scholar]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975. [Google Scholar] [CrossRef] [Green Version]
- Fathy, M.M.; Fahmy, H.M.; Balah, A.M.; Mohamed, F.F.; Elshemey, W.M. Magnetic nanoparticles-loaded liposomes as a novel treatment agent for iron deficiency anemia: In vivo study. Life Sci. 2019, 234, 116787. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Chuang, E.-Y.; Cheng, Y.-H.; Anilkumar, T.; Chen, H.-A.; Chen, J.-P. Thermosensitive magnetic liposomes for alternating magnetic field-inducible drug delivery in dual targeted brain tumor chemotherapy. Chem. Eng. J. 2019, 373, 720–733. [Google Scholar] [CrossRef]
- Karimi, N.; Ghanbarzadeh, B.; Hajibonabi, F.; Hojabri, Z.; Ganbarov, K.; Kafil, H.S.; Hamishehkar, H.; Yousefi, M.; Mokarram, R.R.; Kamounah, F.S. Turmeric extract loaded nanoliposome as a potential antioxidant and antimicrobial nanocarrier for food applications. Food Biosci. 2019, 29, 110–117. [Google Scholar] [CrossRef]
- Kavithaa, K.; Paulpandi, M.; Padma, P.R.; Sumathi, S. Induction of intrinsic apoptotic pathway and cell cycle arrest via baicalein loaded iron oxide nanoparticles as a competent nano-mediated system for triple negative breast cancer therapy. RSC Adv. 2016, 6, 64531–64543. [Google Scholar] [CrossRef]
- Gouda, R.; Baishya, H.; Qing, Z. Application of mathematical models in drug release kinetics of carbidopa and levodopa ER tablets. J. Dev. Drugs 2017, 6, 1–8. [Google Scholar]
- TS, A.; Lu, Y.-J.; Chen, J.-P. Optimization of the preparation of magnetic liposomes for the combined use of magnetic hyperthermia and photothermia in dual magneto-photothermal cancer therapy. Int. J. Mol. Sci. 2020, 21, 5187. [Google Scholar]
- Hardiansyah, A.; Yang, M.-C.; Liu, T.-Y.; Kuo, C.-Y.; Huang, L.-Y.; Chan, T.-Y. Hydrophobic drug-loaded PEGylated magnetic liposomes for drug-controlled release. Nanoscale Res. Lett. 2017, 12, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Najlah, M.; Said Suliman, A.; Tolaymat, I.; Kurusamy, S.; Kannappan, V.; Elhissi, A.M.; Wang, W. Development of injectable PEGylated liposome encapsulating disulfiram for colorectal cancer treatment. Pharmaceutics 2019, 11, 610. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, Y.; Kono, Y.; Ogawara, K.-i.; Kimura, T.; Higaki, K. PEG liposomalization of paclitaxel improved its in vivo disposition and anti-tumor efficacy. Int. J. Pharm. 2011, 412, 132–141. [Google Scholar] [CrossRef]
- Nguyen Thi, P.P.; Nguyen, M.T.; Hai, N.D. Role of collagen concentration in stability of Star-shaped Silver@ Gold nanoparticles. In Proceedings of Journal of Nano Research; Trans Tech Publications, Ltd.: Zurich, Switzerland, 2016; pp. 113–119. [Google Scholar]
- Petros, R.A.; DeSimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Aslan, B.; Ozpolat, B.; Sood, A.K.; Lopez-Berestein, G. Nanotechnology in cancer therapy. J. Drug Target. 2013, 21, 904–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.H.; Lee, J.S.; Bae, J.W.; Choi, J.H.; Lee, Y.; Son, J.Y.; Park, K.D. Targeted doxorubicin nanotherapy strongly suppressing growth of multidrug resistant tumor in mice. Int. J. Pharm. 2015, 495, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Chang, L.; Kuang, T.; Hu, J. PEG/heparin-decorated lipid–polymer hybrid nanoparticles for long-circulating drug delivery. RSC Adv. 2016, 6, 23279–23287. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Zeng, W.; Ge, S.; Lu, H.; Wu, C.; Ge, L.; Liang, D.; Xu, Y. Proniosome-derived niosomes for tacrolimus topical ocular delivery: In vitro cornea permeation, ocular irritation, and in vivo anti-allograft rejection. Eur. J. Pharm. Sci. 2014, 62, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Paini, M.; Daly, S.R.; Aliakbarian, B.; Fathi, A.; Tehrany, E.A.; Perego, P.; Dehghani, F.; Valtchev, P. An efficient liposome based method for antioxidants encapsulation. Colloids Surf. B Biointerfaces 2015, 136, 1067–1072. [Google Scholar] [CrossRef]
- Espinoza, J.T.; Novak, R.S.; Magalhães, C.G.; Budel, J.M.; Justus, B.; Gonçalves, M.M.; Boscardin, P.M.D.; Farago, P.V.; Paula, J.d.F.P.D. Preparation and characterization of liposomes loaded with silver nanoparticles obtained by green synthesis. Braz. J. Pharm. Sci. 2021, 56, e18601. [Google Scholar] [CrossRef]
- Ajitha, B.; Reddy, Y.A.K.; Reddy, P.S. Biogenic nano-scale silver particles by Tephrosia purpurea leaf extract and their inborn antimicrobial activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 121, 164–172. [Google Scholar] [CrossRef]
- Sadhasivam, L.; Durairaj, J. Evaluation profile of silver nanoparticle synthesized by aloe vera extract. Int. J. ChemTech. Res. 2014, 6, 4379–4385. [Google Scholar]
- Zhang, Y.; Cheng, X.; Zhang, Y.; Xue, X.; Fu, Y. Biosynthesis of silver nanoparticles at room temperature using aqueous aloe leaf extract and antibacterial properties. Colloids Surf. A Physicochem. Eng. Asp. 2013, 423, 63–68. [Google Scholar] [CrossRef]
- Jin, H.-H.; Lu, Q.; Jiang, J.-G. Curcumin liposomes prepared with milk fat globule membrane phospholipids and soybean lecithin. J. Dairy Sci. 2016, 99, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Burande, A.S.; Viswanadh, M.K.; Jha, A.; Mehata, A.K.; Shaik, A.; Agrawal, N.; Poddar, S.; Mahto, S.K.; Muthu, M.S. EGFR targeted paclitaxel and piperine co-loaded liposomes for the treatment of triple negative breast cancer. AAPS PharmSciTech 2020, 21, 1–12. [Google Scholar]
- Dash, S.; Murthy, P.N.; Nath, L.; Chowdhury, P. Kinetic modeling on drug release from controlled drug delivery systems. Acta Pol. Pharm. 2010, 67, 217–223. [Google Scholar] [PubMed]
- Singhvi, G.; Singh, M. In-vitro drug release characterization models. Int. J. Pharm. Stud. Res. 2011, 2, 77–84. [Google Scholar]
- Paarakh, M.P.; Jose, P.A.; Setty, C.; Christoper, G. Release kinetics–concepts and applications. Int. J. Pharm. Res. Technol. 2018, 8, 12–20. [Google Scholar]
- Vu, M.T.; Le, N.T.T.; Pham, T.L.-B.; Nguyen, N.H.; Nguyen, D.H. Development and characterization of soy lecithin liposome as potential drug carrier systems for codelivery of letrozole and paclitaxel. J. Nanomater. 2020, 2020, 8896455. [Google Scholar] [CrossRef]
- Haghiralsadat, F.; Amoabediny, G.; Helder, M.N.; Naderinezhad, S.; Sheikhha, M.H.; Forouzanfar, T.; Zandieh-Doulabi, B. A comprehensive mathematical model of drug release kinetics from nano-liposomes, derived from optimization studies of cationic PEGylated liposomal doxorubicin formulations for drug-gene delivery. Artif. Cells Nanomed. Biotechnol. 2018, 46, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Fathi, M.; Varshosaz, J.; Mohebbi, M.; Shahidi, F. Hesperetin-loaded solid lipid nanoparticles and nanostructure lipid carriers for food fortification: Preparation, characterization, and modeling. Food Bioprocess Technol. 2013, 6, 1464–1475. [Google Scholar] [CrossRef]






| Release Exponent (n) | Drug Transport Mechanism |
|---|---|
| n < 0.5 | Quasi-Fickian diffusion |
| 0.5 | Fickian diffusion |
| 0.5 < n < 1. 0 | Anomalous (Non-Fickian transport) |
| 1.0 | Case II transport |
| Higher than1.0 | Super case II transport |
| pH | Equation | Release Exponent (n) | Drug Transport Mechanism |
|---|---|---|---|
| 5.5 | y = 0.821x + 0.551 | 0.821 | Non-Fickian Transport |
| 6.8 | y = 0.666x + 0.241 | 0.666 | |
| 7.4 | y = 0.726x + 0.093 | 0.726 |
| Name of the Mathematical Model | pH 5.5 (Mature Endosomes of Cancer Cells) | pH 6.8 (Cancer Cells) | pH 7.4 (Blood) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | Equation | Slope | Intercept | R2 | Equation | Slope | Intercept | R2 | Equation | slope | Intercept | |
| Zero-order | 0.816 | y = 0.884x + 17.47 | 0.8845 | 17.474 | 0.823 | y = 0.276x + 5.06 | 0.2762 | 5.065 | 0.848 | y = 0.270x + 3.754 | 0.2705 | 3.755 |
| First-order | 0.932 | y = −0.007x + 1.92 | −0.0072 | 1.923798 | 0.848 | y = −0.001x + 1.97 | −0.0013 | 1.977 | 0.869 | y = −0.001x + 1.98 | −0.0013 | 1.983 |
| Higuchi | 0.965 | y = 8.687x + 3.673 | 8.6876 | 3.673744 | 0.967 | y = 2.703x + 0.789 | 2.7038 | 0.789104 | 0.966 | y = 2.608x − 0.283 | 2.6087 | −0.28379 |
| Korsmeyer–Peppas | 0.780 | y = 0.821x + 0.551 | 0.8219 | 0.551602 | 0.911 | y = 0.666x + 0.241 | 0.6663 | 0.241776 | 0.951 | y = 0.726x + 0.093 | 0.7259 | 0.093642 |
| Hixson–Crowell | 0.899 | y = 0.020x + 0.276 | 0.0206 | 0.276325 | 0.840 | y = 0.004x + 0.079 | 0.0046 | 0.079622 | 0.862 | y = 0.004x + 0.058 | 0.0045 | 0.058526 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayachandran, P.; Ilango, S.; Suseela, V.; Nirmaladevi, R.; Shaik, M.R.; Khan, M.; Khan, M.; Shaik, B. Green Synthesized Silver Nanoparticle-Loaded Liposome-Based Nanoarchitectonics for Cancer Management: In Vitro Drug Release Analysis. Biomedicines 2023, 11, 217. https://doi.org/10.3390/biomedicines11010217
Jayachandran P, Ilango S, Suseela V, Nirmaladevi R, Shaik MR, Khan M, Khan M, Shaik B. Green Synthesized Silver Nanoparticle-Loaded Liposome-Based Nanoarchitectonics for Cancer Management: In Vitro Drug Release Analysis. Biomedicines. 2023; 11(1):217. https://doi.org/10.3390/biomedicines11010217
Chicago/Turabian StyleJayachandran, Priyanka, Suganya Ilango, Vivekananthan Suseela, Ramalingam Nirmaladevi, Mohammed Rafi Shaik, Mujeeb Khan, Merajuddin Khan, and Baji Shaik. 2023. "Green Synthesized Silver Nanoparticle-Loaded Liposome-Based Nanoarchitectonics for Cancer Management: In Vitro Drug Release Analysis" Biomedicines 11, no. 1: 217. https://doi.org/10.3390/biomedicines11010217








