Epigenetic Mechanisms and Nephrotic Syndrome: A Systematic Review
Abstract
:1. Introduction
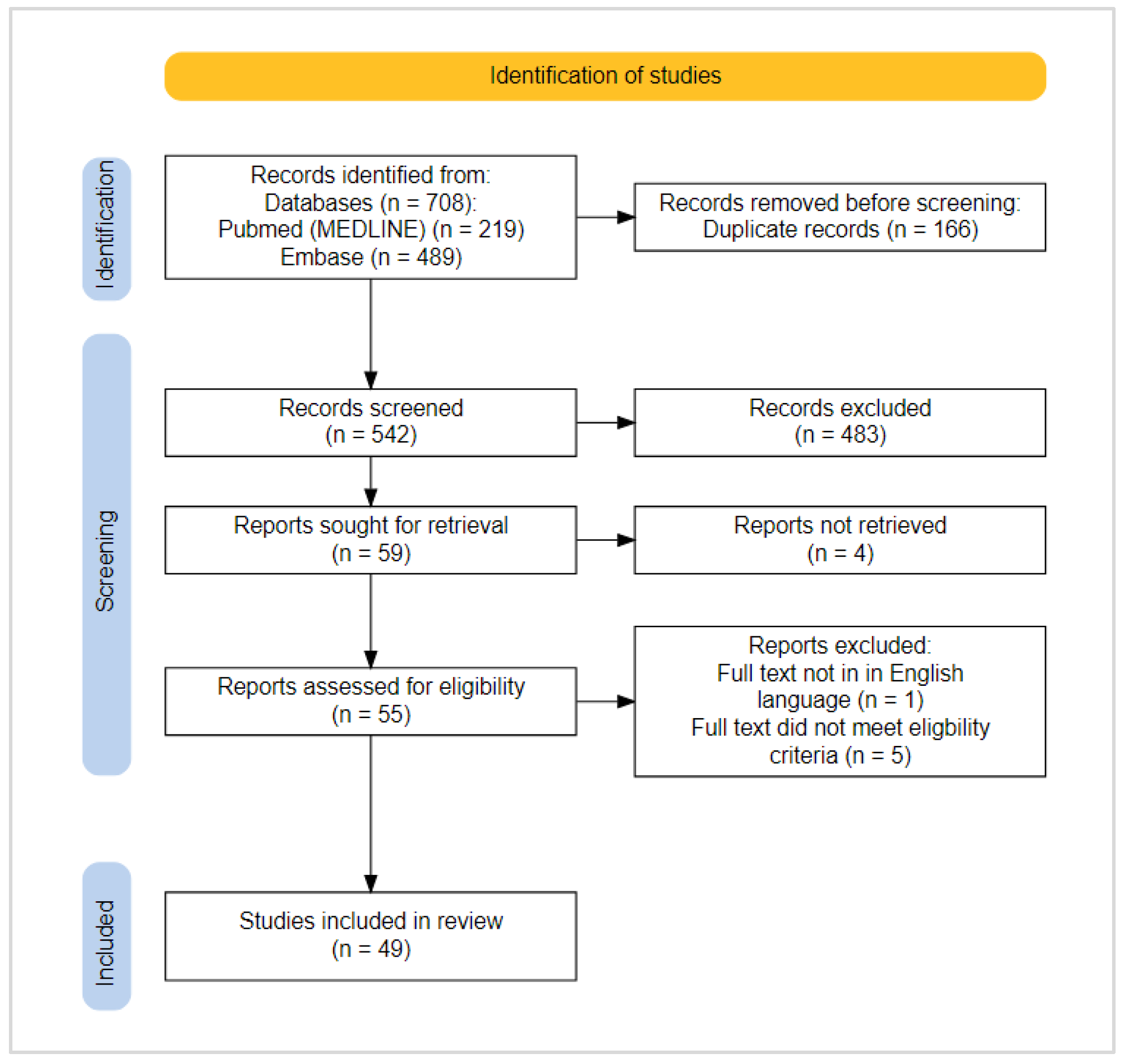
2. Materials and Methods
2.1. Protocol and Registration
2.2. Search Strategy—Eligibility Criteria, Information Sources and Search Terms
2.3. Study Selection and Data Extraction
2.4. Critical Appraisal
3. Results
3.1. Higher Quality Studies—Micro-RNAs
3.2. Higher Quality Studies—DNA Methylation
3.3. Higher Quality Studies—Small RNAs
4. Discussion
5. Conclusions
| Publication Details and Reference | Study Population | Epigenetic Data: Mechanism Studied and COVERAGE | Results: Key Findings | Repeated Epigenetic Measures | Replication | JBI Percentage and Risk of Bias |
|---|---|---|---|---|---|---|
| Micro-RNAs—blood | ||||||
| Xiao, B et al. Cell Death & Disease, 2018. [32] | Aged 16–70. FSGS 102; IgAN 69; MPGN 24; Membranous 26; Healthy controls 129. | Micro-RNA—blood QuantoBio miRNA high-throughput assay—515 miRNAs (Discovery phase); Primer assays for miR-17, miR-451, miR-106a, miR-19b (Validation phase). | MiR-17, miR-451, miR-106a, and miR-19b were significantly downregulated in the plasma of FSGS patients compared with healthy controls, fold changes of 0.55, 0.56, 0.59 and 0.55 respectively (p < 0.05). A 4 miRNA (miR-17, miR-451, miR-106a, and miR-19b) FSGS classification model gave an AUC value of 0.82, p < 0.0001. A 3 miRNA (miR-17, miR-451, and miR-106a) FSGS remission classification model gave an AUC of 0.71, p < 0.01. | No | Yes | 80% Low risk |
| Ardalan, M et al. PeerJ, 2020. [33] | Aged 20–60 FSGS 22; Membranous 30; Healthy controls 24. | Micro-RNA—PBMCs and plasma. MiRNA-135 primer assays. | Lower miR-135a-5p in patients with FSGS compared to controls, median relative expression 0.72 compared to 1.37, p = 0.046. | No | No | 90% Low risk |
| Hejazian, S et al. International Journal of Nephrology and Renovascular Disease, 2020. [14] | Aged 20–60 FSGS 30; Membranous 30; Healthy controls 24. | Micro-RNA—PBMCs and plasma. MiR-30c and miR-186 primer assays. | Increased miR-30c level in PBMCs of patients with FSGS (0.004), compared to controls. Plasma miR-30c levels were not different between FSGS, Membranous or healthy controls. | No | No | 90% Low risk |
| Rahbar Saadat, Y et al. Biofactors, 2020. [34] | Aged 20–60 FSGS 30; Membranous 30; Healthy controls 24 | Micro-RNA—PBMCs MiR-24, miR-30a, and miR-370 primer assays. | Lower miR-24, higher miR-30a and higher miR-370 expression levels were observed in Membranous compared to FSGS (p = 0.040, p = 0.032, p = 0.041, respectively). There was no difference in the levels of these miRNAs between the control group and FSGS patients. | No | No | 90% Low risk |
| Ni, F et al. Frontiers in Pediatrics, 2021. [35] | No specific age restrictions—only children included. Active NS before treatment 20; NS in remission 20; Healthy controls 20. | Micro-RNA—Blood—Th2 cells (CD4 + TCD25- cells) miR-24 and miR-27 primer assays. | Participants with active non-atopic NS had lower levels of miR-24 (mean 21.84 × 10−3) and miR-27 (mean 20.72 × 10−3) compared to healthy controls (46.03 × 10−3, p < 0.05, and 37.83 × 10−3, p < 0.05, respectively). | No | No | 90% Low risk |
| Hejazian, S et al. Biofactors, 2020. [13] | Aged 20–60. NS 60; Healthy controls 24 | Micro-RNA—PBMCs miR-30a, miR-30c, miR-186, miR-193, miR-217 primer assays. | Higher levels of miR-30c-5p (p = 0.005) and miR-193-3p (p = 0.041) were observed between NS patients and healthy controls. There was no difference in mi-RNA196-5p and miR-217 expression levels between NS patients and controls. | No | No | 90% Low risk |
| Bayomy, N et al. Molecular Immunology, 2022. [11] | No specific age restrictions—only children included. SSNS 56; SRNS 24; Healthy controls 100. | Micro-RNA—blood miR-142-5p, miR-191a-5p, miR-181-5p, miR-30a-5p and miR-150a-5p primer assays. | NS patients had higher levels of the 5 studied microRNAs than healthy controls: miR-142-5p (mean expression 14.8 compared to 2.3, p = 7 × 10−5), miR-191a-5p (7.38 compared to 0.65, p < 10−6), miR-181-5p (2.25 compared to 0.41, p < 10−6), miR-30a-5p (1.49 compared to 0.72, p < 1 × 10−6) and miR-150a-5p (7.67 compared to 3.86, p <0.003). SRNS patients had higher levels of the 5 studied microRNAs than SSNS patients: miR-142-5p (mean expression 105.36 compared to 10.87, p < 10−6), miR-191a-5p (22.99 compared to 0.69, p < 10−6), miR-181-5p (4.22 compared to 1.39, p 10−6), miR-30a-5p (3.10 compared to 0.809, p < 10−6) and miR-150a-5p (10.97 compared to 6.26, p < 0.044). MiR-142a-5p had the best discrimination of NS patients from controls (AUC 0.965) and SRNS from SSNS (AUC 1.00) of a single mi-RNA. | No | No | 90% Low risk |
| Zhang, C et al. American Journal of Kidney Diseases, 2015. [36] | Aged 18–69. FSGS nephrotic range proteinuria 78; FSGS remission 35; Membranous 63; DN 59; Healthy controls 69. | Micro-RNA—plasma TaqMan Low Density Array (Applied Biosystems)—384 human microRNAs. | Higher levels of miR-125b, miR-186, and miR-193a-3p were present in patients with FSGS relative to controls, (average fold changes of 5.77 p < 0.001, 3.04 p = 0.006, and 3.44 p < 0.001, respectively). MiR-125b and miR-186 concentrations were significantly lower in patients with FSGS in complete remission (p = 0.02 and p < 0.001) compared to those with nephrotic range proteinuria. In FSGS patients who achieved complete remission with steroid treatment, miR-125b and miR-186 levels declined markedly after they received steroids (SSNS, p = 0.002 and p = 0.002 respectively). There was no change in these miRNA levels in patients who did not respond to steroids (SRNS). Plasma miR-186, but not miR-125b, level was correlated with degree of proteinuria in patients with FSGS (R 0.185, p = 0.02). | No | Yes | 100% Low risk |
| Micro RNAs—urine | ||||||
| Altamemi I et al. Journal of Pharmaceutical Sciences and Research, 2019. [37] | No specific age restrictions—adults and children included. FSGS 24; Healthy controls 24. | Micro-RNA—urine miRNA-193a primer assay. | Urine miR-193a levels were higher in people with FSGS than controls (median fold change 2.125 compared to 0.375, p < 0.001). A fold change of >0.31 miR-193a gave an AUC of 0.829, sensitivity of 100% and specificity of 50% for identifying FSGS from controls patients. | No | No | 80% Low risk |
| Zheng X et al. Experimental and Therapeutic Medicine, 2021. [38] | Aged 11–62FSGS 22; Membranous 36; NS—Nephropathy 22; Healthy controls 60. | Micro-RNA—urine miR-155 assay primers. | Urine miR-155 levels were higher in early NS than in control patients (roughly 4.5-fold compared to 1, p < 0.05). Urine miR-155 levels could distinguish early NS from control patients with an AUC of 0.9548, sensitivity of 93.27% and specificity of 92.58%. | No | No | 90% Low risk |
| Wang N et al. Peer J, 2015. [39] | Aged 20–50 MCD 31; IgAN 120; Membranous 45; Healthy controls 40. | Micro-RNA—urine Affymetrix GeneChip miRNA 4.0 Array—2578 mature human miRNAs. | In the validation cohort the urinary level of miR-3613-3p were lower in IgAN compared with that in healthy controls, membranous and MCD, (relative levels: 0.27, 1.10, 0.58 and 0.61, respectively, all p < 0.01). There were no significant differences in miR-4668-5p between IgAN and patients with Membranous or MCD. | No | Yes | 90% Low risk |
| Zhang W et al. Clinical Journal of The American Society of Nephrology, 2014. [17] | No specific age restrictions—adults included. Active FSGS 107; FSGS remission 103; Healthy controls 105; Membranous active 29; Membranous remission 26; DN with disease activity 23; Incipient DN 27; Validation: Healthy controls 27; FSGS remission 22; Active FSGS 33. | Micro-RNA—urine Taqman Low density Array—754 human miRNAs. | Urinary miR-196a, miR-30a-5p, and miR-490 discriminated patients with active FSGS from those in remission (AUC 0.92, 0.82 and 0.96 respectively). After steroid treatment, the levels of urinary miR-196a, miR-30a-5p, and miR-490 were lower in steroid-responsive FSGS patients (p < 0.001), but were unchanged in steroid-resistant FSGS patients. Urinary miR-30a-5p marginally predicted the response to steroid treatment in patients with active FSGS, (AUC 0.63, p = 0.03). | No | Yes | 100% Low risk |
| Chen T, EBioMedicine, 2019. [16] | Aged ≤ 15 years. 126 NS Healthy controls 126. | Micro-RNA—urinary exosomes Illumina sequencing by synthesis—whole transcriptome. | The Illumina sequencing identified 30 urinary exosomal miRNAs which were increased in NS compared with controls (≥5 fold, p < 0.05), the top 15 proceeded to validation testing. Five mi-RNAs (miR-194-5p, miR-146b-5p, miR-378-3p, miR-23b-3p and miR-30a-5p) were significantly increased in NS in 2 independent cohorts (>3 fold, p < 0.01). During NS remission, all the mi-RNAs except miR-194-5p decreased, almost to the level of controls (p < 0.001). | Yes—before and after treatment | Yes | 70% Low risk |
| Feng D, Translational Andrology and Urology, 2020. [15] | Aged ≤ 12 years NS active disease 68; NS remission 47; Healthy controls 50. | Micro-RNA—urinary exosomes miR-23b-3p, miR-30a-5p and miR-151-3p assay primers. | The levels of miR-23b-3p and miR-30a-5p decreased from the active NS group (10.58 and 18.57 respectively), to the remission NS group (8.63 and 15.62) to the controls (0.56 and 8.62, p all < 0.001). After treatment, levels of exosomal miR-23b-3p and miR-30a-5p in the active NS and remission NS groups decreased significantly (10.58 to 2.68 and 18.57 to 10.48; 8.63 to 2.43 and 15.62 to 9.63 respectively, p < 0.05). Urinary exosomal miR-23b-3p and miR-30a-5p could distinguish between children with NS and healthy children, (AUC 0.711, sensitivity 92.73% and specificity 59.09% for miR-23b-3p and AUC 0.844, sensitivity 85.71% and 63.64% for miR-30a-5p). | Yes—before and after treatment | No | 100% Low risk |
| Micro-RNAs—renal tissue | ||||||
| Yu J, et al. BMC Nephrology, 2019. [40] | No specific age restrictions—only adults included. MCD 4; FSGS 4; DN 4; Healthy controls 4. Validation: MCD 6; FSGS 6; DN 6; Healthy controls 6. | Micro-RNA—Renal—glomeruli and tubules miRCURY LNA Array, which contains 3100 miRNAs (covering all human, mouse and rat miRNAs). | Forty mi-RNAs were increased and 76 decreased in renal tissue from people with MCD, FSGS and DN compared with healthy donor kidney biopsy tissue. In a validation cohort, miR-3607-3p was decreased in people with MCD, FSSG and DN compared to healthy controls (p < 0.05). MiR-4709-3p was increased in people with MCD, FSSG and DN compared to healthy controls (p < 0.05). | No | Yes | 80% Low risk |
| Micro-RNAs—blood and urine | ||||||
| Ramezani A, et al. European Journal of Clinical Investigation, 2015. [12] | No specific age restrictions—adults and children included FSGS 16; MCD 5; healthy controls 5. | Micro-RNA—plasma and urine Affymetrix GeneChip miRNA 3.0 arrays—1733 human miRNAs. | Patients with FSGS had 126 differentially expressed miRNAs in plasma and 155 in urine compared to people with MCD. Plasma levels of miR-30b, miR-30c, miR-34b, miR-34c and miR-342 and urine levels of mir-1225-5p were higher MCD patients than in FSGS and healthy controls, (p < 0.001). Urinary levels of mir-1915 and miR-663 were lower in patients with FSGS compared to MCD and controls (p < 0.001). Urinary levels of miR-155 were higher in patients with FSGS compared to MCD and controls (p < 0.005). | No | No | 90% Low risk |
| Zhang C, et al. Journal of Translational Medicine, 2018. [41] | Aged 16–65.FSGS with nephrotic range proteinuria 100; FSGS complete remission 100; Healthy controls 100; Replication cohort—FSGS 231. | Micro-RNA—plasma and urine TaqMan Human MicroRNA Array v3.0 A and B—754 human miRNAs. | Urinary miR-196a was significantly increased in active FSGS compared with FSGS patients in complete remission and healthy controls (p < 0.0001). There was no difference in plasma miR-196a between these patient groups. Urinary miR-196a was associated with proteinuria (p = 0.003), estimated glomerular filtration rate (p = 0.005), interstitial fibrosis (0.004), tubular atrophy (p = 0.38) and progression to end-stage kidney disease (<0.001). Multivariate Cox analysis confirmed urinary miR-196a as an independent risk factor for FSGS progression after adjusting for age, sex, proteinuria and eGFR (HR = 2.616, 95% CI 1.592–4.301, p < 0.001). | No | Yes | 100% Low risk |
| Luo Y, et al. Clinical Chemistry, 2013. [10] | Aged 1–14 SRNS 24; SSNS 135; Healthy controls 109; Renal disease controls 44 (18 HSP, 15 IgAN, 11 lupus). | Micro-RNA—serum and urine TaqMan Low Density Array—754 human miRNAs. | Serum miR-30a-5p, miR-151-3p, miR-150, miR-191, and miR-19b were higher in NS children compared with healthy controls (median fold change 2.51, 2.56, 2.59, 3.29, 2.43 respectively, all p < 0.0001). Urinary miR-30a-5p was also increased in NS patients compared to healthy controls (median fold change 2.11, p = 0.001). The 5 serum mi-RNAs combined could distinguish NS from healthy controls, OR 40.7 (95% CI, 6.06–103; p < 0.0001). The concentrations of the 5 serum miRNAs and urinary miR-30a-5p declined after steroid treatment (p ≤ 0.002). All patients achieved remission with steroids. | Yes—before and after treatment | Yes | 80% Low risk |
| Li J, et al. BioMed Research International 2017. [42] | Aged < 65 years FSGS 82; Membranous 84; DN 67; Healthy controls 72. | Micro-RNA—plasma and renal tissue MiR-217 assay primer. | There was no difference in miR-217 expression in renal tissue or plasma between healthy controls and people with FSGS. | No | No | 90% Low risk |
| Publication Details and Reference | Study Population | Epigenetic Data: Mechanism Studied and Coverage | Results: Key Findings | Repeated Epigenetic Measures | Replication | JBI Percentage And Risk Of Bias |
|---|---|---|---|---|---|---|
| DNA methylation | ||||||
| Locafo M, et al. Clinical and translational science, 2021. [18] | No specific age restrictions—adults and children included. Adults: SRNS and FSGS 10; SSNS 18 (of which, 13 MCD and 5 FSGS); Validation: Children: FSGS 2; MCD 14. | DNAm—PBMCs Bisulphite conversion and NLRP3 promoter primer assay. | In both adults and paediatric patients, NLRP3 promoter methylation was significantly reduced in SRNS compared with SSNS (median 0.2, and 0.33 respectively, p = 0.00024). NLRP3 methylation distinguished between SRNS and SSNS with AUC 0.736 in children (p = 0.00097) and 0.867 in adults (0.00019). | No | Yes | 90% Low risk |
| Zaorska K, et al. Acta Biochimica Polonica, 2016. [19] | No specific age restrictions—children included. SRNS 40; SSNS 36; Healthy controls 33. | DNAm—blood. Methylation primers (methylation-specific PCR)—CpG islands for SOCS3 and SOCS5. | There was lower DNAm at one region of SOCS3 promoter in SRNS compared to SSNS, 82.5% of SRNS patients were unmethylated in this region compared to 17.7% of SSNS patients (p < 0.0001). | No | No | 100% Low risk |
| Small RNAs | ||||||
| Duan Z, et al. Scientific Reports, 2018. [43] | Aged > 18. FSGS 25; MCD 37; IgAN 330; Healthy controls 130; Membranous 90; HSP 5; Non-IgA MPGN 5; Renal amyloidosis 2. | Small nuclear RNA—urine. U6 primer assay. | No significant difference in the expression of U6 between the patients with IgAN, Membranous, MCD and FSGS. | Yes—before and after treatment in IgAN. | Yes | 100% Low risk |
| Williams A, et al. Kidney international, 2022. [44] | No specific age restrictions—adults included. FSGS 38; Healthy controls 10 | Small RNA—renal—glomeruli and tubules Illumina TruSeq small RNA library preparation kit—whole transcriptome. | Large numbers of small RNAs, including microRNAs, 3′-transfer RNA fragments, 5′- transfer RNA fragments, and mitochondrial transfer RNA fragments, were differentially expressed between histologically indistinguishable tissue regions from patients with FSGS and controls. In FSGS, miR-21-5p progressively increased and miR-192-5p progressively decreased in glomerular and tubulointerstitial regions with increasing levels of histological damage. | No | No | 100% Low risk |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eddy, A.A.; Symons, J.M. Nephrotic syndrome in childhood. Lancet 2003, 362, 629–639. [Google Scholar] [CrossRef]
- Bierzynska, A.; McCarthy, H.J.; Soderquest, K.; Sen, E.S.; Colby, E.; Ding, W.Y.; Nabhan, M.M.; Kerecuk, L.; Hegde, S.; Hughes, D.; et al. Genomic and clinical profiling of a national nephrotic syndrome cohort advocates a precision medicine approach to disease management. Kidney Int. 2017, 91, 937–947. [Google Scholar] [CrossRef]
- Souto, M.F.O.; Teixeira, A.L.; Russo, R.C.; Penido, M.G.M.G.; Silveira, K.D.; Teixeira, M.M. Immune mediators in idiopathic nephrotic syndrome: Evidence for a relation between interleukin 8 and proteinuria. Pediatr. Res. 2008, 64, 637–642. [Google Scholar] [CrossRef]
- Araya, C.; Diaz, L.; Wasserfall, C.; Atkinson, M.; Mu, W.; Johnson, R.; Garin, E. T regulatory cell function in idiopathic minimal lesion nephrotic syndrome. Pediatr. Nephrol. 2009, 24, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Iijima, K.; Sako, M.; Nozu, K.; Mori, R.; Tuchida, N.; Kamei, K.; Miura, K.; Aya, K.; Nakanishi, K.; Ohtomo, Y.; et al. Rituximab for childhood-onset, complicated, frequently relapsing nephrotic syndrome or steroid-dependent nephrotic syndrome: A multicentre, double-blind, randomised, placebo-controlled trial. Lancet 2014, 384, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Coward, R.J.M.; Foster, R.R.; Patton, D.; Ni, L.; Lennon, R.; Bates, D.O.; Harper, S.J.; Mathieson, P.W.; Saleem, M.A. Nephrotic plasma alters slit diaphragm-dependent signaling and translocates nephrin, podocin, and CD2 associated protein in cultured human podocytes. J. Am. Soc. Nephrol. 2005, 16, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.J.; McCarthy, H.J.; Ni, L.; Wherlock, M.; Kang, H.; Wetzels, J.F.; Welsh, G.I.; Saleem, M.A. Active proteases in nephrotic plasma lead to a podocin-dependent phosphorylation of VASP in podocytes via protease activated receptor-1. J. Pathol. 2013, 229, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019, 20, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Critical-Appraisal-Tools—Critical Appraisal Tools. Joanna Briggs Institute. Available online: https://jbi.global/critical-appraisal-tools (accessed on 1 May 2022).
- Luo, Y.; Wang, C.; Chen, X.; Zhong, T.; Cai, X.; Chen, S.; Shi, Y.; Hu, J.; Guan, X.; Xia, Z.; et al. Increased Serum and Urinary MicroRNAs in Children with Idiopathic Nephrotic Syndrome. Clin. Chem. 2013, 59, 658–666. [Google Scholar] [CrossRef]
- Bayomy, N.R.; Alfottoh, W.M.A.; Eldeep, S.A.A.; Mersal, A.M.S.I.M.; El Bary, H.M.A.A.; El Gayed, E.M.A. Mir-142-5p as an indicator of autoimmune processes in childhood idiopathic nephrotic syndrome and as a part of MicroRNAs expression panels for its diagnosis and prediction of response to steroid treatment. Mol. Immunol. 2022, 141, 21–32. [Google Scholar] [CrossRef]
- Ramezani, A.; Devaney, J.M.; Cohen, S.; Wing, M.R.; Scott, R.; Knoblach, S.; Singhal, R.; Howard, L.; Kopp, J.B.; Raj, D.S. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: A pilot study. Eur. J. Clin. Investig. 2015, 45, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Hejazian, S.M.; Saadat, Y.R.; Bahmanpour, Z.; Khatibi, S.M.H.; Ardalan, M.; Vahed, S.Z. Dicer and Drosha expression in patients with nephrotic syndrome. Biofactors 2020, 46, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Hejazian, S.M.; Ardalan, M.; Shoja, M.M.; Samadi, N.; Vahed, S.Z. Expression Levels of miR-30c and miR-186 in Adult Patients with Membranous Glomerulonephritis and Focal Segmental Glomerulosclerosis. Int. J. Nephrol. Renov. Dis. 2020, 13, 193–201. [Google Scholar] [CrossRef]
- Feng, D.; Wu, B.; Pang, Y. Diagnostic value of urinary exosomal miR-23b-3p, miR-30a-5p, and miR-151-3p in children with primary nephrotic syndrome. Transl. Androl. Urol. 2020, 9, 2235–2241. [Google Scholar] [CrossRef]
- Chen, T.; Wang, C.; Yu, H.; Ding, M.; Zhang, C.; Lu, X.; Zhang, C.-Y.; Zhang, C. Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. Ebiomedicine 2019, 39, 552–561. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, C.; Chen, H.; Li, L.; Tu, Y.; Liu, C.; Shi, S.; Zen, K.; Liu, Z. Evaluation of MicroRNAs miR-196a, miR-30a-5P, and miR-490 as Biomarkers of Disease Activity among Patients with FSGS. Clin. J. Am. Soc. Nephrol. 2014, 9, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Lucafò, M.; Granata, S.; Bonten, E.J.; McCorkle, R.; Stocco, G.; Caletti, C.; Selvestrel, D.; Cozzarolo, A.; Zou, C.; Cuzzoni, E.; et al. Hypomethylation of NLRP3 gene promoter discriminates glucocorticoid-resistant from glucocorticoid-sensitive idiopathic nephrotic syndrome patients. Clin. Transl. Sci. 2021, 14, 964–975. [Google Scholar] [CrossRef]
- Zaorska, K.; Zawierucha, P.; Ostalska-Nowicka, D.; Nowicki, M. SOCS3 is epigenetically up-regulated in steroid resistant ne-phrotic children. Acta Biochim. Pol. 2016, 63, 131–138. [Google Scholar] [CrossRef]
- Paugh, S.W.; Bonten, E.J.; Savic, D.; Ramsey, L.B.; E Thierfelder, W.; Gurung, P.; Malireddi, R.K.S.; Actis, M.; Mayasundari, A.; Min, J.; et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat. Genet. 2015, 47, 607–614. [Google Scholar] [CrossRef]
- Ostalska-Nowicka, D.; Smiech, M.; Jaroniec, M.; Zaorska, K.; Zawierucha, P.; Szaflarski, W.; Malinska, A.; Nowicki, M. SOCS3 and SOCS5 mRNA ex-pressions may predict initial steroid response in nephrotic syndrome children. Folia Histochem. Cytobiol. 2011, 49, 719–728. [Google Scholar] [CrossRef] [Green Version]
- Gadegbeku, C.A.; Gipson, D.S.; Holzman, L.B.; Ojo, A.O.; Song, P.X.; Barisoni, L.; Sampson, M.G.; Kopp, J.B.; Lemley, K.V.; Nelson, P.J.; et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013, 83, 749–756. [Google Scholar] [CrossRef] [PubMed]
- NURTuRE—A Unique Kidney Biobank. Available online: https://www.nurturebiobank.org/ (accessed on 6 January 2023).
- Moore, D. Panobinostat (Farydak): A Novel Option for the Treatment of Relapsed Or Relapsed and Refractory Multiple Myeloma. Pharm. Ther. 2016, 41, 296–300. [Google Scholar]
- Kang, J.G.; Park, J.S.; Ko, J.-H.; Kim, Y.-S. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci. Rep. 2019, 9, 11960. [Google Scholar] [CrossRef]
- Kim, Y.Z.; Kim, C.-Y.; Lim, D.H. The Overview of Practical Guidelines for Gliomas by KSNO, NCCN, and EANO. Brain Tumor Res. Treat. 2022, 10, 83–93. [Google Scholar] [CrossRef]
- Peters, T.J.; French, H.J.; Bradford, S.T.; Pidsley, R.; Stirzaker, C.; Varinli, H.; Nair, S.; Qu, W.; Song, J.; A Giles, K.; et al. Evaluation of cross-platform and interlaboratory concordance via consensus modelling of genomic measurements. Bioinformatics 2019, 35, 560–570. [Google Scholar] [CrossRef]
- Wright, C.; Rajpurohit, A.; Burke, E.E.; Williams, C.; Collado-Torres, L.; Kimos, M.; Brandon, N.J.; Cross, A.J.; Jaffe, A.E.; Weinberger, D.R.; et al. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genom. 2019, 20, 513. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; McCoy, C.E.; Rusinova, I.; Saulep, D.; Wang, D.; Xu, D.; Irving, A.T.; Behlke, M.A.; Hertzog, P.J.; Mackay, F.; et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011, 39, 5692–5703. [Google Scholar] [CrossRef]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef]
- Xiao, B.; Wang, L.-N.; Li, W.; Gong, L.; Yu, T.; Zuo, Q.-F.; Zhao, H.-W.; Zou, Q.-M. Plasma microRNA panel is a novel biomarker for focal segmental glomerulosclerosis and associated with podocyte apoptosis. Cell Death Dis. 2018, 9, 533. [Google Scholar] [CrossRef]
- Ardalan, M.; Hejazian, S.M.; Sharabiyani, H.F.; Farnood, F.; Aghdam, A.G.; Bastami, M.; Ahmadian, E.; Vahed, S.Z.; Cucchiarini, M. Dysregulated levels of glycogen synthase kinase-3β (GSK-3β) and miR-135 in peripheral blood samples of cases with nephrotic syndrome. PeerJ 2020, 8, e10377. [Google Scholar] [CrossRef]
- Rahbar Saadat, Y.; Hejazian, S.M.; Nariman-Saleh-Fam, Z.; Bastami, M.; Poursheikhani, A.M.; Shoja, M.; Zununi Vahed, S. Glucocorticoid receptors and their upstream epigenetic regulators in adults with steroid-resistant nephrotic syndrome. BioFactors 2020, 46, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.-F.; Liu, G.-L.; Jia, S.-L.; Chen, R.-R.; Liu, L.-B.; Li, C.-R.; Yang, J.; Gao, X.-J. Function of miR-24 and miR-27 in Pediatric Patients with Idiopathic Nephrotic Syndrome. Front. Pediatr. 2021, 9, 651544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, W.; Chen, H.-M.; Liu, C.; Wu, J.; Shi, S.; Liu, Z.-H. Plasma MicroRNA-186 and Proteinuria in Focal Segmental Glomerulosclerosis. Am. J. Kidney Dis. 2015, 65, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Altamemi, I.A.; Ridha, A. Micro-RNA-193a as a Focal Segmental Glomerulosclerosis Biomarker. J. Pharm. Sci. Res. 2019, 11, 882–885. [Google Scholar]
- Zheng, X.; Zhong, Q.; Lin, X.; Gu, X.; Ling, X.; Liang, Z.; Qin, Q.; Du, X. Transforming growth factor-β1-induced podocyte injury is associated with increased microRNA-155 expression, enhanced inflammatory responses and MAPK pathway activation. Exp. Ther. Med. 2021, 21, 620. [Google Scholar] [CrossRef]
- Wang, N.; Bu, R.; Duan, Z.; Zhang, X.; Chen, P.; Li, Z.; Wu, J.; Cai, G.; Chen, X. Profiling and initial validation of urinary microRNAs as biomarkers in IgA nephropathy. PeerJ 2015, 3, e990. [Google Scholar] [CrossRef]
- Yu, J.; Yu, C.; Feng, B.; Zhan, X.; Luo, N.; Yu, X.; Zhou, Q. Intrarenal microRNA signature related to the fibrosis process in chronic kidney disease: Identification and functional validation of key miRNAs. BMC Nephrol. 2019, 20, 336. [Google Scholar] [CrossRef]
- Zhang, C.; Liang, S.; Cheng, S.; Li, W.; Wang, X.; Zheng, C.; Zeng, C.; Shi, S.; Xie, L.; Zen, K.; et al. Urinary miR-196a predicts disease progression in patients with chronic kidney disease. J. Transl. Med. 2018, 16, 91. [Google Scholar] [CrossRef]
- Li, J.; Liu, B.; Xue, H.; Zhou, Q.Q.; Peng, L. miR-217 Is a Useful Diagnostic Biomarker and Regulates Human Podocyte Cells Apoptosis via Targeting TNFSF11 in Membranous Nephropathy. BioMed Res. Int. 2017, 2017, 2168767. [Google Scholar] [CrossRef]
- Duan, Z.-Y.; Cai, G.-Y.; Li, J.-J.; Bu, R.; Wang, N.; Yin, P.; Chen, X.-M. U6 can be used as a housekeeping gene for urinary sediment miRNA studies of IgA nephropathy. Sci. Rep. 2018, 8, 10875. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.M.; Jensen, D.M.; Pan, X.; Liu, P.; Liu, J.; Huls, S.; Regner, K.R.; Iczkowski, K.A.; Wang, F.; Li, J.; et al. Histologically resolved small RNA maps in primary focal segmental glomerulosclerosis indicate progressive changes within glomerular and tubulointerstitial regions. Kidney Int. 2022, 101, 766–778. [Google Scholar] [CrossRef]
- Sui, W.; Lin, H.; Li, H.; Yan, Q.; Chen, J.; Dai, Y. Circulating microRNAs as potential biomarkers for nephrotic syndrome. Iran. J. Kidney Dis. 2014, 8, 371–376. [Google Scholar]
- Teng, J.; Sun, F.; Yu, P.-F.; Li, J.-X.; Yuan, D.; Chang, J.; Lin, S.-H. Differential microRNA expression in the serum of patients with nephrotic syndrome and clinical correlation analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 7282–7286. [Google Scholar] [PubMed]
- Zapata-Benavides, P.; Arellano-Rodríguez, M.; Bollain-Y-Goytia, J.J.; Franco-Molina, M.A.; Rangel-Ochoa, G.A.; Avalos-Díaz, E.; Herrera-Esparza, R.; Rodríguez-Padilla, C. Cytoplasmic Localization of WT1 and Decrease of miRNA-16-1 in Nephrotic Syndrome. BioMed Res. Int. 2017, 2017, 9531074. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, Y.; Zhou, J.; Zhang, Y. Urinary Exosomal miR-193a Can Be a Potential Biomarker for the Diagnosis of Primary Focal Segmental Glomerulosclerosis in Children. BioMed Res. Int. 2017, 2017, 7298160. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, J.; Wang, Z.; Zhou, J.; Zhang, Y. Higher Urine Exosomal miR-193a Is Associated With a Higher Probability of Primary Focal Segmental Glomerulosclerosis and an Increased Risk of Poor Prognosis Among Children With Nephrotic Syndrome. Front. Cell Dev. Biol. 2021, 9, 727370. [Google Scholar] [CrossRef]
- Konta, T.; Ichikawa, K.; Suzuki, K.; Kudo, K.; Satoh, H.; Kamei, K.; Nishidate, E.; Kubota, I. A microarray analysis of urinary microRNAs in renal diseases. Clin. Exp. Nephrol. 2013, 18, 711–717. [Google Scholar] [CrossRef]
- Wang, G.; Kwan, B.C.-H.; Lai, F.M.-M.; Chow, K.-M.; Li, P.K.-T.; Szeto, C.-C. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin. Chim. Acta 2013, 418, 5–11. [Google Scholar] [CrossRef]
- Baker, M.A.; Davis, S.J.; Liu, P.; Pan, X.; Williams, A.M.; Iczkowski, K.A.; Gallagher, S.T.; Bishop, K.; Regner, K.R.; Liu, Y.; et al. Tissue-Specific MicroRNA Expression Patterns in Four Types of Kidney Disease. J. Am. Soc. Nephrol. 2017, 28, 2985–2992. [Google Scholar] [CrossRef]
- Lu, M.; Wang, C.; Yuan, Y.; Zhu, Y.; Yin, Z.; Xia, Z.; Zhang, C. Differentially expressed microRNAs in kidney biopsies from various subtypes of nephrotic children. Exp. Mol. Pathol. 2015, 99, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, C.A.; Kornauth, C.; Dong, L.; Sierig, R.; Seibler, J.; Reiss, M.; Tauber, S.; Bilban, M.; Wang, S.; Kain, R.; et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat. Med. 2013, 19, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.; Nie, F.; Liu, T.; Yu, X.; Wang, H.; Li, Q.; Peng, R.; Mao, Z.; Zhou, Q.; et al. miR-135 family members mediate podocyte injury through the activation of Wnt/β-catenin signaling. Int. J. Mol. Med. 2015, 36, 669–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Wu, D.; Du, H.; Nie, F.; Pang, X.; Xu, Y. MicroRNA-135a is involved in podocyte injury in a transient receptor potential channel 1-dependent manner. Int. J. Mol. Med. 2017, 40, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Zhou, L.; Zhou, Y.; Zhao, Y.; Li, Q.; Ni, D.; Hu, Y.; Long, Y.; Liu, J.; Lyu, Z.; et al. MiR-30a Inhibits the Epithelial—Mesenchymal Transition of Podocytes through Downregulation of NFATc3. Int. J. Mol. Sci. 2015, 16, 24032–24047. [Google Scholar] [CrossRef]
- Müller-Deile, J.; Dannenberg, J.; Schroder, P.; Lin, M.-H.; Miner, J.H.; Chen, R.; Bräsen, J.-H.; Thum, T.; Nyström, J.; Staggs, L.B.; et al. Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int. 2017, 92, 836–849. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, C.; Fan, Y.; Zeng, C.; Chen, Z.; Qin, W.; Zhang, C.; Zhang, W.; Wang, X.; Zhu, X.; et al. Downregulation of MicroRNA-30 Facilitates Podocyte Injury and Is Prevented by Glucocorticoids. J. Am. Soc. Nephrol. 2014, 25, 92–104. [Google Scholar] [CrossRef]
- Wang, N.; Zhou, Y.; Jiang, L.; Li, D.; Yang, J.; Zhang, C.-Y.; Zen, K. Urinary MicroRNA-10a and MicroRNA-30d Serve as Novel, Sensitive and Specific Biomarkers for Kidney Injury. PLoS ONE 2012, 7, e51140. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Aizawa, A.; Takizawa, T.; Igarashi, K.; Hatada, I.; Arakawa, H. Changes in DNA methylation in naïve T helper cells regulate the pathophysiological state in minimal-change nephrotic syndrome. BMC Res. Notes 2017, 10, 480. [Google Scholar] [CrossRef]
- Zaorska, K.; Zawierucha, P.; Świerczewska, M.; Ostalska-Nowicka, D.; Zachwieja, J.; Nowicki, M. Prediction of steroid resistance and steroid dependence in nephrotic syndrome children. J. Transl. Med. 2021, 19, 130. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Aizawa, A.; Takizawa, T.; Yoshizawa, C.; Horiguchi, H.; Ikeuchi, Y.; Kakegawa, S.; Watanabe, T.; Maruyama, K.; Morikawa, A. DNA methylation changes between relapse and remission of minimal change nephrotic syndrome. Pediatr. Nephrol. 2012, 27, 2233–2241. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, Y.; Peng, W.; Lu, J.; Zhang, Y.; Wang, L. Genome-Wide Analysis of Histone H3 Lysine 4 Trimethylation in Pe-ripheral Blood Mononuclear Cells of Minimal Change Nephrotic Syndrome Patients. Am. J. Nephrol. 2009, 30, 505–513. [Google Scholar] [CrossRef]
- Majumder, S.; Thieme, K.; Batchu, S.N.; Alghamdi, T.A.; Bowskill, B.B.; Kabir, M.G.; Liu, Y.; Advani, S.L.; White, K.E.; Geldenhuys, L.; et al. Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J. Clin. Investig. 2017, 128, 483–499. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Hu, S.; Qin, W.; Shi, J.; Zeng, C.; Bao, H.; Liu, Z. Upregulated long noncoding RNA LOC105375913 induces tubulointerstitial fibrosis in focal segmental glomerulosclerosis. Sci. Rep. 2019, 9, 716. [Google Scholar] [CrossRef]
- Hu, S.; Han, R.; Shi, J.; Zhu, X.; Qin, W.; Zeng, C.; Bao, H.; Liu, Z. The long noncoding RNA LOC105374325 causes podocyte injury in individuals with focal segmental glomerulosclerosis. J. Biol. Chem. 2018, 293, 20227–20239. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Torres, F.; Medina-Perez, M.; Melo, Z.; Mendoza-Cerpa, C.; Echavarria, R. Urinary expression of long non-coding RNA TUG1 in non-diabetic patients with glomerulonephritides. Biomed. Rep. 2020, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ge, T.; Zhou, H.; Zhang, L.; Zhao, L. Absence of Long Noncoding RNA H19 Promotes Childhood Nephrotic Syndrome through Inhibiting ADCK4 Signal. Experiment 2020, 26, e922090. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Fu, J.; Luan, J.; Qi, H.; Jiao, C.; Ran, M.; Wang, D.; Hao, X.; Zhang, Y.; Kopp, J.B.; et al. CircZNF609 is involved in the pathogenesis of focal segmental glomerulo-sclerosis by sponging miR-615-5p. Biochem. Biophys. Res. Commun. 2020, 531, 341–349. [Google Scholar] [CrossRef] [PubMed]


| Data | Comments | |
|---|---|---|
| Study Design | E.g., Case-control, cohort study etc. | |
| Study population | Sample size | Total number of participants and number with NS |
| Diagnosis and control group | E.g., SRNS v age matched controls | |
| Age | E.g., 0–18 years only | |
| Epigenetic data | Mechanism studied | E.g., DNA methylation |
| Tissue studied | E.g., Lymphocytes | |
| Data generation approach and coverage | E.g., 450 k Illumina array | |
| Research objectives | Directly as stated in the article | |
| Analysis | Exposure | E.g., The epigenetic mechanism |
| Outcome | E.g., Treatment response, disease subgroup | |
| Confounders | E.g., Age, sex, cell type proportions | |
| Mediation analysis | Yes/No and details if Yes | |
| Mendelian randomization | Yes/No and details if Yes | |
| Machine learning | Yes/No and details if Yes | |
| Other ‘omics’ data incorporated | Yes/No and details if Yes | |
| Repeated measures of epigenetic data | Yes/No and details if Yes | |
| Replication or validation attempted | Yes/No and details if Yes | |
| Results | Key findings (state effect size and statistics) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayward, S.; Parmesar, K.; Welsh, G.I.; Suderman, M.; Saleem, M.A. Epigenetic Mechanisms and Nephrotic Syndrome: A Systematic Review. Biomedicines 2023, 11, 514. https://doi.org/10.3390/biomedicines11020514
Hayward S, Parmesar K, Welsh GI, Suderman M, Saleem MA. Epigenetic Mechanisms and Nephrotic Syndrome: A Systematic Review. Biomedicines. 2023; 11(2):514. https://doi.org/10.3390/biomedicines11020514
Chicago/Turabian StyleHayward, Samantha, Kevon Parmesar, Gavin I. Welsh, Matthew Suderman, and Moin A. Saleem. 2023. "Epigenetic Mechanisms and Nephrotic Syndrome: A Systematic Review" Biomedicines 11, no. 2: 514. https://doi.org/10.3390/biomedicines11020514






