Pregnancy Zone Protein Serves as a Prognostic Marker and Favors Immune Infiltration in Lung Adenocarcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Patient and Sample
2.3. Gene Set Enrichment Analysis
2.4. Evaluation of TIICs
2.5. Immunohistochemistry (IHC) Staining
2.6. Cell Culture
2.7. Quantitative Real-Time PCR (qRT-PCR)
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
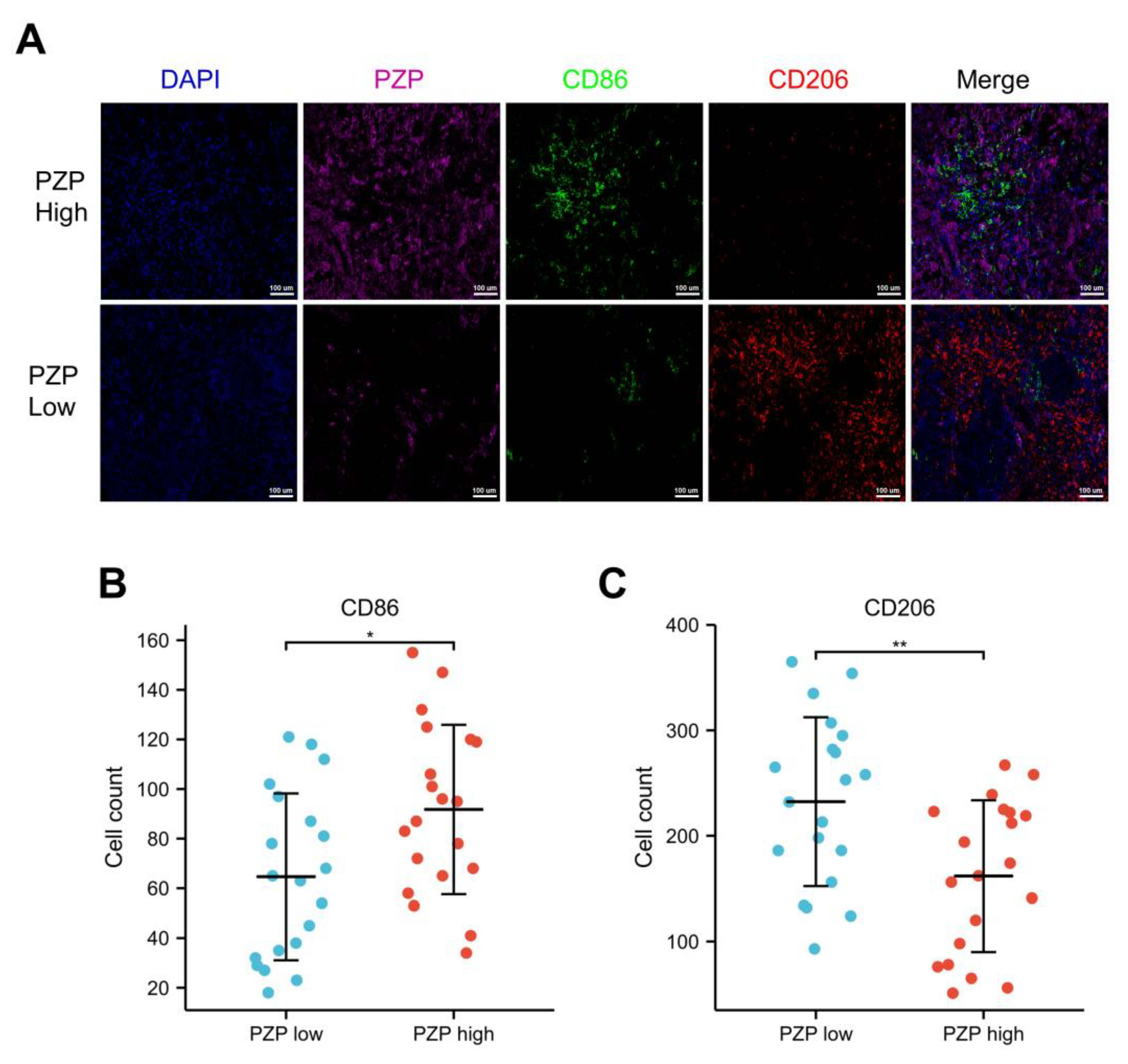
2.9. Multiplex Immunohistochemistry (mIHC) Staining
2.10. Statistical Analysis
3. Results
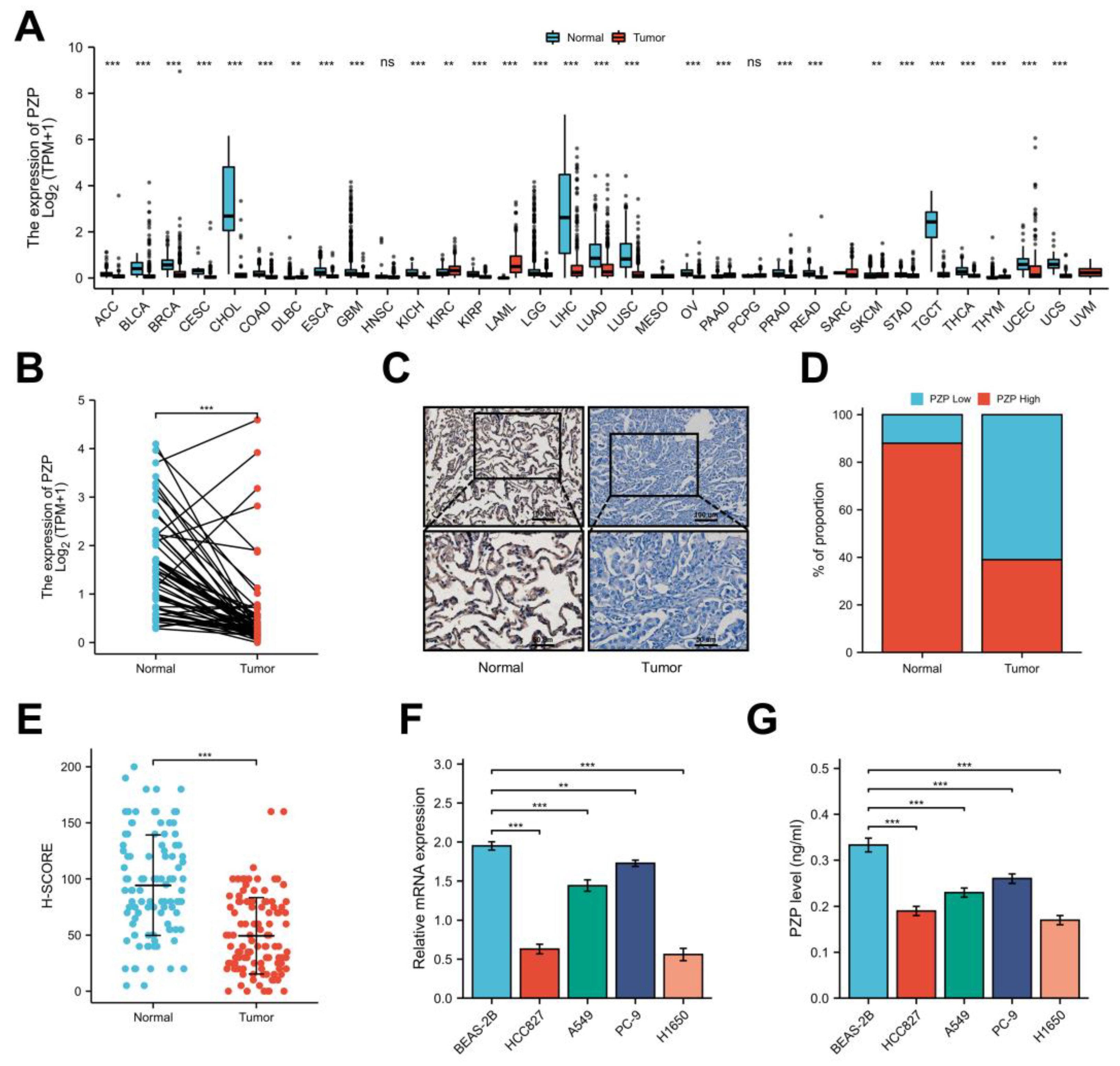
3.1. A Notable Decrease in PZP Expression Was Observed in Both Databases and Clinical Samples
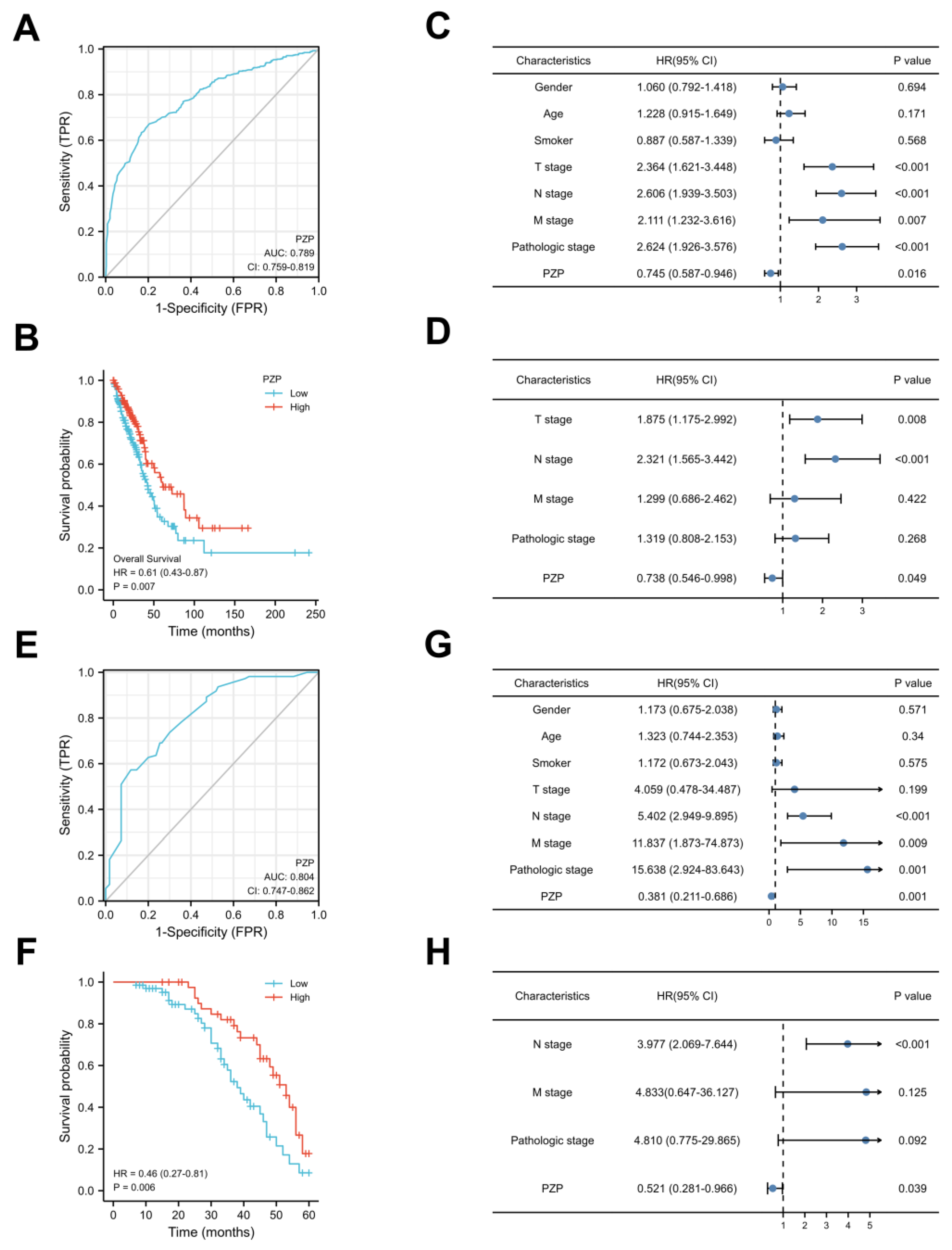
3.2. PZP Has a High Diagnostic Value and Can Be Used as an Independent Prognostic Predictor
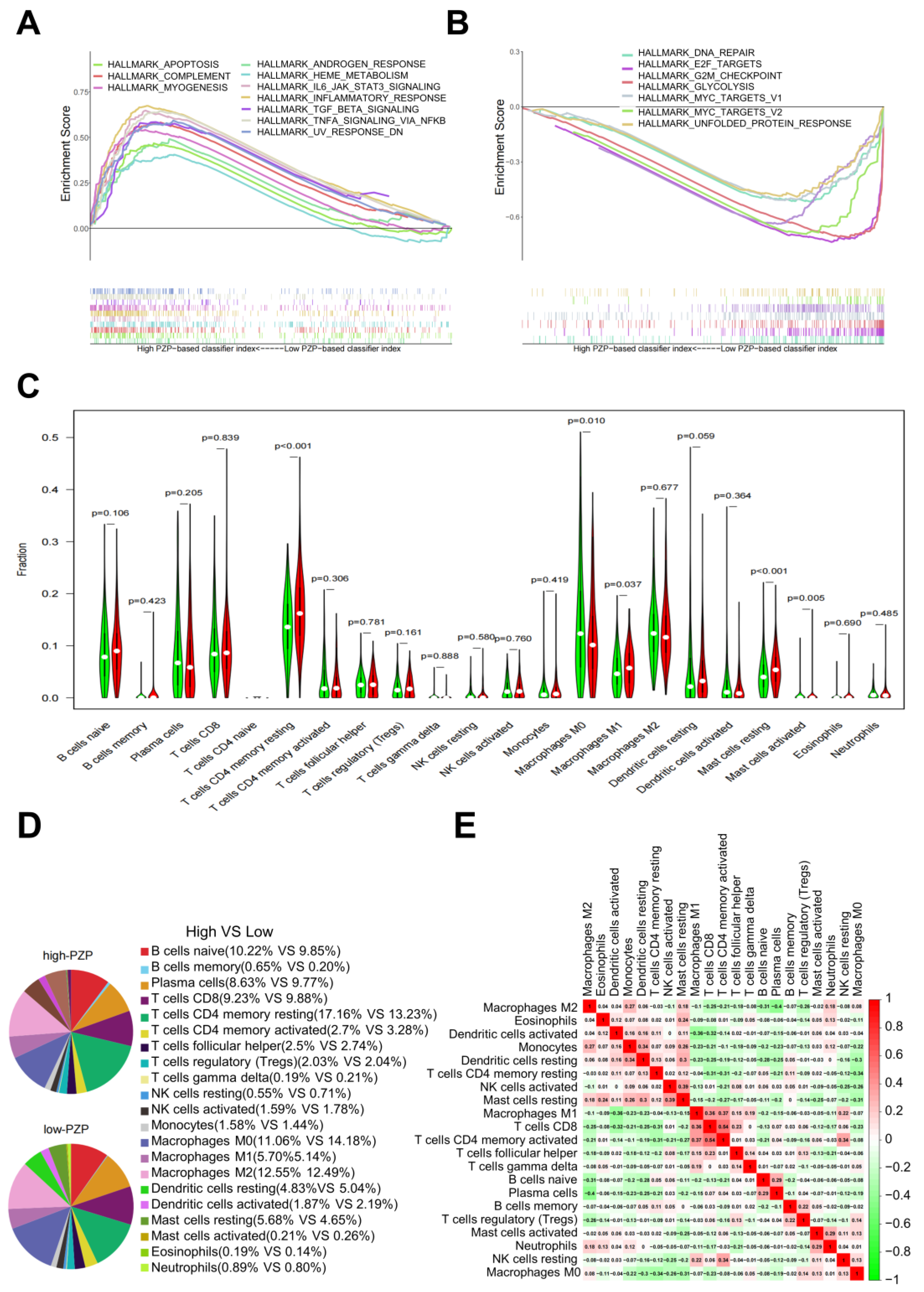
3.3. PZP Expression Was Associated with TME Modulation in LUAD
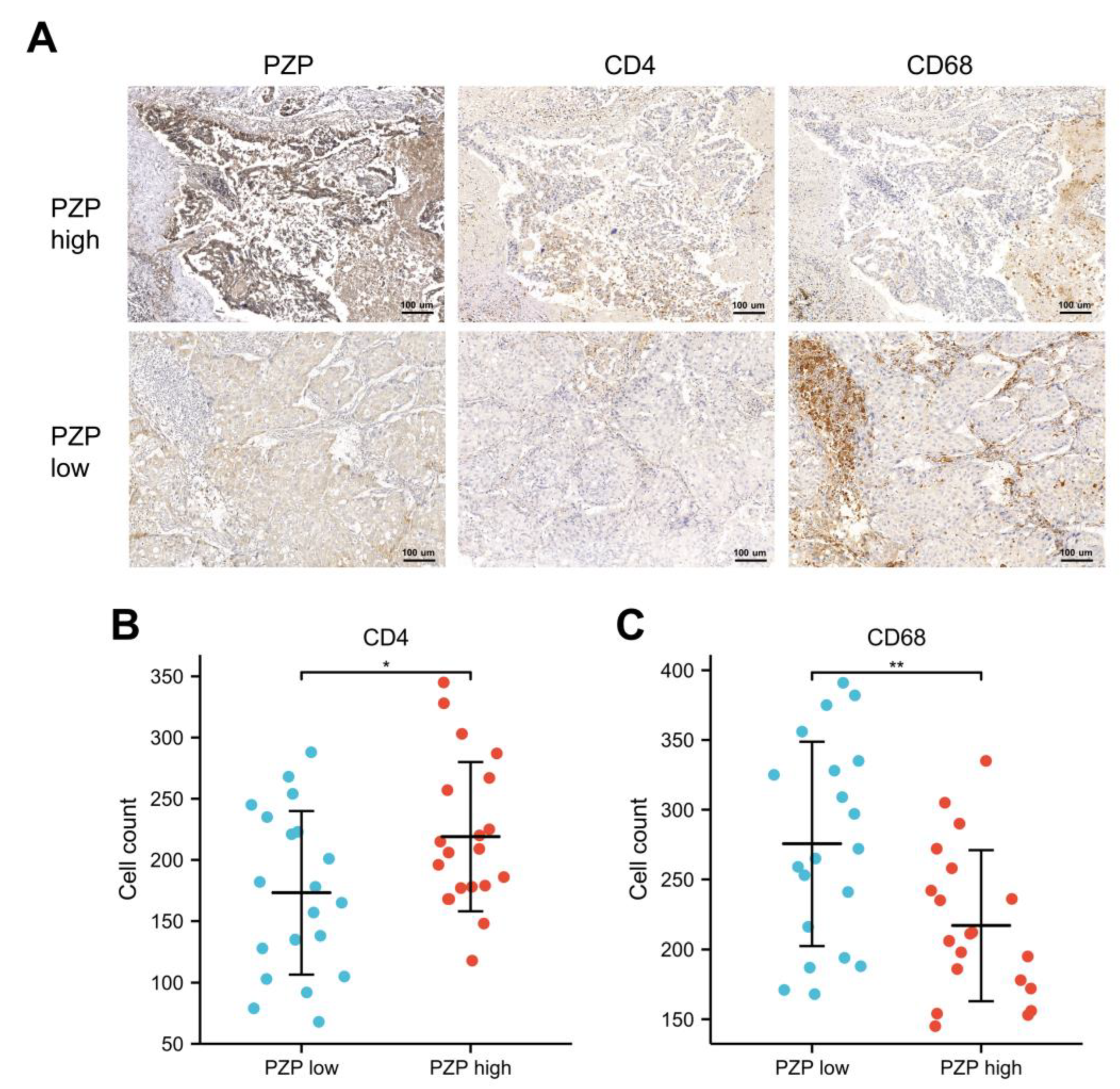
3.4. Relationship between PZP and Immune Cells in Clinical Samples
3.5. Association between PZP and the Polarization State of Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Lv, J.; Zhu, M.; Wang, Y.; Qin, N.; Ma, H.; He, Y.Q.; Zhang, R.; Tan, W.; Fan, J.; et al. Identification of risk loci and a polygenic risk score for lung cancer: A large-scale prospective cohort study in Chinese populations. Lancet Respir. Med. 2019, 7, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Kim, T.H.; Shin, S.; Kim, H.K.; Choi, Y.S.; Kim, J.; Zo, J.I.; Shim, Y.M.; Cho, J.H. Recommended Change in the N Descriptor Proposed by the International Association for the Study of Lung Cancer: A Validation Study. J. Thorac. Oncol. 2019, 14, 1962–1969. [Google Scholar] [CrossRef]
- Xu, J.Y.; Zhang, C.; Wang, X.; Zhai, L.; Ma, Y.; Mao, Y.; Qian, K.; Sun, C.; Liu, Z.; Jiang, S.; et al. Integrative Proteomic Characterization of Human Lung Adenocarcinoma. Cell 2020, 182, 245–261.e17. [Google Scholar] [CrossRef]
- Liu, X.S.; Zhou, L.M.; Yuan, L.L.; Gao, Y.; Kui, X.Y.; Liu, X.Y.; Pei, Z.J. NPM1 Is a Prognostic Biomarker Involved in Immune Infiltration of Lung Adenocarcinoma and Associated With m6A Modification and Glycolysis. Front. Immunol. 2021, 12, 724741. [Google Scholar] [CrossRef]
- Liu, X.S.; Zeng, J.; Zhang, Y.H.; Zhang, Y.; Gao, Y.; Liu, C.; Pei, Z.J. DARS2 is a prognostic biomarker and correlated with immune infiltrates and cuproptosis in lung adenocarcinoma. Am. J. Cancer Res. 2023, 13, 818–834. [Google Scholar] [PubMed]
- Skornicka, E.L.; Kiyatkina, N.; Weber, M.C.; Tykocinski, M.L.; Koo, P.H. Pregnancy zone protein is a carrier and modulator of placental protein-14 in T-cell growth and cytokine production. Cell Immunol. 2004, 232, 144–156. [Google Scholar] [CrossRef]
- Teng, H.; Zhang, W.Y.; Zhu, F.Q. A study on the serum pregnancy zone protein levels in pregnant women and patients with gynecological tumors. Chin. Med. J. 1994, 107, 910–914. [Google Scholar]
- Löb, S.; Vattai, A.; Kuhn, C.; Schmoeckel, E.; Mahner, S.; Wöckel, A.; Kolben, T.; Keil, C.; Jeschke, U.; Vilsmaier, T. Pregnancy Zone Protein (PZP) is significantly upregulated in the decidua of recurrent and spontaneous miscarriage and negatively correlated to Glycodelin A (GdA). J. Reprod. Immunol. 2021, 143, 103267. [Google Scholar] [CrossRef]
- Svendsen, P.; Stigbrand, T.; Teisner, B.; Folkersen, J.; Damber, M.G.; von Schoultz, B.; Kemp, E.; Svehag, S.E. Immunosuppressive effect of human pregnancy zone protein on H-2 incompatible mouse heart allografts. Acta Pathol. Microbiol. Scand. C 1978, 86, 199–201. [Google Scholar] [CrossRef]
- Jensen, P.H.; Davidsen, O.; Gliemann, J.; Petersen, C.M. Cell association and degradation of pregnancy zone protein-chymotrypsin complex in cultured human monocytes. Scand. J. Clin. Lab. Investig. 1988, 48, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.; Bostedt, L.; Stigbrand, T.; O’Connor-McCourt, M.D. Binding of transforming growth factor-beta (TGF-beta) to pregnancy zone protein (PZP). Comparison to the TGF-beta-alpha 2-macroglobulin interaction. Eur. J. Biochem. 1994, 221, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kuligina, E.; Sokolenko, A.; Siddiqui, Q.; Gardi, N.; Gupta, S.; Varma, A.K.; Hasan, S.K. Genetic ablation of pregnancy zone protein promotes breast cancer progression by activating TGF-beta/SMAD signaling. Breast Cancer Res. Treat. 2021, 185, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Y.; Zhao, S.; Zheng, Z.; Shen, C.; An, L.; Yuan, Y. Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 6079–6096. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Zhang, G.; Kong, X. Prognostic Significance of Pregnancy Zone Protein and Its Correlation with Immune Infiltrates in Hepatocellular Carcinoma. Cancer Manag. Res. 2020, 12, 9883–9891. [Google Scholar] [CrossRef]
- Su, L.; Zhang, G.; Kong, X. A Novel Five-Gene Signature for Prognosis Prediction in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 642563. [Google Scholar] [CrossRef]
- Wang, Z.; Pan, L.; Guo, D.; Luo, X.; Tang, J.; Yang, W.; Zhang, Y.; Luo, A.; Gu, Y.; Pan, Y. A novel five-gene signature predicts overall survival of patients with hepatocellular carcinoma. Cancer Med. 2021, 10, 3808–3821. [Google Scholar] [CrossRef]
- Wu, M.; Lan, H.; Ye, Z.; Wang, Y. Hypermethylation of the PZP gene is associated with hepatocellular carcinoma cell proliferation, invasion and migration. FEBS Open Bio 2021, 11, 826–832. [Google Scholar] [CrossRef]
- Zhang, C.; Leighl, N.B.; Wu, Y.L.; Zhong, W.Z. Emerging therapies for non-small cell lung cancer. J. Hematol. Oncol. 2019, 12, 45. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wan, Y.Y.; Zhu, B. Immune Cell Metabolism in Tumor Microenvironment. Adv. Exp. Med. Biol. 2017, 1011, 163–196. [Google Scholar] [CrossRef]
- Vafadar, S. Immunotherapy for non-small cell lung cancer. Jaapa 2019, 32, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longato, E.; Vettoretti, M.; Di Camillo, B. A practical perspective on the concordance index for the evaluation and selection of prognostic time-to-event models. J. Biomed. Inform. 2020, 108, 103496. [Google Scholar] [CrossRef]
- Guo, Q.K.; Yang, H.S.; Shan, S.C.; Chang, D.D.; Qiu, L.J.; Luo, H.H.; Li, H.P.; Ke, Z.F.; Zhu, Y. A radiomics nomogram prediction for survival of patients with “driver gene-negative” lung adenocarcinomas (LUAD). Radiol. Med. 2023, 128, 714–725. [Google Scholar] [CrossRef]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Qiao, B.; Huang, J.; Mei, Z.; Lam, A.K.; Zhao, J.; Ying, L. Analysis of Immune Microenvironment by Multiplex Immunohistochemistry Staining in Different Oral Diseases and Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 555757. [Google Scholar] [CrossRef]
- Heynemann, S.; Yu, H.; Churilov, L.; Rivalland, G.; Asadi, K.; Mosher, R.; Hirsch, F.; Rivard, C.; Mitchell, P. NaPi2b expression in a large surgical Non-Small Cell Lung Cancer (NSCLC) cohort. Clin. Lung Cancer 2022, 23, e90–e98. [Google Scholar] [CrossRef]
- Chen, P.; Qin, Z.; Sun, X.; Yang, J.; Lv, J.; Diao, M. Expression and Clinical Significance of lncRNA OSER1-AS1 in Peripheral Blood of Patients with Non-Small Cell Lung Cancer. Cells Tissues Organs 2022, 211, 589–600. [Google Scholar] [CrossRef]
- Parra, E.R.; Villalobos, P.; Behrens, C.; Jiang, M.; Pataer, A.; Swisher, S.G.; William, W.N., Jr.; Zhang, J.; Lee, J.; Cascone, T.; et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in non-small cell lung carcinomas as determined by multiplex immunofluorescence and image analysis approaches. J. Immunother. Cancer 2018, 6, 48. [Google Scholar] [CrossRef]
- Liu, X.; Wu, S.; Yang, Y.; Zhao, M.; Zhu, G.; Hou, Z. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed. Pharmacother. 2017, 95, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Von Schoultz, B.; Stigbrand, T. Characterization of the “pregnancy zone protein” in relation to other alpha 2-globulins of pregnancy. Biochim. Biophys. Acta 1974, 359, 303–310. [Google Scholar] [CrossRef]
- Tayade, C.; Esadeg, S.; Fang, Y.; Croy, B.A. Functions of alpha 2 macroglobulins in pregnancy. Mol. Cell Endocrinol. 2005, 245, 60–66. [Google Scholar] [CrossRef]
- Finch, S.; Shoemark, A.; Dicker, A.J.; Keir, H.R.; Smith, A.; Ong, S.; Tan, B.; Choi, J.Y.; Fardon, T.C.; Cassidy, D.; et al. Pregnancy Zone Protein Is Associated with Airway Infection, Neutrophil Extracellular Trap Formation, and Disease Severity in Bronchiectasis. Am. J. Respir. Crit. Care Med. 2019, 200, 992–1001. [Google Scholar] [CrossRef]
- Bastos-Amador, P.; Royo, F.; Gonzalez, E.; Conde-Vancells, J.; Palomo-Diez, L.; Borras, F.E.; Falcon-Perez, J.M. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J. Proteom. 2012, 75, 3574–3584. [Google Scholar] [CrossRef] [PubMed]
- Cater, J.H.; Kumita, J.R.; Zeineddine Abdallah, R.; Zhao, G.; Bernardo-Gancedo, A.; Henry, A.; Winata, W.; Chi, M.; Grenyer, B.S.F.; Townsend, M.L.; et al. Human pregnancy zone protein stabilizes misfolded proteins including preeclampsia- and Alzheimer’s-associated amyloid beta peptide. Proc. Natl. Acad. Sci. USA 2019, 116, 6101–6110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiabrando, G.A.; Sanchez, M.C.; Skornicka, E.L.; Koo, P.H. Low-density lipoprotein receptor-related protein mediates in PC12 cell cultures the inhibition of nerve growth factor-promoted neurite outgrowth by pregnancy zone protein and alpha2-macroglobulin. J. Neurosci. Res. 2002, 70, 57–64. [Google Scholar] [CrossRef]
- Lin, J.; Jiang, X.; Dong, M.; Liu, X.; Shen, Q.; Huang, Y.; Zhang, H.; Ye, R.; Zhou, H.; Yan, C.; et al. Hepatokine Pregnancy Zone Protein Governs the Diet-Induced Thermogenesis Through Activating Brown Adipose Tissue. Adv. Sci. 2021, 8, e2101991. [Google Scholar] [CrossRef]
- Hwang, I.; Kim, J.W.; Ylaya, K.; Chung, E.J.; Kitano, H.; Perry, C.; Hanaoka, J.; Fukuoka, J.; Chung, J.Y.; Hewitt, S.M. Tumor-associated macrophage, angiogenesis and lymphangiogenesis markers predict prognosis of non-small cell lung cancer patients. J. Transl. Med. 2020, 18, 443. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Weigert, A.; Reu, S.; Guenther, S.; Mansouri, S.; Bassaly, B.; Gattenlöhner, S.; Grimminger, F.; Pullamsetti, S.; Seeger, W.; et al. Spatial Density and Distribution of Tumor-Associated Macrophages Predict Survival in Non-Small Cell Lung Carcinoma. Cancer Res. 2020, 80, 4414–4425. [Google Scholar] [CrossRef]
- Pan, Y.; Sha, Y.; Wang, H.; Zhuang, H.; Ren, X.; Zhu, X.; Wei, X. Comprehensive analysis of the association between tumor-infiltrating immune cells and the prognosis of lung adenocarcinoma. J. Cancer Res. Ther. 2020, 16, 320–326. [Google Scholar] [CrossRef]
- Gliemann, J.; Moestrup, S.; Henning Jensen, P.; Sottrup-Jensen, L.; Busk Andersen, H.; Munck Petersen, C.; Sonne, O. Evidence for binding of human pregnancy zone protein-proteinase complex to alpha 2-macroglobulin receptors. Biochim. Biophys. Acta 1986, 883, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Karnevi, E.; Andersson, R.; Rosendahl, A.H. Tumour-educated macrophages display a mixed polarisation and enhance pancreatic cancer cell invasion. Immunol. Cell Biol. 2014, 92, 543–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Liang, Z.W.; Ge, X.X.; Xu, M.D.; Qin, H.; Wu, M.Y.; Shen, M.; Zhang, Y.; Liu, X.M.; Chen, K.; Li, W.; et al. Tumor-associated macrophages promote the metastasis and growth of non-small-cell lung cancer cells through NF-κB/PP2Ac-positive feedback loop. Cancer Sci. 2021, 112, 2140–2157. [Google Scholar] [CrossRef]
- Sedighzadeh, S.S.; Khoshbin, A.P.; Razi, S.; Keshavarz-Fathi, M.; Rezaei, N. A narrative review of tumor-associated macrophages in lung cancer: Regulation of macrophage polarization and therapeutic implications. Transl. Lung Cancer Res. 2021, 10, 1889–1916. [Google Scholar] [CrossRef]
| PZP Expression | ||||
|---|---|---|---|---|
| Characteristics | NO. of Cases(%) | Low NO.Cases | High NO.Cases | p Value |
| Age (y) | ||||
| ≤65 | 46 (41.8%) | 28 | 18 | 0.994 |
| >65 | 64 (58.2%) | 39 | 25 | |
| Gender | ||||
| Female | 59 (53.6%) | 33 | 26 | 0.250 |
| Male | 51 (46.4%) | 34 | 17 | |
| Smoker | ||||
| No | 63 (57.3%) | 37 | 26 | 0.588 |
| Yes | 47 (42.7%) | 30 | 17 | |
| T classification | ||||
| T1&T2 | 104 (94.5%) | 62 | 42 | 0.247 |
| T3&T4 | 6 (5.5%) | 5 | 1 | |
| N classification | ||||
| N0 | 71 (64.5%) | 38 | 33 | 0.032 |
| N1&N2&N3 | 39 (35.5%) | 29 | 10 | |
| M classification | ||||
| M0 | 97 (88.2%) | 56 | 41 | 0.062 |
| M1 | 13 (11.8%) | 11 | 2 | |
| Clinical stage | ||||
| I&II | 82 (74.5%) | 43 | 39 | 0.002 |
| III&IV | 28 (25.5%) | 24 | 4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Zheng, T.; Chen, C.; Liu, L.; Guo, Z.; Peng, Y.; Zhang, X.; Yang, Z. Pregnancy Zone Protein Serves as a Prognostic Marker and Favors Immune Infiltration in Lung Adenocarcinoma. Biomedicines 2023, 11, 1978. https://doi.org/10.3390/biomedicines11071978
Chen K, Zheng T, Chen C, Liu L, Guo Z, Peng Y, Zhang X, Yang Z. Pregnancy Zone Protein Serves as a Prognostic Marker and Favors Immune Infiltration in Lung Adenocarcinoma. Biomedicines. 2023; 11(7):1978. https://doi.org/10.3390/biomedicines11071978
Chicago/Turabian StyleChen, Kehong, Taihao Zheng, Cai Chen, Liangzhong Liu, Zhengjun Guo, Yuan Peng, Xiaoyue Zhang, and Zhenzhou Yang. 2023. "Pregnancy Zone Protein Serves as a Prognostic Marker and Favors Immune Infiltration in Lung Adenocarcinoma" Biomedicines 11, no. 7: 1978. https://doi.org/10.3390/biomedicines11071978




