Identifying Microbiome Dynamics in Pediatric IBD: More than a Family Matter
Abstract
:1. Introduction
2. Results
2.1. IBD versus HC Overall
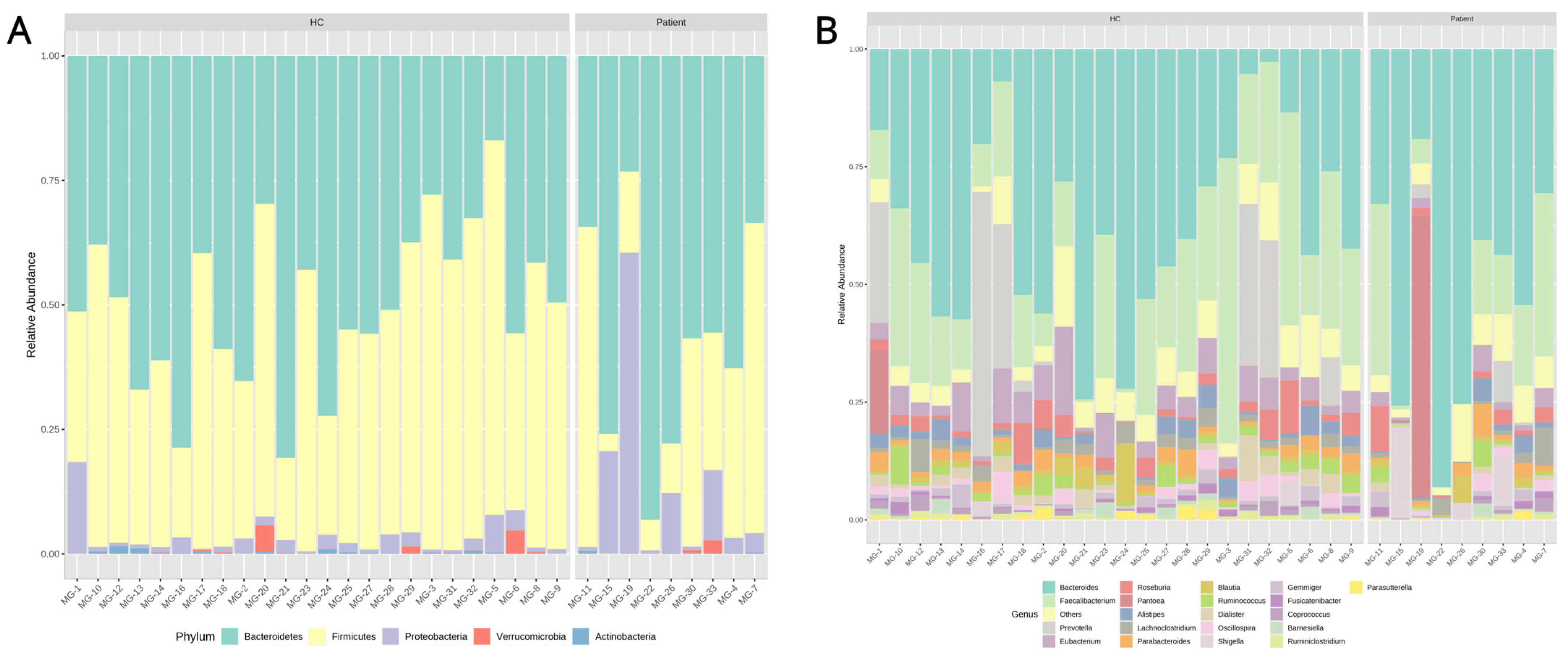
2.1.1. Microbiota Composition and Univariate Analysis
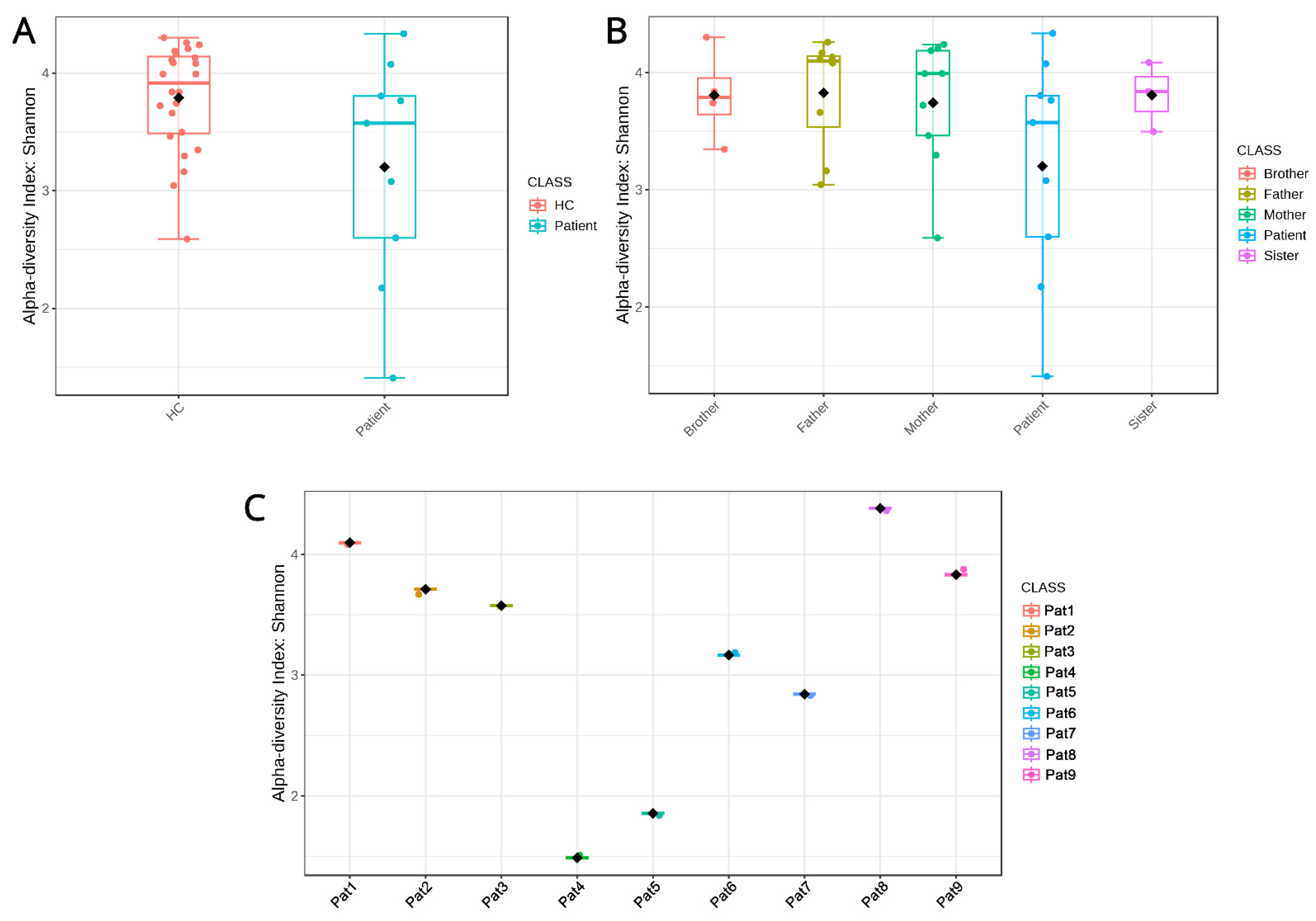
2.1.2. Diversity Metrics
2.2. Intrafamilial Microbiome Changes
2.2.1. Biodiversity
2.2.2. Similarity Clustering
2.2.3. Microbial Composition
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. DNA Extraction and 16S rRNA Amplicon Sequencing
4.3. 16S rRNA Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Peloquin, J.M.; Goel, G.; Villablanca, E.J.; Xavier, R.J. Mechanisms of pediatric inflammatory bowel disease. Annu. Rev. Immunol. 2016, 34, 31–64. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.A.; Rosh, J.R. Pediatric inflammatory bowel disease. Pediatr. Clin. 2017, 64, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Corica, D.; Romano, C. Biological therapy in pediatric inflammatory bowel disease. J. Clin. Gastroenterol. 2017, 51, 100–110. [Google Scholar] [CrossRef]
- Miele, E.; Shamir, R.; Aloi, M.; Assa, A.; Braegger, C.; Bronsky, J.; De Ridder, L.; Escher, J.C.; Hojsak, I.; Kolaček, S. Nutrition in pediatric inflammatory bowel disease: A position paper on behalf of the Porto Inflammatory Bowel Disease Group of the European Society of Pediatric Gastroenterology, Hepatology and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2018, 66, 687–708. [Google Scholar] [CrossRef] [Green Version]
- Ricciuto, A.; Aardoom, M.; Orlanski-Meyer, E.; Navon, D.; Carman, N.; Aloi, M.; Bronsky, J.; Däbritz, J.; Dubinsky, M.; Hussey, S. Predicting outcomes in pediatric crohn’s disease for management optimization: Systematic review and consensus statements from the pediatric inflammatory bowel disease–ahead program. Gastroenterology 2021, 160, 403–436.e426. [Google Scholar] [CrossRef]
- Goldstein-Leever, A.; Bass, J.A.; Goyal, A.; Maddux, M.H. Health-related quality of life predicts psychology referral in youth with inflammatory bowel disease. J. Pediatr. Nurs. 2019, 47, 73–77. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Gazouli, M.; Dovrolis, N.; Franke, A.; Spyrou, G.M.; Sechi, L.A.; Kolios, G. Differential genetic and functional background in inflammatory bowel disease phenotypes of a Greek population: A systems bioinformatics approach. Gut Pathog. 2019, 11, 31. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Kumar, A.; Al-Hassi, H.O.; Steed, H.; Phipps, O.; Brookes, M.J. Bile acids and the microbiome: Making sense of this dynamic relationship in their role and management in Crohn’s disease. Can. J. Gastroenterol. Hepatol. 2022, 2022, 8416578. [Google Scholar] [CrossRef]
- Russell, R.K.; Satsangi, J. Does IBD run in families? Inflamm. Bowel Dis. 2008, 14 (Suppl. 2), S20–S21. [Google Scholar] [CrossRef] [PubMed]
- Spencer, E.A.; Helmus, D.; Telesco, S.; Colombel, J.-F.; Dubinsky, M.C. Inflammatory bowel disease clusters within affected sibships in Ashkenazi Jewish multiplex families. Gastroenterology 2020, 159, 381–382. [Google Scholar] [CrossRef] [PubMed]
- Colombel, J.F.; Grandbastien, B.; Gower-Rousseau, C.; Plegat, S.; Evrard, J.P.; Dupas, J.L.; Gendre, J.P.; Modigliani, R.; Belaiche, J.; Hostein, J. Clinical characteristics of Crohn’s disease in 72 families. Gastroenterology 1996, 111, 604–607. [Google Scholar] [CrossRef]
- Moller, F.T.; Andersen, V.; Wohlfahrt, J.; Jess, T. Familial risk of inflammatory bowel disease: A population-based cohort study 1977–2011. Off. J. Am. Coll. Gastroenterol. 2015, 110, 564–571. [Google Scholar] [CrossRef]
- Sharma, A.; Junge, O.; Szymczak, S.; Rühlemann, M.C.; Enderle, J.; Schreiber, S.; Laudes, M.; Franke, A.; Lieb, W.; Krawczak, M. Network-based quantitative trait linkage analysis of microbiome composition in inflammatory bowel disease families. Front. Genet. 2023, 14, 1048312. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.P.; Goudarzi, M.; Singh, N.; Tong, M.; McHardy, I.H.; Ruegger, P.; Asadourian, M.; Moon, B.H.; Ayson, A.; Borneman, J.; et al. A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 750–766. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S. Cohabiting family members share microbiota with one another and with their dogs. Elife 2013, 2, e00458. [Google Scholar] [CrossRef]
- Mosites, E.; Sammons, M.; Otiang, E.; Eng, A.; Noecker, C.; Manor, O.; Hilton, S.; Thumbi, S.M.; Onyango, C.; Garland-Lewis, G. Microbiome sharing between children, livestock and household surfaces in western Kenya. PLoS ONE 2017, 12, e0171017. [Google Scholar] [CrossRef] [Green Version]
- Valles-Colomer, M.; Blanco-Míguez, A.; Manghi, P.; Asnicar, F.; Dubois, L.; Golzato, D.; Armanini, F.; Cumbo, F.; Huang, K.D.; Manara, S. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 2023, 614, 125–135. [Google Scholar] [CrossRef]
- Schloss, P.D.; Iverson, K.D.; Petrosino, J.F.; Schloss, S.J. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome 2014, 2, 25. [Google Scholar] [CrossRef] [Green Version]
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [Green Version]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Chang, E.B. Inflammatory bowel diseases (IBD) and the microbiome—Searching the crime scene for clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Olbjørn, C.; Småstuen, M.C.; Moen, A.E.F. Targeted analysis of the gut microbiome for diagnosis, prognosis and treatment individualization in pediatric inflammatory bowel disease. Microorganisms 2022, 10, 1273. [Google Scholar] [CrossRef] [PubMed]
- Riesenfeld, C.S.; Schloss, P.D.; Handelsman, J. Metagenomics: Genomic analysis of microbial communities. Annu. Rev. Genet. 2004, 38, 525–552. [Google Scholar] [CrossRef] [Green Version]
- Calle, M.L. Statistical analysis of metagenomics data. Genom. Inform. 2019, 17, e6. [Google Scholar] [CrossRef]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef]
- Loh, G.; Blaut, M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 2012, 3, 544–555. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.T.; Amos, G.C.; Murphy, A.R.; Murch, S.; Wellington, E.M.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. Biomed Res. Int. 2016, 2016, 5828959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomura, K.; Ishikawa, D. Bacteroidetes Species Are Correlated with Disease Activity in Ulcerative Colitis. 2021, 10, 1749. J. Clin. Med. 2021, 10, 1749. [Google Scholar] [CrossRef]
- Vester-Andersen, M.; Mirsepasi-Lauridsen, H.; Prosberg, M.; Mortensen, C.; Träger, C.; Skovsen, K.; Thorkilgaard, T.; Nøjgaard, C.; Vind, I.; Krogfelt, K.A. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci. Rep. 2019, 9, 13473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Eppinga, H.; Sperna Weiland, C.J.; Thio, H.B.; van der Woude, C.J.; Nijsten, T.E.; Peppelenbosch, M.P.; Konstantinov, S.R. Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J. Crohn’s Colitis 2016, 10, 1067–1075. [Google Scholar] [CrossRef] [Green Version]
- Hansen, R.; Russell, R.K.; Reiff, C.; Louis, P.; McIntosh, F.; Berry, S.H.; Mukhopadhya, I.; Bisset, W.M.; Barclay, A.R.; Bishop, J.; et al. Microbiota of de-novo pediatric IBD: Increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn’s but not in ulcerative colitis. Am. J. Gastroenterol. 2012, 107, 1913–1922. [Google Scholar] [CrossRef] [Green Version]
- Dovrolis, N.; Drygiannakis, I.; Filidou, E.; Kandilogiannakis, L.; Arvanitidis, K.; Tentes, I.; Kolios, G.; Valatas, V. Gut Microbial Signatures Underline Complicated Crohn’s Disease but Vary Between Cohorts; An In Silico Approach. Inflamm. Bowel Dis. 2019, 25, 217–225. [Google Scholar] [CrossRef]
- Dovrolis, N.; Michalopoulos, G.; Theodoropoulos, G.E.; Arvanitidis, K.; Kolios, G.; Sechi, L.A.; Eliopoulos, A.G.; Gazouli, M. The interplay between mucosal microbiota composition and host gene-expression is linked with infliximab response in inflammatory bowel diseases. Microorganisms 2020, 8, 438. [Google Scholar] [CrossRef] [Green Version]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Rausch, P.; Rehman, A.; Künzel, S.; Häsler, R.; Ott, S.J.; Schreiber, S.; Rosenstiel, P.; Franke, A.; Baines, J.F. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. USA 2011, 108, 19030–19035. [Google Scholar] [CrossRef]
- Zhuang, X.; Liu, C.; Zhan, S.; Tian, Z.; Li, N.; Mao, R.; Zeng, Z.; Chen, M. Gut microbiota profile in pediatric patients with inflammatory bowel disease: A systematic review. Front. Pediatr. 2021, 9, 626232. [Google Scholar] [CrossRef]
- Kowalska-Duplaga, K.; Gosiewski, T.; Kapusta, P.; Sroka-Oleksiak, A.; Wędrychowicz, A.; Pieczarkowski, S.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Fyderek, K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci. Rep. 2019, 9, 18880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Huang, Y.; Shi, P.; Wang, Y.; Zhang, Y.; Xue, A.; Tang, Z.; Hu, W.; Sun, H.; Zhang, P. Effect of exclusive enteral nutrition on the disease process, nutrition status, and gastrointestinal microbiota for Chinese children with Crohn’s disease. J. Parenter. Enter. Nutr. 2021, 45, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, U.Z.; Quince, C.; Hanske, L.; Loman, N.; Calus, S.T.; Bertz, M.; Edwards, C.A.; Gaya, D.R.; Hansen, R.; McGrogan, P. The distinct features of microbial ‘dysbiosis’ of Crohn’s disease do not occur to the same extent in their unaffected, genetically-linked kindred. PLoS ONE 2017, 12, e0172605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Kaplan, J.L.; Gold, B.D.; Bhasin, M.K.; Ward, N.L.; Kellermayer, R.; Kirschner, B.S.; Heyman, M.B.; Dowd, S.E.; Cox, S.B. Detecting microbial dysbiosis associated with pediatric Crohn disease despite the high variability of the gut microbiota. Cell Rep. 2016, 14, 945–955. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Cheng, S.; Yao, J.; Lin, X.; Li, Y.; Wang, W.; Weng, J.; Zou, Y.; Zhu, L.; Zhi, M. Correlation between altered gut microbiota and elevated inflammation markers in patients with Crohn’s disease. Front. Immunol. 2022, 13, 947313. [Google Scholar] [CrossRef]
- Xiong, R.-G.; Zhou, D.-D.; Wu, S.-X.; Huang, S.-Y.; Saimaiti, A.; Yang, Z.-J.; Shang, A.; Zhao, C.-N.; Gan, R.-Y.; Li, H.-B. Health benefits and side effects of short-chain fatty acids. Foods 2022, 11, 2863. [Google Scholar] [CrossRef]
- El Mouzan, M.I.; Winter, H.S.; Assiri, A.A.; Korolev, K.S.; Al Sarkhy, A.A.; Dowd, S.E.; Al Mofarreh, M.A.; Menon, R. Microbiota profile in new-onset pediatric Crohn’s disease: Data from a non-Western population. Gut Pathog. 2018, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Sila, S.; Jelic, M.; Trivic, I.; Andraševic, A.T.; Hojsak, I.; Kolacek, S. Altered gut microbiota is present in newly diagnosed pediatric patients with inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 497–502. [Google Scholar] [CrossRef]
- Malham, M.; Lilje, B.; Houen, G.; Winther, K.; Andersen, P.S.; Jakobsen, C. The microbiome reflects diagnosis and predicts disease severity in paediatric onset inflammatory bowel disease. Scand. J. Gastroenterol. 2019, 54, 969–975. [Google Scholar] [CrossRef]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augmentin vitroutilization of mucin by other bacteria. Off. J. Am. Coll. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef]
- Höyhtyä, M.; Korpela, K.; Saqib, S.; Junkkari, S.; Nissilä, E.; Nikkonen, A.; Dikareva, E.; Salonen, A.; de Vos, W.M.; Kolho, K.-L. Quantitative fecal microbiota profiles relate to therapy response during induction with tumor necrosis factor α antagonist infliximab in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2023, 29, 116–124. [Google Scholar] [CrossRef]
- Sonnenberg, G.F.; Monticelli, L.A.; Alenghat, T.; Fung, T.C.; Hutnick, N.A.; Kunisawa, J.; Shibata, N.; Grunberg, S.; Sinha, R.; Zahm, A.M. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 2012, 336, 1321–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gao, X.; Ghozlane, A.; Hu, H.; Li, X.; Xiao, Y.; Li, D.; Yu, G.; Zhang, T. Characteristics of faecal microbiota in paediatric Crohn’s disease and their dynamic changes during infliximab therapy. J. Crohn’s Colitis 2018, 12, 337–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assa, A.; Butcher, J.; Li, J.; Elkadri, A.; Sherman, P.M.; Muise, A.M.; Stintzi, A.; Mack, D. Mucosa-associated ileal microbiota in new-onset pediatric Crohn’s disease. Inflamm. Bowel Dis. 2016, 22, 1533–1539. [Google Scholar] [CrossRef] [Green Version]
- Santana, P.T.; Rosas, S.L.B.; Ribeiro, B.E.; Marinho, Y.; de Souza, H.S. Dysbiosis in inflammatory bowel disease: Pathogenic role and potential therapeutic targets. Int. J. Mol. Sci. 2022, 23, 3464. [Google Scholar] [CrossRef]
- Sultan, S.; El-Mowafy, M.; Elgaml, A.; Ahmed, T.A.; Hassan, H.; Mottawea, W. Metabolic influences of gut microbiota dysbiosis on inflammatory bowel disease. Front. Physiol. 2021, 12, 715506. [Google Scholar] [CrossRef]
- Stinson, L.F. Establishment of the early-life microbiome: A DOHaD perspective. J. Dev. Orig. Health Dis. 2020, 11, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Ríos-Covian, D.; Langella, P.; Martín, R. From short-to long-term effects of c-section delivery on microbiome establishment and host health. Microorganisms 2021, 9, 2122. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; Lieber, A.D.; Wu, F.; Perez-Perez, G.I.; Chen, Y. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casas, L.; Karvonen, A.M.; Kirjavainen, P.V.; Täubel, M.; Hyytiäinen, H.; Jayaprakash, B.; Lehmann, I.; Standl, M.; Pekkanen, J.; Heinrich, J. Early life home microbiome and hyperactivity/inattention in school-age children. Sci. Rep. 2019, 9, 17355. [Google Scholar] [CrossRef] [Green Version]
- Flannery, J.; Callaghan, B.; Sharpton, T.; Fisher, P.; Pfeifer, J. Is adolescence the missing developmental link in Microbiome–Gut–Brain axis communication? Dev. Psychobiol. 2019, 61, 783–795. [Google Scholar] [CrossRef]
- Chen, B.; Ye, D.; Luo, L.; Liu, W.; Peng, K.; Shu, X.; Gu, W.; Wang, X.; Xiang, C.; Jiang, M. Adhesive bacteria in the terminal ileum of children correlates with increasing Th17 cell activation. Front. Pharmacol. 2020, 11, 588560. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. The role of butyrate in attenuating pathobiont-induced hyperinflammation. Immune Netw. 2020, 20, e15. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current sampling methods for gut microbiota: A call for more precise devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Svolos, V.; Hansen, R.; Nichols, B.; Quince, C.; Ijaz, U.Z.; Papadopoulou, R.T.; Edwards, C.A.; Watson, D.; Alghamdi, A.; Brejnrod, A. Treatment of active Crohn’s disease with an ordinary food-based diet that replicates exclusive enteral nutrition. Gastroenterology 2019, 156, 1354–1367.e1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamji, M.; Day, A.S. An overview of the role of exclusive enteral nutrition for complicated Crohn’s disease. Intest. Res. 2019, 17, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Kansal, S.; Catto-Smith, A.G.; Boniface, K.; Thomas, S.; Cameron, D.J.; Oliver, M.; Alex, G.; Kirkwood, C.D.; Wagner, J. The microbiome in paediatric crohn’s disease—A longitudinal, prospective, single-centre study. J. Crohn’s Colitis 2019, 13, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.; Li, L.; Zhang, L.; Genco, R.J.; Falkner, K.L.; Tettelin, H.; Rowsam, A.M.; Smiraglia, D.J.; Novak, J.M.; Diaz, P.I. Periodontal disease is associated with increased gut colonization of pathogenic Haemophilus parainfluenzae in patients with Crohn’s disease. Cell Rep. 2023, 42, 112120. [Google Scholar] [CrossRef]
- Mavragani, C.P.; Nezos, A.; Dovrolis, N.; Andreou, N.-P.; Legaki, E.; Sechi, L.A.; Bamias, G.; Gazouli, M. Type I and II interferon signatures can predict the response to anti-TNF agents in inflammatory bowel disease patients: Involvement of the microbiota. Inflamm. Bowel Dis. 2020, 26, 1543–1553. [Google Scholar] [CrossRef]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y. QIIME 2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.molbiotools.com/listcompare.php (accessed on 1 July 2023).




| Genera More Abundant in IBD | Genera Less Abundant in IBD | Species More Abundant in IBD | Species Less Abundant in IBD |
|---|---|---|---|
| Veillonella | Candidatus Soleaferrea | Veillonella_parvula | Ruminococcus__flavefaciens |
| Haemophilus | Prevotella | Haemophilus_parainfluenzae | Bacteroides__massiliensis |
| Granulicatella | Holdemanella | Erysipelatoclostridium_ramosum | Prevotella__copri |
| Sutterella | Anaerobacterium | Granulicatella_paradiacens | Bacteroides__stercoris |
| Shigella | Lactobacillus | Streptococcus_parasanguinis | Dialister__succinatiphilus |
| Erysipelatoclostridium | Holdemania | Bacteroides_caccae | Lachnoclostridium__xylanolyticum |
| Pantoea | Catenibacterium | Shigella_sonnei | Eubacterium__hallii |
| Streptococcus | Lachnospira | Sutterella_wadsworthensis | Alistipes__indistinctus |
| Butyricimonas | Dorea | Bacteroides_fragilis | Holdemanella__biforme |
| Alcaligenes | Anaerovorax | Bacteroides_acidofaciens | Anaerobacterium__chartisolvens |
| Family Identifier | Age at Diagnosis | Gender | IBD Phenotype | Fecal Calprotectin (μg/fecal gr) | CRP 1 (mg/dL) | ESR 2 (mm/h) | PUCAI/PUCDAI 3 | Birth Mode | Prematurity |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 13 | F | CD | 250 | <3 | 1 | 25 (Mild) | Cesarean | Yes |
| 2 | 12 | M | CD | 530 | <3 | 40 | 30 (Moderate) | Cesarean | No |
| 3 | 14 | F | UC | 600 | 8 | 50 | 20 (Mild) | Vaginal | No |
| 4 | 4.5 | F | CD | 95 | 96 | 120 | >40 (Severe) | Vaginal | No |
| 5 | 16 | M | CD | 1660 | 110 | 65 | >40 (Severe) | Vaginal | No |
| 6 | 9 | M | CD | 1200 | 100 | 40 | >40 (Severe) | Cesarean | No |
| 7 | 13 | M | CD | 1500 | 40 | 30 | 37.5 (Moderate) | Cesarean | Yes |
| 8 | 16 | M | CD | 1200 | 20 | 55 | >40 (Severe) | Cesarean | No |
| 9 | 13.5 | M | UC | 1270 | <3 | 15 | 50 (Moderate) | Vaginal | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dovrolis, N.; Moschoviti, A.; Fessatou, S.; Karamanolis, G.; Kolios, G.; Gazouli, M. Identifying Microbiome Dynamics in Pediatric IBD: More than a Family Matter. Biomedicines 2023, 11, 1979. https://doi.org/10.3390/biomedicines11071979
Dovrolis N, Moschoviti A, Fessatou S, Karamanolis G, Kolios G, Gazouli M. Identifying Microbiome Dynamics in Pediatric IBD: More than a Family Matter. Biomedicines. 2023; 11(7):1979. https://doi.org/10.3390/biomedicines11071979
Chicago/Turabian StyleDovrolis, Nikolas, Anastasia Moschoviti, Smaragdi Fessatou, George Karamanolis, George Kolios, and Maria Gazouli. 2023. "Identifying Microbiome Dynamics in Pediatric IBD: More than a Family Matter" Biomedicines 11, no. 7: 1979. https://doi.org/10.3390/biomedicines11071979






