Characterization of the Outer Membrane Vesicles of Pseudomonas aeruginosa Exhibiting Growth Inhibition against Acinetobacter baumannii
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection of GNBs
2.2. Modified Cross-Streak Assay
2.3. Preparation and Quantification of OMVs
2.4. Transmission Electron Microscope
2.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
2.6. Nanoparticle Tracking Analysis of OMVs
2.7. Modified Time-Kill Assay
2.8. Proteomics
2.8.1. Proteomic Sample Preparation
2.8.2. Mass Spectrometry Analysis
2.8.3. Processing, Quantification, and Statistical Analysis of MS Data
2.9. Statistical Analysis
3. Results
3.1. Interaction of GNBs with P. aeruginosa in Cross-Streak Assay
3.2. OMV Production of PA022 Compared with PA ATCC 25783
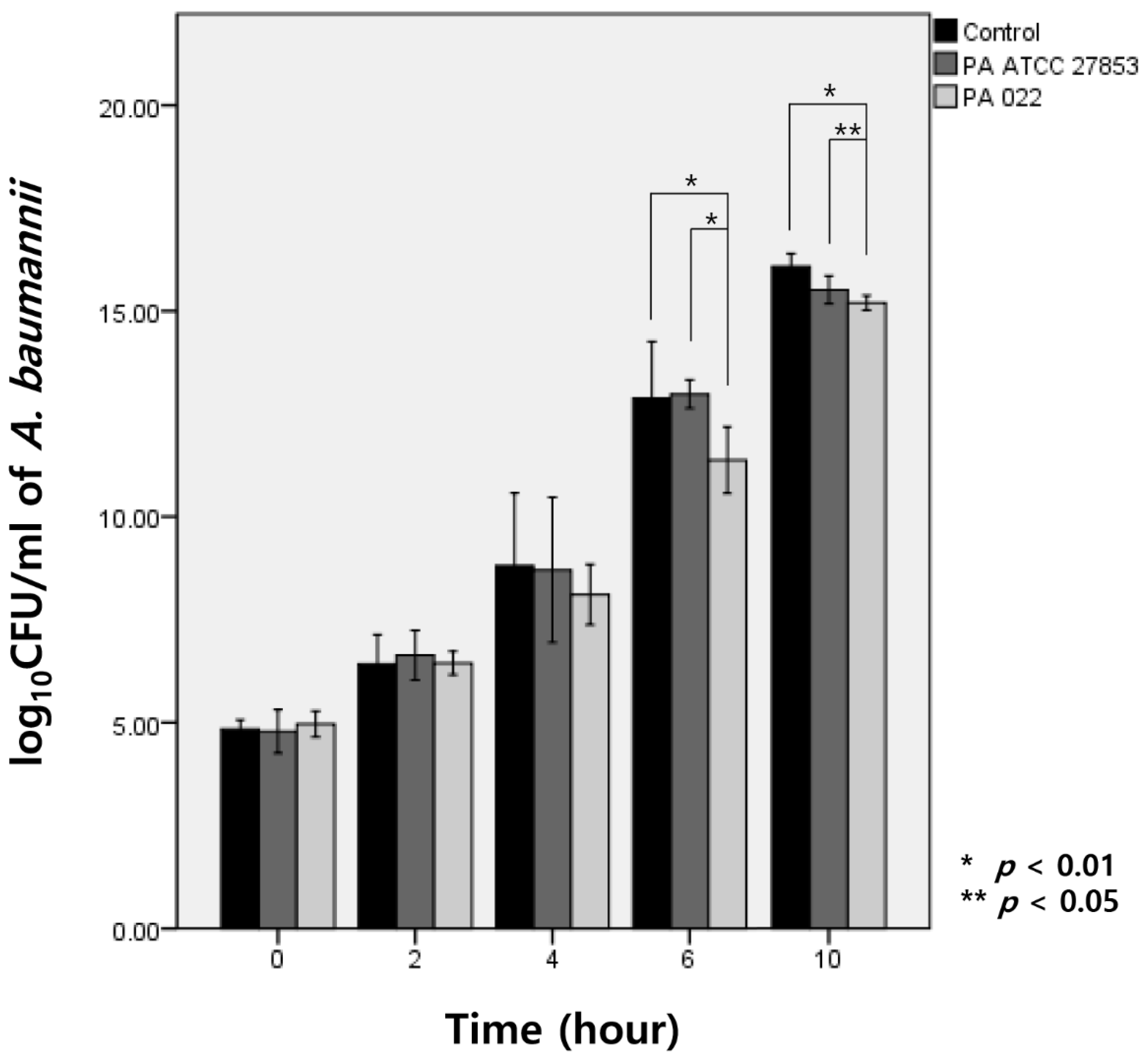
3.3. Modified Time-Kill Assay
3.4. OMV Proteomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Bladen, H.A.; Waters, J.F. Electron microscopic study of some strains of bacteroides. J. Bacteriol. 1963, 86, 1339–1344. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Timothy Prior, J.; Davitt, C.; Kurtz, J.; Gellings, P.; McLachlan, J.B.; Morici, L.A. Bacterial-derived outer membrane vesicles are potent adjuvants that drive humoral and cellular immune responses. Pharmaceutics 2021, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Klimentova, J.; Rehulka, P.; Pavkova, I.; Kubelkova, K.; Bavlovic, J.; Stulik, J. Cross-species proteomic comparison of outer membrane vesicles and membranes of Francisella tularensis subsp. tularensis versus subsp. holarctica. J. Proteome Res. 2021, 20, 1716–1732. [Google Scholar] [CrossRef]
- Higgins, P.G.; Dammhayn, C.; Hackel, M.; Seifert, H. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 2009, 65, 233–238. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Bomberger, J.M.; MacEachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef]
- Hachani, A.; Lossi, N.S.; Hamilton, A.; Jones, C.; Bleves, S.; Albesa-Jové, D.; Filloux, A. Type VI secretion system in Pseudomonas aeruginosa: Secretion and multimerization of VgrG proteins. J. Biol. Chem. 2011, 286, 12317–12327. [Google Scholar] [CrossRef]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef]
- CLSI. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. In CLSI Document M100-S25. Vol. 32, Performance Standards for Antimicrobial Susceptibility Testing, 25th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; pp. 1–184. [Google Scholar]
- Kaur, J.; Pethani, B.P.; Kumar, S.; Kim, M.; Sunna, A.; Kautto, L.; Penesyan, A.; Paulsen, I.T.; Nevalainen, H. Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 2015, 6, 866. [Google Scholar] [CrossRef]
- Mendez, J.A.; Soares, N.C.; Mateos, J.; Gayoso, C.; Rumbo, C.; Aranda, J.; Tomas, M.; Bou, G. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J. Proteome Res. 2012, 11, 5678–5694. [Google Scholar] [CrossRef]
- Burghardt, R.C.; Droleskey, R. Transmission electron microscopy. Curr. Protoc. Microbiol. 2006, 3, 2B-1. [Google Scholar] [CrossRef]
- Gerritzen, M.J.H.; Martens, D.E.; Wijffels, R.H.; Stork, M. High throughput nanoparticle tracking analysis for monitoring outer membrane vesicle production. J. Extracell. Vesicles 2017, 6, 1333883. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hoffmann, J.P.; Baker, S.M.; Bentrup, K.H.Z.; Wimley, W.C.; Fuselier, J.A.; Bitoun, J.P.; Morici, L.A. Inhibition of Streptococcus mutans biofilms with bacterial-derived outer membrane vesicles. BMC Microbiol. 2021, 21, 234. [Google Scholar] [CrossRef]
- Meier, F.; Beck, S.; Grassl, N.; Lubeck, M.; Park, M.A.; Raether, O.; Mann, M. Parallel accumulation-serial fragmentation (PASEF): Multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. J. Proteome Res. 2015, 14, 5378–5387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell Proteom. 2012, 11, 1–8. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Choi, D.S.; Kim, K.P.; Gho, Y.S. Proteomics in gram-negative bacterial outer membrane vesicles. Mass. Spectrom. Rev. 2008, 27, 535–555. [Google Scholar] [CrossRef] [PubMed]
- Database of Clusters of Orthologous Genes (COGs). Available online: http://www.ncbi.nlm.nih.gov/research/cog/ (accessed on 20 January 2024).
- Huang, C.-W.; Lin, Y.-S.; Huang, W.-C.; Lai, C.-C.; Chien, H.-J.; Hu, N.-J.; Chen, J.-H. Inhibition of the clinical isolates of Acinetobacter baumannii by Pseudomonas aeruginosa: In vitro assessment of a case-based study. J. Microbiol. Immunol. Infect. 2020, 55, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Beaume, M.; Köhler, T.; Fontana, T.; Tognon, M.; Renzoni, A.; van Delden, C. Metabolic pathways of Pseudomonas aeruginosa involved in competition with respiratory bacterial pathogens. Front. Microbiol. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, N.; Sharma, P.; Capalash, N. N-acyl homoserine lactone mediated interspecies interactions between A. baumannii and P. aeruginosa. Biofouling 2012, 28, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Kragh, K.N.; Su, Y.C.; Sandblad, L.; Singh, B.; Bjarnsholt, T.; Riesbeck, K. Peptidoglycan-binding anchor is a Pseudomonas aeruginosa OmpA family lipoprotein with importance for outer membrane vesicles, biofilms, and the periplasmic shape. Front. Microbiol. 2021, 12, 639582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, S.Q.; Zhang, J.; Sun, Y.; Xie, Y.L.; Liu, Y.B.; Ma, C.C.; Jiang, B.G.; Liao, X.Y.; Li, W.F.; et al. Proteomic analysis of vesicle-producing Pseudomonas aeruginosa PAO1 exposed to x-ray irradiation. Front. Microbiol. 2020, 11, 558233. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Cañas, M.A.; Giménez, R.; Badia, J.; Mercade, E.; Baldomà, L.; Aguilera, L. Membrane vesicles released by a hypervesiculating Escherichia coli Nissle 1917 tolR mutant are highly heterogeneous and show reduced capacity for epithelial cell interaction and entry. PLoS ONE 2016, 11, e0169186. [Google Scholar] [CrossRef]
- McBroom, A.J.; Kuehn, M.J. Outer membrane vesicles. EcoSal Plus 2005, 1, 10–1128. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Delgado, L.; López-Iglesias, C.; Mercade, E. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS ONE 2015, 10, e0116896. [Google Scholar] [CrossRef]
- Sauvage, S.; Hardouin, J. Exoproteomics for better understanding Pseudomonas aeruginosa virulence. Toxins 2020, 12, 571. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Choi, S.J.; Lee, J.; Choi, J.P.; Rho, S.; Park, S.H.; Kim, Y.K.; Hwang, D.; Gho, Y.S. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 2011, 11, 3424–3429. [Google Scholar] [CrossRef]
- Orange, N. Growth temperature regulates the induction of β-lactamase in Pseudomonas fluorescens through modulation of the outer membrane permeation of a β-lactam-inducing antibiotic. Microbiology 1994, 140, 3125–3130. [Google Scholar] [CrossRef]
- McPhee, J.B.; Tamber, S.; Bains, M.; Maier, E.; Gellatly, S.; Lo, A.; Benz, R.; Hancock, R.E. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol. Lett. 2009, 296, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Mohd Zaki, N.A.; Latif, Z.A.; Suratman, M.N. Study of allometric equations for estimating the above-ground biomass and carbon stock in tropical lowland dipterocarp at Royal Belum Forest reserve. Natl. Conf. R. Belum 2014, 302, 1–13. [Google Scholar]
- Modi, N.; Ganguly, S.; Bárcena-Uribarri, I.; Benz, R.; Van Den Berg, B.; Kleinekathöfer, U. Structure, dynamics, and substrate specificity of the OprO porin from Pseudomonas aeruginosa. Biophys. J. 2015, 109, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Jeon, J.; Lee, J.H.; Jin, S.H.U. Pseudomonas aeruginosa GroEL stimulates production of PTX3 by activating the NF-κB pathway and simultaneously downregulating MicroRNA-9. Infect. Immun. 2017, 85, e00935-16. [Google Scholar] [CrossRef] [PubMed]
- Susanne, J.; Bauman, M.J.K. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006, 8, 2400–2408. [Google Scholar]
- Renelli, M.; Matias, V.; Lo, R.Y.; Beveridge, T.J. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004, 150, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Ichikawa, S.; Shimizu, M.; Toyofuku, M.; Takaya, N.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010, 76, 3732–3739. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.N.; Leiman, S.A.; Kuehn, M.J. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 2010, 78, 3822–3831. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Narasaki, C.T.; Schweizer, H.P. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 2002, 184, 5036–5044. [Google Scholar] [CrossRef]
- Yonehara, R.; Yamashita, E.; Nakagawa, A. Crystal structures of OprN and OprJ, outer membrane factors of multidrug tripartite efflux pumps of Pseudomonas aeruginosa. Proteins Struct. Funct. Bioinforma 2016, 84, 759–769. [Google Scholar] [CrossRef]
- Ntreh, A.T.; Weeks, J.W.; Nickels, L.M.; Zgurskaya, H.I. Opening the channel: The two functional interfaces of Pseudomonas aeruginosa OpmH with the triclosan efflux pump TriABC. J. Bacteriol. 2016, 198, 3176–3185. [Google Scholar] [CrossRef]
- Gaviard, C.; Cosette, P.; Jouenne, T.; Hardouin, J. LasB and CbpD virulence factors of Pseudomonas aeruginosa carry multiple post-translational modifications on their lysine residues. J. Proteome Res. 2019, 18, 923–933. [Google Scholar] [CrossRef]
- Ramel, C.; Baechler, N.; Hildbrand, M.; Meyer, M.; Schädeli, D.; Dudler, R. Regulation of biosynthesis of syringolin A, a Pseudomonas syringae virulence factor targeting the host proteasome. Mol. Plant-Microbe Interact. 2012, 25, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Patel, S. Pathogenicity-associated protein domains: The fiercely-conserved evolutionary signatures. Gene Rep. 2017, 7, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Bardiaux, B.; Francetic, O.; Izadi-Pruneyre, N.; Nilges, M. Structure and function of minor pilins of type IV pili. Med. Microbiol. Immunol. 2020, 209, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Ben Haj Khalifa, A.; Moissenet, D.; Vu Thien, H.K.M. Virulence factors in Pseudomonas aeruginosa: Mechanisms and modes of regulation. Ann. Biol. Clin. 2011, 69, 393–403. [Google Scholar] [CrossRef]
- Palma, V.; Gutiérrez, M.S.; Vargas, O.; Parthasarathy, R.; Navarrete, P. Methods to evaluate bacterial motility and its role in bacterial–host interactions. Microorganisms 2022, 10, 563. [Google Scholar] [CrossRef]
- Booth, S.C.; Rice, S.A. Influence of interspecies interactions on the spatial organization of dual species bacterial communities. Biofilm 2020, 2, 100035. [Google Scholar] [CrossRef] [PubMed]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Ouidir, T.; Jouenne, T.; Hardouin, J. Post-translational modifications in Pseudomonas aeruginosa revolutionized by proteomic analysis. Biochimie 2016, 125, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Ferro, C.; Hentzer, M.; Reil, G.; Görg, A.; Kjelleberg, S.; Givskov, M.; Riedel, K.; Eberl, L. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 2003, 5, 1350–1369. [Google Scholar] [CrossRef]
| UniProt Accession | Protein Name | No. | Average Mass (kDa) | Gene Name | Subcellular Location |
|---|---|---|---|---|---|
| PA ATCC 27853 OMVs | |||||
| A0A8D9JJ04 | Outer membrane protein assembly complex protein | 2 | 87,678.00 | yaeT | Outer membrane |
| A0A246FDS1 | Peptidoglycan-associated protein | 14 | 17,963.58 | pal | Outer membrane |
| A0A069PYF8 | Lipid A deacylase | 4 | 18,399.60 | pagL | Outer membrane |
| A0A8D9JLC9 | Lipid A 3-O-deacylase | 3 | 18,394.00 | pagL | Outer membrane |
| A0A8H2EWC8 | Multidrug efflux RND transporter outer membrane protein | 24 | 51,163.11 | oprN | Outer membrane |
| I1SY51 | Peptidoglycan-associated lipoprotein | 7 | 16,841.40 | oprL | Outer membrane |
| A0A8G3E6L1 | Transport protein | 3 | 45,130.33 | IPC1518_14570 | Outer membrane |
| A0A0C6ELC4 | Long-chain fatty acid transport protein | 3 | 45,566.67 | fadL1 | Outer membrane |
| A0A8G2ZED3 | Outer membrane protein assembly factor protein | 11 | 88,435.73 | bamA | Outer membrane |
| A0A8G5T8N0 | TolC family protein | 1 | 47,611.00 | IPC183_04225 | Outer membrane |
| A0A2R3J2M9 | Efflux transporter outer membrane factor lipo NodT family protein | 1 | 51,290.00 | CSB93_4029 | Outer membrane |
| A0A241XQD4 | RND transport protein | 1 | 51,161.00 | CAZ10_13505 | outer membrane |
| A0A069Q437 | Outer membrane protein transport protein | 3 | 45,562.00 | ALP65_01642 | Outer membrane |
| A0A0A8RDW0 | Porin D | 1 | 46,900 | oprD3 | Outer membrane |
| A0A8G6ZVZ6 | Phage tail protein | 2 | 72,190.00 | IPC65_03235 | Extracellular |
| PA 022 OMVs | |||||
| A0A127MNT2 | Glycine zipper 2TM domain-containing protein | 7 | 15,660.67 | slyB_1 | Outer membrane |
| A0A1C7BTT8 | Rick_17kDa_Anti domain-containing protein | 5 | 15,649.00 | Q058_03352 | Outer membrane |
| A0A1C7BQ09 | TonB-dependent copper receptor | 26 | 79,249.31 | Q058_00426 | Outer membrane |
| A6VD89 | Outer membrane efflux protein | 4 | 53,298.00 | PSPA7_5705 | Outer membrane |
| A6V9E3 | Lipoprotein | 5 | 14,798.00 | PSPA7_4325 | Outer membrane |
| A0A1C7BA19 | Lipid A deacylase | 7 | 18,403.86 | pagL | Outer membrane |
| Q51487 | Outer membrane protein OprM | 28 | 52,607.55 | oprM | Outer membrane |
| V6ALH3 | Outer membrane protein oprJ | 12 | 51,955.83 | oprJ | Outer membrane |
| V5UUX5 | Outer membrane porin protein OprD | 10 | 47,550.67 | oprD | Outer membrane |
| A0A8D9JM75 | Copper transport outer membrane porin OprC | 5 | 78,644.00 | oprC | Outer membrane |
| G3XCK3 | Channel protein TolC | 21 | 53,345.61 | opmH | Outer membrane |
| A0A8G3VVE4 | YgdI/YgdR family lipoprotein | 2 | 8095.00 | IPC1338_27820 | Outer membrane |
| A0A072ZKU5 | 17 kDa surface antigen | 1 | 15,649.00 | slyB | Outer membrane |
| A6V457 | Putative outer membrane protein | 1 | 46,879.00 | PSPA7_2476 | Outer membrane |
| A0A8G1KGY7 | YadA-like family protein | 1 | 90,268.00 | K3T18_31340 | Outer membrane |
| A0A1B1SN37 | Pilin | 3 | 15,958.33 | pilA | Extracellular |
| V6AAB7 | Elastase | 19 | 51,771.00 | lasB | Extracellular |
| A7LI11 | Peptidase M4 family protein | 10 | 53,672.10 | IPC113_02740 | Extracellular |
| Q9KW03 | Phage tail protein | 17 | 68,141.85 | hr1 | Extracellular |
| A6V997 | Flagellin | 30 | 31,929.25 | fliC | Extracellular |
| A0A8G3YS22 | Pseudolysin | 1 | 53,687.00 | Q041_00565 | Extracellular |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suh, J.-W.; Kang, J.-S.; Kim, J.-Y.; Kim, S.-B.; Yoon, Y.-K.; Sohn, J.-W. Characterization of the Outer Membrane Vesicles of Pseudomonas aeruginosa Exhibiting Growth Inhibition against Acinetobacter baumannii. Biomedicines 2024, 12, 556. https://doi.org/10.3390/biomedicines12030556
Suh J-W, Kang J-S, Kim J-Y, Kim S-B, Yoon Y-K, Sohn J-W. Characterization of the Outer Membrane Vesicles of Pseudomonas aeruginosa Exhibiting Growth Inhibition against Acinetobacter baumannii. Biomedicines. 2024; 12(3):556. https://doi.org/10.3390/biomedicines12030556
Chicago/Turabian StyleSuh, Jin-Woong, Jae-Seong Kang, Jeong-Yeon Kim, Sun-Bean Kim, Young-Kyung Yoon, and Jang-Wook Sohn. 2024. "Characterization of the Outer Membrane Vesicles of Pseudomonas aeruginosa Exhibiting Growth Inhibition against Acinetobacter baumannii" Biomedicines 12, no. 3: 556. https://doi.org/10.3390/biomedicines12030556