R-α-Lipoic Acid and 4-Phenylbutyric Acid Have Distinct Hypolipidemic Mechanisms in Hepatic Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Trichloroacetic Acid (TCA) Precipitation of Proteins Secreted in Conditioned Media
2.4. Whole Cell Lysate and Nuclear Fraction
2.5. Western Blotting
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Chromatin Immunoprecipitation (ChIP) Assay
2.8. Statistical Analysis
3. Results
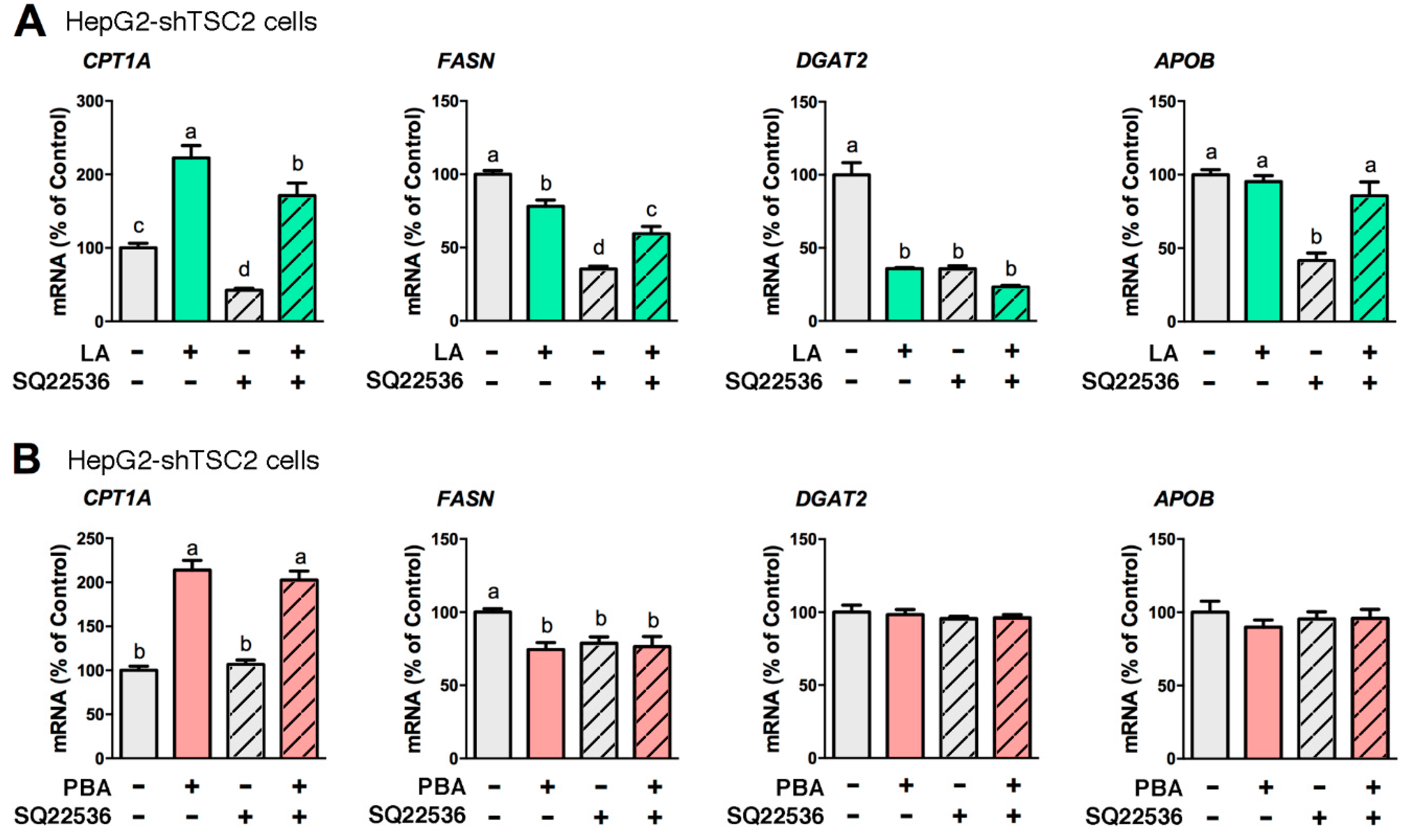
3.1. LA and PBA Regulate Lipid Metabolism-Related Genes Irrespective of mTORC1 Activity Level
3.2. cAMP Mediates the Lipid-Lowering Action of LA
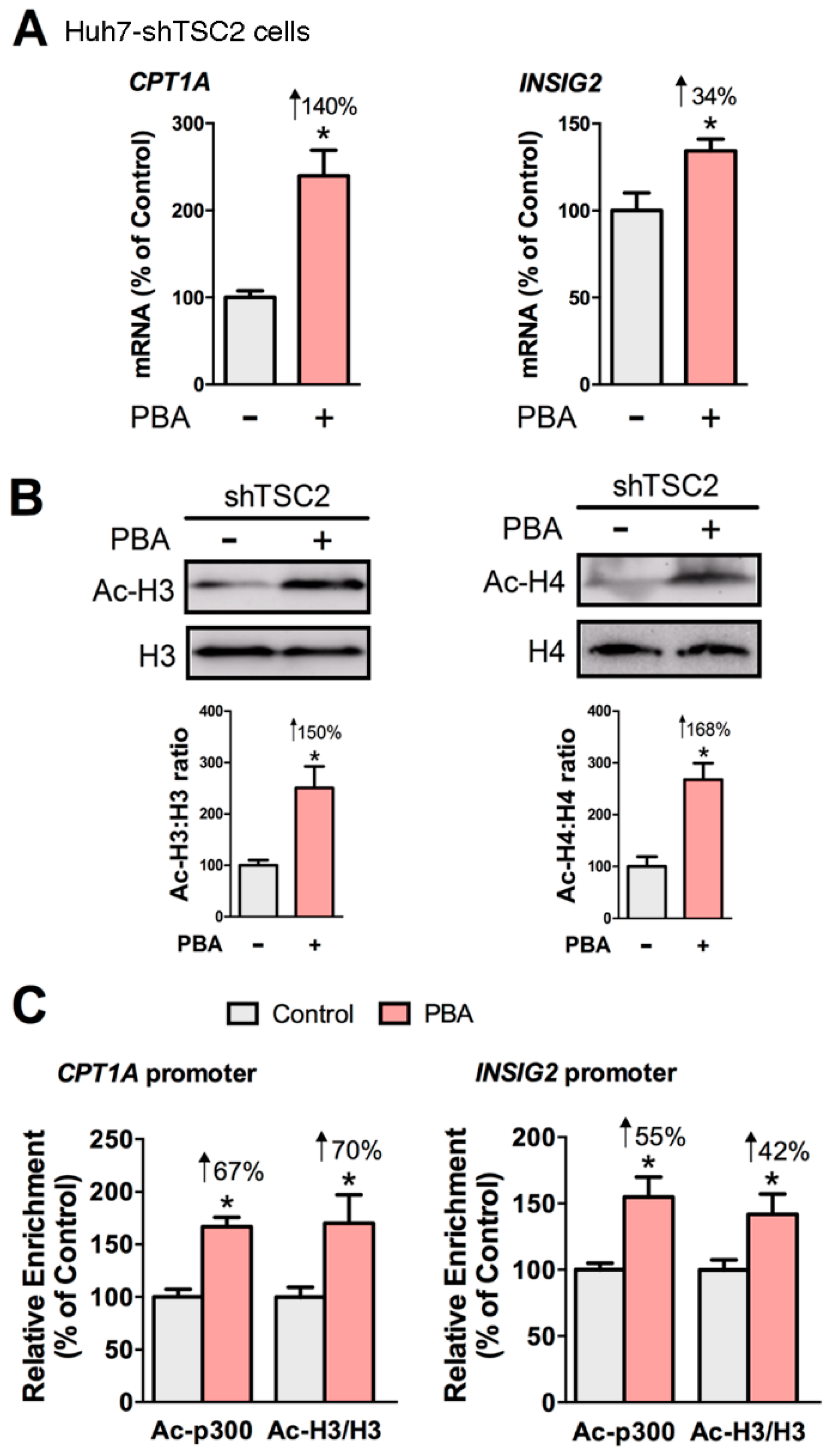
3.3. PBA Acts as HDAC Inhibitor to Drive CPT1A and INSIG2 Expression
3.4. LA, But Not PBA, Suppresses PCSK9 Secretion
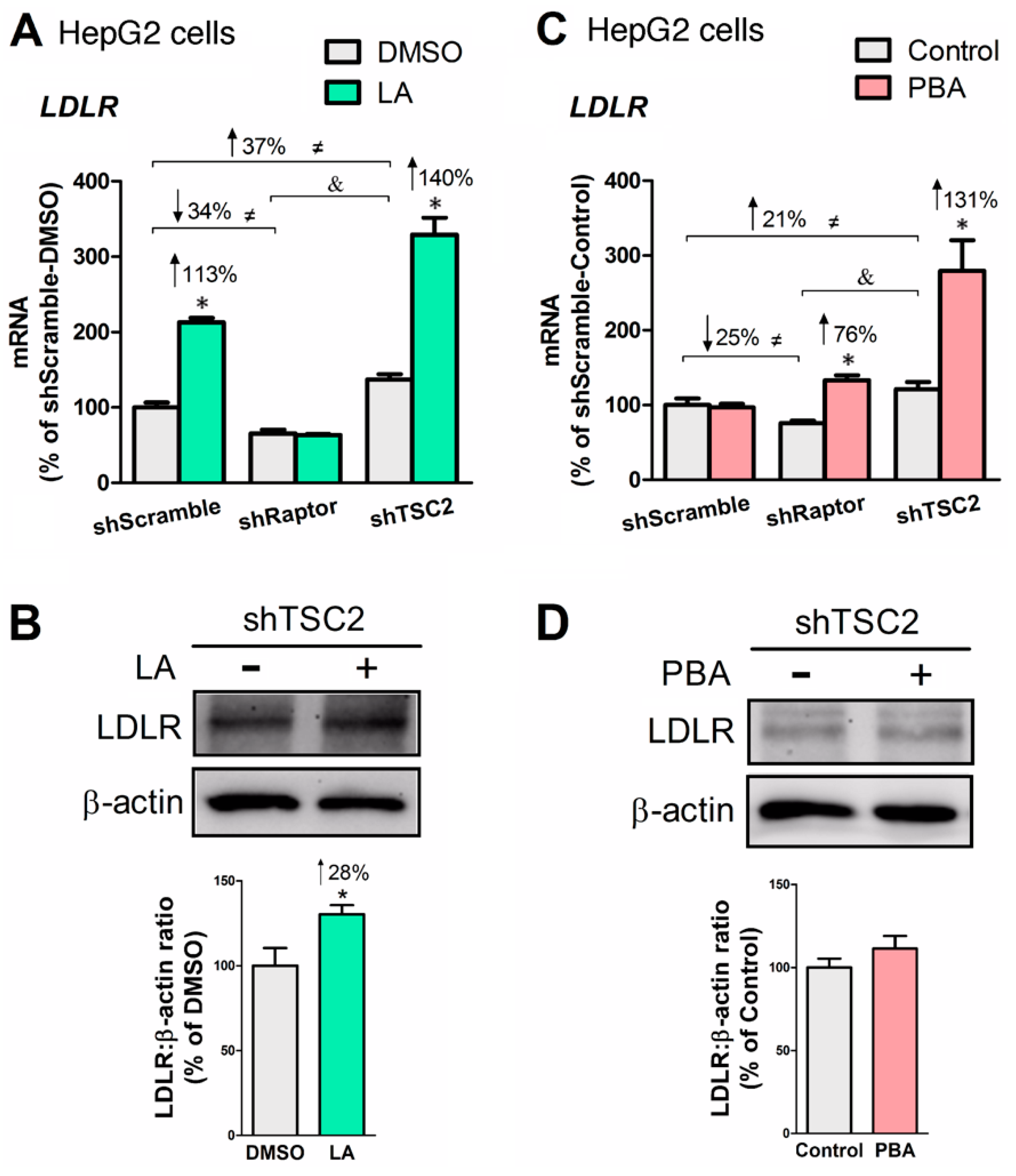
3.5. LA or PBA Induces LDLR Gene Expression, But Only LA Upregulates LDLR Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Morrison, A.; Hokanson, J.E. The independent relationship between triglycerides and coronary heart disease. Vasc. Health Risk Manag. 2009, 5, 89–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manninen, V.; Tenkanen, L.; Koskinen, P.; Huttunen, J.K.; Manttari, M.; Heinonen, O.P.; Frick, M.H. Joint effects of serum triglyceride and LDL cholesterol and HDL cholesterol concentrations on coronary heart disease risk in the Helsinki Heart Study. Implications for treatment. Circulation 1992, 85, 37–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarwar, N.; Danesh, J.; Eiriksdottir, G.; Sigurdsson, G.; Wareham, N.; Bingham, S.; Boekholdt, S.M.; Khaw, K.T.; Gudnason, V. Triglycerides and the risk of coronary heart disease: 10,158 incident cases among 262,525 participants in 29 Western prospective studies. Circulation 2007, 115, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Ghandehari, H.; Le, V.; Kamal-Bahl, S.; Bassin, S.L.; Wong, N.D. Abdominal obesity and the spectrum of global cardiometabolic risks in US adults. Int. J. Obes. (Lond.) 2009, 33, 239–248. [Google Scholar] [CrossRef] [Green Version]
- NCEP, Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). Jama 2001, 285, 2486–2497. [CrossRef]
- Adiels, M.; Olofsson, S.O.; Taskinen, M.R.; Boren, J. Overproduction of very low-density lipoproteins is the hallmark of the dyslipidemia in the metabolic syndrome. Arter. Thromb. Vasc. Biol. 2008, 28, 1225–1236. [Google Scholar] [CrossRef]
- Herbert, A.A.; Guest, J.R. Lipoic acid content of Escherichia coli and other microorganisms. Arch. Microbiol. 1975, 106, 259–266. [Google Scholar] [CrossRef]
- Yasuno, R.; Wada, H. The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. Febs Lett. 2002, 517, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Breithaupt-Grogler, K.; Niebch, G.; Schneider, E.; Erb, K.; Hermann, R.; Blume, H.H.; Schug, B.S.; Belz, G.G. Dose-proportionality of oral thioctic acid--coincidence of assessments via pooled plasma and individual data. Eur. J. Pharm. Sci. 1999, 8, 57–65. [Google Scholar] [CrossRef]
- Carlson, D.A.; Smith, A.R.; Fischer, S.J.; Young, K.L.; Packer, L. The plasma pharmacokinetics of R-(+)-lipoic acid administered as sodium R-(+)-lipoate to healthy human subjects. Altern. Med. Rev. 2007, 12, 343–351. [Google Scholar]
- Shay, K.P.; Moreau, R.F.; Smith, E.J.; Smith, A.R.; Hagen, T.M. Alpha-lipoic acid as a dietary supplement: Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta 2009, 1790, 1149–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, D. Thioctic acid for patients with symptomatic diabetic polyneuropathy: A critical review. Treat. Endocrinol. 2004, 3, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.V.; Gille, L.; Staniek, K.; Nohl, H. Dihydrolipoic acid maintains ubiquinone in the antioxidant active form by two-electron reduction of ubiquinone and one-electron reduction of ubisemiquinone. Arch. Biochem. Biophys. 1999, 363, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Ansar, H.; Mazloom, Z.; Kazemi, F.; Hejazi, N. Effect of alpha-lipoic acid on blood glucose, insulin resistance and glutathione peroxidase of type 2 diabetic patients. Saudi Med. J. 2011, 32, 584–588. [Google Scholar]
- Heinisch, B.B.; Francesconi, M.; Mittermayer, F.; Schaller, G.; Gouya, G.; Wolzt, M.; Pleiner, J. Alpha-lipoic acid improves vascular endothelial function in patients with type 2 diabetes: A placebo-controlled randomized trial. Eur. J. Clin. Investig. 2010, 40, 148–154. [Google Scholar] [CrossRef]
- Pashaj, A.; Xia, M.; Moreau, R. α-Lipoic acid as a triglyceride-lowering nutraceutical. Can. J. Physiol. Pharmacol. 2015, 93, 1029–1041. [Google Scholar] [CrossRef]
- Huong, D.T.; Ide, T. Dietary lipoic acid-dependent changes in the activity and mRNA levels of hepatic lipogenic enzymes in rats. Br. J. Nutr. 2008, 100, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.L.; Kang, C.H.; Wang, S.G.; Lee, H.M. alpha-Lipoic acid regulates lipid metabolism through induction of sirtuin 1 (SIRT1) and activation of AMP-activated protein kinase. Diabetologia 2012, 55, 1824–1835. [Google Scholar] [CrossRef] [Green Version]
- Valdecantos, M.P.; Perez-Matute, P.; Gonzalez-Muniesa, P.; Prieto-Hontoria, P.L.; Moreno-Aliaga, M.J.; Martinez, J.A. Lipoic acid administration prevents nonalcoholic steatosis linked to long-term high-fat feeding by modulating mitochondrial function. J. Nutr. Biochem. 2012, 23, 1676–1684. [Google Scholar] [CrossRef]
- Yi, X.; Pashaj, A.; Xia, M.; Moreau, R. Reversal of obesity-induced hypertriglyceridemia by (R)-α-lipoic acid in ZDF (fa/fa) rats. Biochem. Biophys. Res. Commun. 2013, 439, 390–395. [Google Scholar] [CrossRef]
- Pashaj, A.; Yi, X.; Xia, M.; Canny, S.; Riethoven, J.J.; Moreau, R. Characterization of genome-wide transcriptional changes in liver and adipose tissues of ZDF (fa/fa) rats fed R-alpha-lipoic acid by next-generation sequencing. Physiol. Genom. 2013, 45, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.A.; Hagen, T.M.; Moreau, R. Lipoic acid improves hypertriglyceridemia by stimulating triacylglycerol clearance and downregulating liver triacylglycerol secretion. Arch. Biochem. Biophys. 2009, 485, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.-S.; Park, J.-Y.; Namkoong, C.; Jang, P.-G.; Ryu, J.-W.; Song, H.-S.; Yun, J.-Y.; Namgoong, I.-S.; Ha, J.; Park, I.-S.; et al. Anti-obesity effects of alpha-lipoic acid mediated by suppression of hypothalamic AMP-activated protein kinase. Nat. Med. 2004, 10, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Lee, I.K.; Kim, H.S.; Kim, Y.M.; Koh, E.H.; Won, J.C.; Han, S.M.; Kim, M.S.; Jo, I.; Oh, G.T.; et al. Alpha-lipoic acid prevents endothelial dysfunction in obese rats via activation of AMP-activated protein kinase. Arter. Thromb. Vasc. Biol. 2005, 25, 2488–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, X.; Christian, P.; Zhao, M.; Wang, H.; Moreau, R.; Su, Q. Activation of hepatic CREBH and Insig signaling in the anti-hypertriglyceridemic mechanism of R-alpha-lipoic acid. J. Nutr. Biochem. 2015, 26, 921–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Erickson, A.; Yi, X.; Moreau, R. Mapping the response of human fibroblast growth factor 21 (FGF21) promoter to serum availability and lipoic acid in HepG2 hepatoma cells. Biochim. Biophys. Acta 2016, 1860, 498–507. [Google Scholar] [CrossRef]
- Salinthone, S.; Schillace, R.V.; Marracci, G.H.; Bourdette, D.N.; Carr, D.W. Lipoic acid stimulates cAMP production via the EP2 and EP4 prostanoid receptors and inhibits IFN gamma synthesis and cellular cytotoxicity in NK cells. J. Neuroimmunol. 2008, 199, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Maestri, N.E.; Brusilow, S.W.; Clissold, D.B.; Bassett, S.S. Long-term treatment of girls with ornithine transcarbamylase deficiency. N. Engl. J. Med. 1996, 335, 855–859. [Google Scholar] [CrossRef]
- Yu, K.H.; Weng, L.J.; Fu, S.; Piantadosi, S.; Gore, S.D. Augmentation of phenylbutyrate-induced differentiation of myeloid leukemia cells using all-trans retinoic acid. Leukemia 1999, 13, 1258–1265. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Ding, S.; Li, Y.; Wang, L.; Chen, W.; Bo, T.; Wu, K.; Li, C.; Liu, X.; Zhao, J.; et al. Endoplasmic Reticulum Stress May Play a Pivotal Role in Lipid Metabolic Disorders in a Novel Mouse Model of Subclinical Hypothyroidism. Sci. Rep. 2016, 6, 31381. [Google Scholar] [CrossRef]
- Ren, L.P.; Song, G.Y.; Hu, Z.J.; Zhang, M.; Peng, L.; Chen, S.C.; Wei, L.; Li, F.; Sun, W. The chemical chaperon 4-phenylbutyric acid ameliorates hepatic steatosis through inhibition of de novo lipogenesis in high-fructose-fed rats. Int. J. Mol. Med. 2013, 32, 1029–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.T.; Meng, M.; Zhang, J.C.; Jin, C.J.; Jiang, J.J.; Ren, H.S.; Jiang, J.M.; Qin, C.Y.; Yu, D.Q. Growth inhibition and gene induction in human hepatocellular carcinoma cell exposed to sodium 4-phenylbutanoate. Chin. Med. J. (Engl.) 2008, 121, 1707–1711. [Google Scholar] [CrossRef]
- Svechnikova, I.; Almqvist, P.M.; Ekstrom, T.J. HDAC inhibitors effectively induce cell type-specific differentiation in human glioblastoma cell lines of different origin. Int. J. Oncol. 2008, 32, 821–827. [Google Scholar] [PubMed] [Green Version]
- Kusaczuk, M.; Kretowski, R.; Bartoszewicz, M.; Cechowska-Pasko, M. Phenylbutyrate-a pan-HDAC inhibitor-suppresses proliferation of glioblastoma LN-229 cell line. Tumour Biol. 2016, 37, 931–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Li, L.; Wang, C.; He, H.; Mao, K.; Ma, X.; Shi, R.; Oh, Y.; Zhang, F.; Lu, Y.; et al. 4-Phenylbutyric acid increases GLUT4 gene expression through suppression of HDAC5 but not endoplasmic reticulum stress. Cell Physiol. Biochem. 2014, 33, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Moreau, R. Lipid-regulating properties of butyric acid and 4-phenylbutyric acid: Molecular mechanisms and therapeutic applications. Pharm. Res. 2019, 144, 116–131. [Google Scholar] [CrossRef] [Green Version]
- Allard, D.; Amsellem, S.; Abifadel, M.; Trillard, M.; Devillers, M.; Luc, G.; Krempf, M.; Reznik, Y.; Girardet, J.P.; Fredenrich, A.; et al. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum. Mutat. 2005, 26, 497. [Google Scholar] [CrossRef]
- Berge, K.E.; Ose, L.; Leren, T.P. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arter. Thromb. Vasc. Biol. 2006, 26, 1094–1100. [Google Scholar] [CrossRef] [Green Version]
- Carrier, B.; Wen, S.; Zigouras, S.; Browne, R.W.; Li, Z.; Patel, M.S.; Williamson, D.L.; Rideout, T.C. Alpha-lipoic acid reduces LDL-particle number and PCSK9 concentrations in high-fat fed obese Zucker rats. PLoS ONE 2014, 9, e90863. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.L.; He, B.; Erickson, A.; Moreau, R. Improvement of mTORC1-driven overproduction of apoB-containing triacylglyceride-rich lipoproteins by short-chain fatty acids, 4-phenylbutyric acid and (R)-alpha-lipoic acid, in human hepatocellular carcinoma cells. Biochim. Biophys. Acta 2016, 1861, 166–176. [Google Scholar] [CrossRef]
- Inoki, K.; Corradetti, M.N.; Guan, K.L. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 2005, 37, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, J.L.; Zhang, Y.; Bae, S.H.; Farooqi, M.S.; Liang, G.; Hammer, R.E.; Goldstein, J.L.; Brown, M.S. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. USA 2012, 109, 16184–16189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [Green Version]
- Kenerson, H.L.; Yeh, M.M.; Yeung, R.S. Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation. PLoS ONE 2011, 6, e18075. [Google Scholar] [CrossRef]
- Yecies, J.L.; Zhang, H.H.; Menon, S.; Liu, S.; Yecies, D.; Lipovsky, A.I.; Gorgun, C.; Kwiatkowski, D.J.; Hotamisligil, G.S.; Lee, C.H.; et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011, 14, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [Green Version]
- Vander Haar, E.; Lee, S.I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef]
- Thompson, P.R.; Wang, D.; Wang, L.; Fulco, M.; Pediconi, N.; Zhang, D.; An, W.; Ge, Q.; Roeder, R.G.; Wong, J.; et al. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004, 11, 308–315. [Google Scholar] [CrossRef]
- Goldberg, I.J. Clinical review 124: Diabetic dyslipidemia: Causes and consequences. J. Clin. Endocrinol. Metab. 2001, 86, 965–971. [Google Scholar] [CrossRef]
- Lewis, G.F.; Carpentier, A.; Adeli, K.; Giacca, A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr. Rev. 2002, 23, 201–229. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Uffelman, K.; Naples, M.; Szeto, L.; Haidari, M.; Adeli, K. Intestinal lipoprotein overproduction, a newly recognized component of insulin resistance, is ameliorated by the insulin sensitizer rosiglitazone: Studies in the fructose-fed Syrian golden hamster. Endocrinology 2005, 146, 247–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravnskjaer, K.; Madiraju, A.; Montminy, M. Role of the cAMP Pathway in Glucose and Lipid Metabolism. Handb. Exp. Pharm. 2016, 233, 29–49. [Google Scholar]
- Brown, K.M.; Lee, L.C.; Findlay, J.E.; Day, J.P.; Baillie, G.S. Cyclic AMP-specific phosphodiesterase, PDE8A1, is activated by protein kinase A-mediated phosphorylation. Febs Lett. 2012, 586, 1631–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Munoz, M.D.; Osma-Garcia, I.C.; Fresno, M.; Iniguez, M.A. Involvement of PGE2 and the cAMP signalling pathway in the up-regulation of COX-2 and mPGES-1 expression in LPS-activated macrophages. Biochem. J. 2012, 443, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Viollet, B.; Foretz, M.; Guigas, B.; Horman, S.; Dentin, R.; Bertrand, L.; Hue, L.; Andreelli, F. Activation of AMP-activated protein kinase in the liver: A new strategy for the management of metabolic hepatic disorders. J. Physiol. 2006, 574, 41–53. [Google Scholar] [CrossRef]
- Postic, C.; Girard, J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: Lessons from genetically engineered mice. J. Clin. Investig. 2008, 118, 829–838. [Google Scholar] [CrossRef] [Green Version]
- Ruderman, N.; Prentki, M. AMP kinase and malonyl-CoA: Targets for therapy of the metabolic syndrome. Nat. Rev. Drug Discov. 2004, 3, 340–351. [Google Scholar] [CrossRef]
- Chatelain, F.; Kohl, C.; Esser, V.; McGarry, J.D.; Girard, J.; Pegorier, J.P. Cyclic AMP and fatty acids increase carnitine palmitoyltransferase I gene transcription in cultured fetal rat hepatocytes. Eur. J. Biochem. 1996, 235, 789–798. [Google Scholar] [CrossRef]
- Fox, K.E.; Fankell, D.M.; Erickson, P.F.; Majka, S.M.; Crossno, J.T., Jr.; Klemm, D.J. Depletion of cAMP-response element-binding protein/ATF1 inhibits adipogenic conversion of 3T3-L1 cells ectopically expressing CCAAT/enhancer-binding protein (C/EBP) alpha, C/EBP beta, or PPAR gamma 2. J. Biol. Chem. 2006, 281, 40341–40353. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar] [PubMed]
- Zhang, J.W.; Klemm, D.J.; Vinson, C.; Lane, M.D. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 2004, 279, 4471–4478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antony, N.; Weir, J.R.; McDougall, A.R.; Mantamadiotis, T.; Meikle, P.J.; Cole, T.J.; Bird, A.D. cAMP response element binding protein1 is essential for activation of steroyl co-enzyme a desaturase 1 (Scd1) in mouse lung type II epithelial cells. PLoS ONE 2013, 8, e59763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erion, D.M.; Ignatova, I.D.; Yonemitsu, S.; Nagai, Y.; Chatterjee, P.; Weismann, D.; Hsiao, J.J.; Zhang, D.; Iwasaki, T.; Stark, R.; et al. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metab. 2009, 10, 499–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.J.; Moon, H.E.; Moini, H.; Packer, L.; Yoon, D.Y.; Chung, A.S. Alpha-lipoic acid inhibits adipocyte differentiation by regulating pro-adipogenic transcription factors via mitogen-activated protein kinase pathways. J. Biol. Chem. 2003, 278, 34823–34833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahm, J.R.; Noh, H.S.; Ha, J.H.; Roh, G.S.; Kim, D.R. Alpha-lipoic acid attenuates adipocyte differentiation and lipid accumulation in 3T3-L1 cells via AMPK-dependent autophagy. Life Sci. 2014, 100, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Eberharter, A.; Becker, P.B. Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. Embo Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [Green Version]
- Feng, D.; Liu, T.; Sun, Z.; Bugge, A.; Mullican, S.E.; Alenghat, T.; Liu, X.S.; Lazar, M.A. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011, 331, 1315–1319. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Peng, C.; Ai, Y.; Wang, H.; Xiao, X.; Li, J. Docosahexaenoic Acid Ameliorates Fructose-Induced Hepatic Steatosis Involving ER Stress Response in Primary Mouse Hepatocytes. Nutrients 2016, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Samid, D.; Wells, M.; Greene, M.E.; Shen, W.; Palmer, C.N.; Thibault, A. Peroxisome proliferator-activated receptor gamma as a novel target in cancer therapy: Binding and activation by an aromatic fatty acid with clinical antitumor activity. Clin. Cancer Res. 2000, 6, 933–941. [Google Scholar]
- Liu, J.; Ma, K.L.; Zhang, Y.; Wu, Y.; Hu, Z.B.; Lv, L.L.; Tang, R.N.; Liu, H.; Ruan, X.Z.; Liu, B.C. Activation of mTORC1 disrupted LDL receptor pathway: A potential new mechanism for the progression of non-alcoholic fatty liver disease. Int. J. Biochem. Cell Biol. 2015, 61, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Costet, P.; Cariou, B.; Lambert, G.; Lalanne, F.; Lardeux, B.; Jarnoux, A.L.; Grefhorst, A.; Staels, B.; Krempf, M. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J. Biol. Chem. 2006, 281, 6211–6218. [Google Scholar] [CrossRef] [Green Version]
- Tveten, K.; Holla, O.L.; Ranheim, T.; Berge, K.E.; Leren, T.P.; Kulseth, M.A. 4-Phenylbutyrate restores the functionality of a misfolded mutant low-density lipoprotein receptor. Febs J. 2007, 274, 1881–1893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, P.; Wu, N.; He, B.; Lu, Y.; Li, S.; Liu, Y.; Zhao, S.; Liu, L.; Li, Y. Amelioration of lipid abnormalities by alpha-lipoic acid through antioxidative and anti-inflammatory effects. Obesity (Silver Spring) 2011, 19, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Okanovic, A.; Prnjavorac, B.; Jusufovic, E.; Sejdinovic, R. Alpha-lipoic acid reduces body weight and regulates triglycerides in obese patients with diabetes mellitus. Med. Glas. (Zenica) 2015, 12, 122–127. [Google Scholar] [PubMed]
- Bobe, G.; Michels, A.J.; Zhang, W.J.; Purnell, J.Q.; Woffendin, C.; Pereira, C.; Vita, J.A.; Thomas, N.O.; Traber, M.G.; Frei, B.; et al. A Randomized Controlled Trial of Long-Term (R)-alpha-Lipoic Acid Supplementation Promotes Weight Loss in Overweight or Obese Adults without Altering Baseline Elevated Plasma Triglyceride Concentrations. J. Nutr. 2020. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, B.; Moreau, R. R-α-Lipoic Acid and 4-Phenylbutyric Acid Have Distinct Hypolipidemic Mechanisms in Hepatic Cells. Biomedicines 2020, 8, 289. https://doi.org/10.3390/biomedicines8080289
He B, Moreau R. R-α-Lipoic Acid and 4-Phenylbutyric Acid Have Distinct Hypolipidemic Mechanisms in Hepatic Cells. Biomedicines. 2020; 8(8):289. https://doi.org/10.3390/biomedicines8080289
Chicago/Turabian StyleHe, Bo, and Régis Moreau. 2020. "R-α-Lipoic Acid and 4-Phenylbutyric Acid Have Distinct Hypolipidemic Mechanisms in Hepatic Cells" Biomedicines 8, no. 8: 289. https://doi.org/10.3390/biomedicines8080289




