Reliability of IL-6 Alone and in Combination for Diagnosis of Late Onset Sepsis: A Systematic Review
Abstract
:1. Introduction
2. Material and Methods
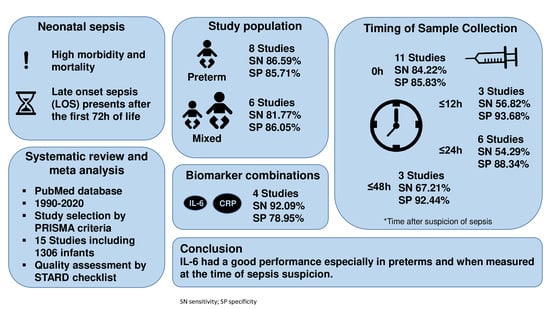
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, Y.; Speer, C.P. Late-onset neonatal sepsis: Recent developments. Arch. Dis. Child. Fetal Neonatal Ed. 2015, 100, F257–F263. [Google Scholar] [CrossRef] [PubMed]
- Raynor, L.L.; Saucerman, J.J.; Akinola, M.O.; Lake, D.E.; Moorman, J.R.; Fairchild, K.D. Cytokine screening identifies NICU patients with Gram-negative bacteremia. Pediatr. Res. 2012, 71, 261–266. [Google Scholar] [CrossRef]
- Verboon-Maciolek, M.A.; Thijsen, S.F.; Hemels, M.A.; Menses, M.; van Loon, A.M.; Krediet, T.G.; Gerards, L.J.; Fleer, A.; Voorbij, H.A.M.; Rijkers, G.T. Inflammatory mediators for the diagnosis and treatment of sepsis in early infancy. Pediatr. Res. 2006, 59, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Korang, S.K.; Safi, S.; Nava, C.; Greisen, G.; Gupta, M.; Lausten-Thomsen, U.; Jakobsen, J.C. Antibiotic regimens for late-onset neonatal sepsis. Cochrane Database Syst. Rev. 2021, 5, Cd013836. [Google Scholar] [PubMed]
- Bakhuizen, S.E.; de Haan, T.R.; Teune, M.J.; van Wassenaer-Leemhuis, A.G.; van der Heyden, J.L.; van der Ham, D.P.; Mol, B.W.J. Meta-analysis shows that infants who have suffered neonatal sepsis face an increased risk of mortality and severe complications. Acta Paediatr. 2014, 103, 1211–1218. [Google Scholar] [CrossRef]
- Silveira, R.C.; Procianoy, R.S. Evaluation of interleukin-6, tumour necrosis factor-alpha and interleukin-1beta for early diagnosis of neonatal sepsis. Acta Paediatr. 1999, 88, 647–650. [Google Scholar] [PubMed]
- Kurul, Ş.; Simons, S.H.; Ramakers, C.R.B.; De Rijke, Y.B.; Kornelisse, R.F.; Reiss, I.K.M.; Taal, H.R. Association of inflammatory biomarkers with subsequent clinical course in suspected late onset sepsis in preterm neonates. Crit. Care. 2021, 25, 12. [Google Scholar] [CrossRef] [PubMed]
- Gilfillan, M.; Bhandari, V. Biomarkers for the diagnosis of neonatal sepsis and necrotizing enterocolitis: Clinical practice guidelines. Early Hum. Dev. 2017, 105, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Küster, H.; Weiss, M.; Willeitner, A.E.; Detlefsen, S.; Jeremias, I.; Zbojan, J.; Geiger, R.; Lipowsky, G.; Simbruner, G. Interleukin-1 receptor antagonist and interleukin-6 for early diagnosis of neonatal sepsis 2 days before clinical manifestation. Lancet 1998, 352, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Cheng, S.H.; Chui, K.M.; Fok, T.F.; Wong, M.Y.; Wong, W.; Wong, R.P.; Cheung, K.L. Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C-reactive protein in preterm very low birthweight infants. Arch. Dis. Child. Fetal Neonatal Ed. 1997, 77, F221–F227. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, L.; Tong, Y.; Qu, Y.; Wang, H.; Mu, D. Interleukin-6 for early diagnosis of neonatal sepsis with premature rupture of the membranes: A meta-analysis. Medicine 2018, 97, e13146. [Google Scholar] [CrossRef]
- Chiesa, C.; Pacifico, L.; Natale, F.; Hofer, N.; Osborn, J.F.; Resch, B. Fetal and early neonatal interleukin-6 response. Cytokine 2015, 76, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.; Rutjes, A.W.; Reitsma, J.B.; Bossuyt, P.M.; Kleijnen, J. The development of QUADAS: A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med. Res. Methodol. 2003, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Rosman, A.S.; Korsten, M.A. Application of summary receiver operating characteristics (sROC) analysis to diagnostic clinical testing. Adv. Med. Sci. 2007, 52, 76–82. [Google Scholar]
- Lee, J.; Kim, K.W.; Choi, S.H.; Huh, J.; Park, S.H. Systematic Review and Meta-Analysis of Studies Evaluating Diagnostic Test Accuracy: A Practical Review for Clinical Researchers-Part II. Statistical Methods of Meta-Analysis. Korean J Radiol. 2015, 16, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Hotoura, E.; Giapros, V.; Kostoula, A.; Spyrou, P.; Andronikou, S. Pre-inflammatory mediators and lymphocyte subpopulations in preterm neonates with sepsis. Inflammation 2012, 35, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Lusyati, S.; Hulzebos, C.V.; Zandvoort, J.; Sukandar, H.; Sauer, P.J. Cytokines patterns in newborn infants with late onset sepsis. J. Neonat-Perinat. Med. 2013, 6, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Sarafidis, K.; Soubasi-Griva, V.; Piretzi, K.; Thomaidou, A.; Agakidou, E.; Taparkou, A.; Diamanti, E.; Drossou-Agakidou, V. Diagnostic utility of elevated serum soluble triggering receptor expressed on myeloid cells (sTREM)-1 in infected neonates. Intensive Care Med. 2010, 36, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Tunc, T.; Cekmez, F.; Cetinkaya, M.; Kalayci, T.; Fidanci, K.; Saldir, M.; Babacan, O.; Sari, E.; Erdem, G.; Cayci, T.; et al. Diagnostic value of elevated CXCR4 and CXCL12 in neonatal sepsis. J. Matern. Fetal Neonat. Med. 2015, 28, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Saldir, M.; Tunc, T.; Cekmez, F.; Cetinkaya, M.; Kalayci, T.; Fidanci, K.; Babacan, O.; Erdem, G.; Kocak, N.; Sari, E.; et al. Endocan and Soluble Triggering Receptor Expressed on Myeloid Cells-1 as Novel Markers for Neonatal Sepsis. Pediatr. Neonatol. 2015, 56, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Li, K.; Wong, R.P.; Chui, K.M.; Wong, E.; Fok, T.F. Neutrophil CD64 expression: A sensitive diagnostic marker for late-onset nosocomial infection in very low birthweight infants. Pediatr. Res. 2002, 51, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Li, K.; Chui, K.M.; Leung, T.F.; Wong, R.P.; Chu, W.C.; Wong, E.; Fok, T.F. IP-10 is an early diagnostic marker for identification of late-onset bacterial infection in preterm infants. Pediatr. Res. 2007, 61, 93–98. [Google Scholar] [CrossRef]
- Değirmencioğlu, H.; Ozer Bekmez, B.; Derme, T.; Öncel, M.Y.; Canpolat, F.E.; Tayman, C. Presepsin and fetuin-A dyad for the diagnosis of proven sepsis in preterm neonates. BMC Infect. Dis. 2019, 19, 695. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, B.E.; Mercado, C.K.; Johnson, L.; Brodsky, N.L.; Bhandari, V. Early markers of late-onset sepsis in premature neonates: Clinical, hematological and cytokine profile. J. Perinat. Med. 2003, 31, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Dillenseger, L.; Langlet, C.; Iacobelli, S.; Lavaux, T.; Ratomponirina, C.; Labenne, M.; Astruc, D.; Severac, F.; Gouyon, J.B.; Kuhn, P. Early Inflammatory Markers for the Diagnosis of Late-Onset Sepsis in Neonates: The Nosodiag Study. Front. Pediatr. 2018, 6, 346. [Google Scholar] [CrossRef] [PubMed]
- Panero, A.; Pacifico, L.; Rossi, N.; Mancuso, G.; Stegagno, M.; Chiesa, C. Interleukin 6 in neonates with early and late onset infection. Pediatr. Infect. Dis. J. 1997, 16, 370–375. [Google Scholar] [CrossRef]
- Arnon, S.; Litmanovitz, I.; Regev, R.; Bauer, S.; Lis, M.; Shainkin-Kestenbaum, R.; Dolfin, T. Serum amyloid A protein is a useful inflammatory marker during late-onset sepsis in preterm infants. Biol. Neonate 2005, 87, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C. Diagnostic markers of infection in neonates. Arch. Dis. Child Fetal. Neonatal Ed. 2004, 89, F229–F235. [Google Scholar] [CrossRef] [PubMed]
- Obuchowski, N.A.; Lieber, M.L.; Wians, F.H., Jr. ROC curves in clinical chemistry: Uses, misuses, and possible solutions. Clin. Chem. 2004, 50, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Liang, L.F.; Li, J.; Yang, D.; Zhao, X.B.; Zhang, K.G. A meta-analysis of interleukin-6 as a valid and accurate index in diagnosing early neonatal sepsis. Int. Wound J. 2019, 16, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Eichberger, J.; Resch, B. Reliability of Interleukin-6 Alone and in Combination for Diagnosis of Early Onset Neonatal Sepsis: Syst. Review. Front. Pediatr. 2022, 10, 840778. [Google Scholar] [CrossRef] [PubMed]
- Dierig, A.; Berger, C.; Agyeman, P.K.A.; Bernhard-Stirnemann, S.; Giannoni, E.; Stocker, M.; Posfay-Barbe, K.M.; Niederer-Loher, A.; Kahlert, C.R.; Donas, A.; et al. Swiss Pediatric Sepsis Study. Time-to-Positivity of Blood Cultures in Children with Sepsis. Front. Pediatr. 2018, 6, 222. [Google Scholar] [CrossRef] [PubMed]




| Author, Year, Country, Reference | LOS Definition | Recruitment | Reference Standard in Infected Neonates | Reference Standard in Control Neonates | Sample Studied, Time of Sample Collection | Test | IL-6 Cut-Off (pg/mL) | Sens, % (95% CI) | Spec, % (95% CI) | AUC (95% CI) | PPV, % | NPV, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Değirmencioğlu H, 2019, Turkey [23] | >72 h | 55 very preterm NICU infants (≤32 weeks): 26 infected (PS = 100%), 29 uninfected | Positive blood culture in addition to clinical signs and abnormal acute phase reactants | GA, birth-weight- and gender-matched infants with no signs or symptoms of sepsis | Neonatal serum, day 0 (after SS, at enrollment) | Solid phase, enzyme labeled, chemi-luminescent sequential immunometric assay | 23.22 (ROC, Youden) | 94.4 | 78.2 | 95.9 | 75 | 95.4 |
| Saldir M, 2015, Turkey [20] | >72 h | 50 near-term (>34 weeks) and term NICU infants: 30 infected (PS = 20%), 20 uninfected | (1) Positive blood/CSF culture or (2) negative culture, but >3 clinical signs of sepsis and abnormal laboratory results (CRP > 5 mg/dL) | Suspected sepsis, which was not supported by clinical or laboratory findings | Venous blood, 0 h (after SS) | NS | 7 (ROC, NS) | 93.3 | 95 | 0.96 (0.908–0.998) | 96.6 | 90.5 |
| Tunc T, 2015, Turkey [19] | >72 h | 50 near-term (>34 weeks) and term NICU infants: 30 infected (PS = 17%), 20 uninfected | (1) Positive blood/CSF culture or (2) negative culture, but >3 clinical signs of sepsis and abnormal laboratory results (CRP > 5 mg/dL) | Suspected sepsis, which was not supported by clinical or laboratory findings | Venous blood, 0 h (after SS) | NS | 7 (ROC, NS) | 96.7 | 95 | 0.97 (0.918–0.998) | 96.7 | 95 |
| Lusyati S, 2013, Indonesia [17] | >72 h | 52 preterm and term NICU infants: 18 infected (PS = 100%), 34 uninfected | Positive culture | Negative blood culture, clinically stable and no signs of infection, except mild respiratory problems treated with CPAP in the first 2 days after birth | Peripheral blood, 0 h (after SS) | Multiplex bead immunoassay | 93 (ROC, NS) | 72.22 (46.5–90.3) | 72.22 (46.5–90.3) | NA | NA | NA |
| Peripheral blood, 12 h (after SS) | 25 | 100 (76.8–100) | 80 (56.3–94.3) | NA | ||||||||
| Peripheral blood, 24 h (after SS) | 40 | 82.35 (56.6–96.2) | 80 (56.3–94.3) | NA | ||||||||
| Peripheral blood, 48 h (after SS) | 88 | 64.71 (38.3–85.8) | 100 (84.6–100) | NA | ||||||||
| 59 preterm and term NICU infants: 25 infected (PS = 0%), 34 uninfected | Negative culture, but ≥2 clinical signs of sepsis | Negative blood culture, clinically stable and no signs of infection, except mild respiratory problems treated with CPAP in the first 2 days after birth | Peripheral blood, 0 h (after SS) | 28 (ROC, NS) | 81.48 (61.9–93.6) | 61.11 (35.8–82.6) | NA | |||||
| Peripheral blood, 12 h (after SS) | 10 | 70.00 (45.7–88.0) | 60.00 (36.1–80.8) | NA | ||||||||
| Peripheral blood, 24 h (after SS) | 13 | 57.14 (39.4–73.7) | 70.00 (45.7–88.0) | NA | ||||||||
| Peripheral blood, 48 h (after SS) | 3 | 100.00 (89.0–100.0) | 31.82 (13.9–54.9) | NA | ||||||||
| Raynor LL, 2012, USA [2] | >72 h | 226 samples from 163 preterm and term NICU infants: 128 infected (PS = 26%), 98 uninfected | (1) Positive blood culture for Gram-positive bacteria or Candida in a patient with signs of sepsis or (2) positive blood culture for Gram-negative bacteria in a patient with signs of sepsis or (3) negative blood culture but antibiotics continued ≥5 d | Negative blood culture and antibiotics for <5 d | Peripheral blood, ≤6 h (after taking the blood culture) | Multiplex antibody-coated bead array with dual-laser fluorometric detection | 130 (ROC, sens = 100%) | 100 | 28 | NA | 52 | 100 |
| Hotoura E, 2012, Greece [16] | >72 h | 82 preterm infants: 42 infected (PS = 41%), 40 healthy controls | (1) Positive blood culture and compatible signs and symptoms or (2) negative blood culture, but signs and symptoms of infection | Infection-free controls, without clinical findings or maternal risk factors for infection | Peripheral blood, 0 h (after SS), for controls at the respective days | ELISA | 60 (ROC, NS) | 67 (41–85) | 96 (89–99) | 0.95 | 80 (51–94) | 89 (78–94) |
| 30 | 100 (78–100) | 74 (63–83) | 0.95 | 40 (30–50) | 100 (90–100) | |||||||
| Sarafidis K, 2010, Greece [18] | >72 h | 52 preterm and term NICU infants with suspected LOS: 31 infected (PS = 71%), 21 uninfected | (1) Positive blood culture (for microbes or fungi) or (2) negative blood culture, but clinical and laboratory (metabolic acidosis, thrombocytopenia, leukopenia/leukocytosis, I:T ratio ≤ 0.2 and CRP ≤ 10 mg/L) evidence of sepsis | Negative blood culture and no laboratory evidence of infection | Peripheral blood, 0 h (after SS) | ELISA | 65.98 (ROC, NS) | 80 (61–92) | 81 (58–94) | 0.892 (0.808– 0.976) | 86 (67–95) | 74 (59–89) |
| Ng PC, 2007, China [22] | >72 h | 155 preterm and VLBW infants with suspected sepsis or NEC: 44 infected (PS = 59%), 111 uninfected | Confirmed episode of septicemia, meningitis, pneumonia, peritonitis, systemic fungal infection, or NEC | Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious and improvement after antibiotic treatment was stopped between 24 and 96 h after initiation | Peripheral blood, 0 h (after SS) | Cytometric bead array (flow cytometry) | 26.1 (ROC, sensitivity approaching 100% and specificity >85% or if not possible sensitivity and specificity approaching 75%) | 82 | 82 | 0.88 | 64 | 92 |
| Peripheral blood, 24 h (after SS) | 26.1 | 48 | 82 | 0.69 | 50 | 81 | ||||||
| Verboon-Maciolek MA, 2006, The Netherlands [3] | NS, all infants older ≥3 days | 92 preterm and term NICU infants: 66 infected (PS = 56%), 26 uninfected | (1) Positive blood culture or (2) negative blood culture but clinical sepsis | No symptoms of infection | Venous blood, 0 h (after SS) | Fully automated chemi-luminescence assay (Immulite) | 60 (ROC, NS) | 68 (50–82) | 76 (56 –90) | NA | 78 (60–91) | 65 (46–80) |
| Arnon S, 2005, Israel [27] | NS, all infants older ≥4 days | 116 preterm infants: 38 infected (PS = 61%), 78 uninfected | (1) Positive blood/CSF/urine culture (in the case of CNS 2, positive blood cultures were required) and ≥1 clinical signs of sepsis or (2) negative cultures, but ≥1 clinical signs of sepsis and 2 abnormal laboratory results persisting for >24 h | (1) Not fulfilling sepsis criteria or (2) blood taken for other reasons than infection | Peripheral blood, 0 h (after SS) | ELISA | 31 (ROC, NS) | 78 (65–85) | 89 (79–95) | 0.65 (0.35–0.76) | 64 (52–76) | 88 (79–95) |
| Peripheral blood, 8 h (after SS) | 31 | 47(39–51) | 100 (97–100) | 0.65 (0.35–0.76) | 100 (93–100) | 80 (68–88) | ||||||
| Peripheral blood, 24 h (after SS) | 31 | 19 (10–30) | 97 (93–99) | NA | 78 (67–86) | 69 (57–77) | ||||||
| Gonzalez BE, 2003, USA [24] | >72 h | 27 preterm NICU infants: 8 infected (PS = 100%), 19 uninfected | Positive blood culture | Negative blood culture | Peripheral blood, day 0 (after SS) | Quantikine kit | 18 (by inspection) | 75 | 68 | NA | 50 | 87 |
| Peripheral blood, day 1 (after SS) | 18 | 75 | 90 | NA | 50 | 90 | ||||||
| Ng PC, 2002, China [21] | >72 h | 80 preterm and VLBW infants with 127 episodes of suspected sepsis: 32 infected (PS = 69%), 58 noninfected and 20 healthy controls | Confirmed episode of septicemia, meningitis, pneumonia, peritonitis, systemic fungal infection, or NEC | (1) Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious or (2) healthy infant with 1–5 weeks neonatal age | Peripheral blood, 0 h (after SS) | ELISA | 31 (ROC, sensitivity approaching 100% and specificity >85% or if not possible, sensitivity and specificity approaching 75%) | 78 | 92 | NA | 81 | 91 |
| Peripheral blood, 12 h (after SS) | 31 | 44 | 93 | NA | 72 | 81 | ||||||
| Peripheral blood, 24 h (after SS) | 31 | 46 | 91 | NA | 68 | 80 | ||||||
| Küster H, 1998, Germany, Slovakia, Austria [9] | >48 h | 41 preterm and VLBW NICU infants: 21 infected (PS = 100%), 20 uninfected | Subjective clinical suspicion of sepsis, followed within 2 days by objective clinical evidence and sampling of specimens for positive cultures | Neither positive cultures, nor objective clinical evidence, nor subjective clinical suspicion of sepsis | Peripheral blood, day − 4 to day − 1 (diagnosis of sepsis on day 0) | ELISA | 25 (ROC, maximum sens + spec) | 57.1 | 82.9 | 0.94 | NA | NA |
| Peripheral blood, day − 4 to day 0 (diagnosis of sepsis on day 0) | 25 | 85.7 | 82.9 | 0.94 | NA | NA | ||||||
| Peripheral blood, day − 4 to day + 1 (diagnosis of sepsis on day 0) | 25 | 89.3 | 82.9 | 0.94 | NA | NA | ||||||
| Ng PC, 1997, China [10] | >72 h | 68 preterm and VLBW infants with 101 episodes of clinical suspected sepsis: 35 infected (PS = NA), 46 uninfected, 20 healthy controls | Positive blood culture or confirmed infection other than septicemia (pneumonia, peritonitis, meningitis, systemic fungal infection, and NEC) with or without positive blood culture | (1) Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious and improvement after antibiotic treatment was stopped or (2) healthy infant with 1–8 weeks neonatal age | Peripheral blood, day 0 (after SS) | ELISA | 31 (ROC, minimizing the number of misclassified episodes) | 89 | 96 | NA | 95 | 91 |
| Peripheral blood, day 1 (after SS) | 31 | 67 | 89 | NA | 84 | 77 | ||||||
| Panero A, 1997, Italy [26] | >72 h | 68 preterm and term NICU infants: 17 infected (PS = 82%), 51 uninfected | (1) Positive blood culture (septicemia) or (2) meningitis or (3) NEC | Uninfected controls matched for neonatal age and duration of hospital stay | Peripheral blood, 0 h (after SS) | Solid phase sandwich enzyme-amplified sensitivity immunoassay (Medgenix) | 15 (NA) | 100 | 100 | NA | NA | NA |
| Author, Year, Country, Reference | LOS Definition | Recruitment | Reference Standard in Infected Neonates | Reference Standard in Control Neonates | Sample Studied, Time of Sample Collection | Test | Biomarker Combination | Cut-Offs: IL-6 (pg/mL), sTREM-1 (pg/mL), IP-10 (pg/mL), IL-10 (pg/mL), CRP (mg/L), CD64 (Phycoerythrin-Molecules Bound Per Cell), TNF-α (pg/mL) | Sens, % (95% CI) | Spec, % (95% CI) | AUC | PPV, % | NPV, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dillenseger L, 2018, France [25] | >72 h | 130 preterm and term NICU infants with suspected sepsis: 34 infected (PS = 53%), 96 uninfected | (1) Positive blood culture alone, or in combination with clinical signs of infection and a CRP >10 mg/L (in the case of typical skin contaminants), or meningitis (>10 cells/mL in lumbar puncture), or pneumonia (>104 bacteria/mL in BAL/tracheal aspiration, positive chest radiographs, ventilator support, ≥4 clinical signs), or pyelonephritis (clinical signs of sepsis, CRP > 10 and >106 cells/L and >105 bacteria/mL in the urine) or (2) clinical signs and CRP ≥ 10 mg/L, no alternative diagnosis and improvement upon antibiotic treatment | (1) Clinical signs or elevated CRP explained by alternative diagnosis or positive culture, but no clinical or biological signs of infection, or positive blood culture but CRP < 4 mg/L, or antibiotic treatment <5 days or (2) clinical improvement and normalization of CRP levels without antibiotics | Peripheral blood, 0 h (after SS) | Fully automated chemiluminescence assay (Immulite) | IL-6 + CRP | IL-6: 21.7, CRP: 4.05 | 78.12 (60.03–90.72) | 76.34 (66.40–84.54) | 84.80 (75.03–96.58) | 53.19 (38.08–67.89) | 91.03 (82.38–96.32) |
| Hotoura E, 2012, Greece [16] | >72 h | 82 preterm infants: 42 infected (PS = 41%), 40 healthy controls | (1) Positive blood culture and compatible signs and symptoms or (2) negative blood culture, but signs and symptoms of infection | Infection-free controls, without clinical findings or maternal risk factors for infection | Peripheral blood, 0 h (after SS), for controls at the respective days | ELISA | IL-6 + CRP | IL-6: 30, CRP: 10 | 100 (79–100) | 96 (89–99) | NA | NA | NA |
| Sarafidis K, 2010, Greece [18] | >72 h | 52 preterm and term NICU infants with suspected LOS: 31 infected (PS = 71%), 21 uninfected | (1) Positive blood culture (for microbes or fungi) or (2) negative blood culture, but clinical and laboratory (metabolic acidosis, thrombocytopenia, leukopenia/leukocytosis, I:T ratio ≤ 0.2 and CRP ≤ 10 mg/L) evidence of sepsis | Negative blood culture and no laboratory evidence of infection | Peripheral blood, 0 h (after SS) | ELISA | IL-6 + sTREM-1 (NS) | IL-6: 66, sTREM-1: 144 | 90 (73–98) | 62 (38–82) | NA | 77 (59-89) | 81 (54–96) |
| Ng PC, 2007, China [22] | >72 h | 155 preterm VLBW infants with suspected sepsis or NEC: 44 infected (PS = 59%), 111 uninfected | Confirmed episode of septicemia, meningitis, pneumonia, peritonitis, systemic fungal infection, or NEC | Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious and improvement after antibiotic treatment was stopped between 24 and 96 h after initiation | Peripheral blood, 0 h (after SS) | Cytometric bead array (flow cytometry) | IL-6 + IP-10 | IL-6: 26.1, IP-10: 1250 (ROC, sensitivity approaching 100% and specificity >85% or if not possible sensitivity and specificity approaching 75%) | 98 | 72 | NA | 58 | 99 |
| IL-6 + IP-10 + IL-10 | IL-6: 26.1, IP-10: 1250, IL-10: 7.6 | 98 | 61 | NA | 50 | 99 | |||||||
| Verboon-Maciolek MA, 2006, The Netherlands [3] | NS, all infants ≥ 3 days | 92 preterm and term NICU infants: 66 infected (PS = 56%), 26 uninfected | (1) Positive blood culture or (2) negative blood culture but clinical sepsis | No symptoms of infection | Venous blood, 0 h (after SS) | IL-6: fully automated chemiluminescence assay (Immulite), CRP: rate nephelometry | IL-6 + CRP | IL-6: 60, CRP: 14 | 92 (78–98) | 41 (24–61) | NA | 67 (54–80) | 80 (52–96) |
| Ng PC, 2002, China [21] | >72 h | 80 preterm VLBW infants with 127 episodes of suspected sepsis: 32 infected (PS = 69%), 58 noninfected and 20 healthy controls | Confirmed episode of septicemia, meningitis, pneumonia, peritonitis, systemic fungal infection, or NEC (stage II or above in Bell’s classification) | (1) Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious or (2) healthy infant with 1–5 weeks neonatal age | Peripheral blood, 0 h (IL-6) and 24 h (CD64) after SS | IL-6: ELISA, CD64: flow cytometry | IL-6 + CD64 | IL-6: 31, CD64: 4000 (ROC, sensitivity approaching 100% and specificity >85% or if not possible sensitivity and specificity approaching 75%) | 100 | 86 | NA | 74 | 100 |
| Peripheral blood, 24 h (after SS) | IL-6 + CD64 | 97 | 86 | NA | 73 | 99 | |||||||
| peripheral blood, 48 h (IL-6) and 24 h (CD64) after SS | IL-6 + CD64 | 95 | 83 | NA | 70 | 97 | |||||||
| Ng PC, 1997, China [10] | >72 h | 68 preterm VLBW infants with 101 episodes of clinical suspected sepsis: 35 infected (PS = NA), 46 uninfected, 20 healthy controls | Positive blood culture or confirmed infection other than septicemia (pneumonia, peritonitis, meningitis, systemic fungal infection, and NEC) with or without positive blood culture | (1) Episode meeting the screening criteria for suspected clinical sepsis, subsequently proven not to be infectious and improvement after antibiotic treatment was stopped or (2) healthy infant with 1–8 weeks neonatal age | Peripheral blood, day 0 (after SS) | IL-6+TNF-α: ELISA, CRP: turbidity assay | IL-6 + CRP | IL-6: 31, CRP: 12 (ROC, sensitivity approaching 100% and specificity >85% or if not possible sensitivity and specificity approaching 75%) | 93 | 96 | NA | 95 | 95 |
| Peripheral blood, day 1 (after SS) | IL-6 + CRP | 93 | 88 | NA | 86 | 94 | |||||||
| Peripheral blood, day 0 (after SS) | IL-6 + TNF-α | 95 | 84 | NA | 83 | 96 | |||||||
| Peripheral blood, day 1 (after SS) | IL-6 + TNF-α | 91 | 84 | NA | 82 | 92 | |||||||
| Peripheral blood, day 0 (after SS) | IL-6 + CRP + TNF-α | 95 | 84 | NA | 82 | 96 | |||||||
| Peripheral blood, day 1 (after SS) | IL-6 + CRP + TNF-α | 98 | 80 | NA | 80 | 98 | |||||||
| Peripheral blood, day 0 (IL-6+CRP) and day 1 (TNF-α) after SS | IL-6 + CRP + TNF-α | 98 | 91 | NA | 90 | 98 | |||||||
| Peripheral blood, day 0 (IL-6+CRP) and day 2 (CRP) after SS | IL-6 + CRP | 98 | 91 | NA | 90 | 98 |
| Subgroup | No. Studies | Pooled Sensitivity, % | Pooled Specificity, % | |
|---|---|---|---|---|
| Study population | Preterm | 8 | 86.59 | 85.71 |
| Preterm and term | 6 | 81.77 | 86.05 | |
| Timing | 0 h * | 11 | 84.22 | 85.83 |
| ≤12 h * | 3 | 56.82 | 93.68 | |
| ≤24 h * | 6 | 54.29 | 88.34 | |
| ≤48 h * | 3 | 67.21 | 92.44 | |
| Sepsis definition | Culture proven only | 3 | 84.62 | 74.36 |
| Study design | Blinding | 2 | 80.77 | 80.00 |
| Biomarker combinations | IL-6 + CRP | 4 | 92.09 | 78.95 |
| Quality of Reporting of IL-6 Accuracy Studies for Diagnosing Late (>72 h) Onset Infection | ||
|---|---|---|
| Category and Item No. | YES | NO |
| Methods: participants | ||
| Describe the study population: | ||
| 1A. The inclusion and exclusion criteria | 10 | 6 |
| 1B. Setting, and locations where data were collected | 15 | 1 |
| Describe participant recruitment: | ||
| 2A. Was enrollment of patients based only on clinical signs suggesting infection? | 12 | 4 |
| 2B. Were such patients consecutively enrolled? | 2 | 10 |
| 2C. Was enrollment of patients based only on maternal risk factors for infection? | 0 | 16 |
| 2D. Were such patients consecutively enrolled? | 0 | 0 |
| 2E. Were patients identified by searching hospital records? | 0 | 16 |
| 2F. Did the study include both patients already diagnosed with sepsis and participants in whom sepsis had been excluded? | 2 | 14 |
| Describe data collection: | ||
| 3. Was data collection planned before the index test and reference standard were performed (prospective study)? | 14 | 2 |
| Test methods | ||
| Methods pertaining to the reference standard and the index test: | ||
| 4A. Was a composite reference standard used to identify all newborns with sepsis, and verify index test results in infected babies? | 13 | 3 |
| 4B. Was a reference standard used to exclude sepsis? | 14 | 2 |
| 4C. Was a composite reference standard used to identify all newborns without sepsis, and verify index test results in uninfected babies? | 4 | 10 |
| 4D. Did the index test or its comparator form part of the reference standard? | 2 | 14 |
| 5. Were categories of results of the index test (including cut-offs) and the reference standard defined after obtaining results? | 16 | 0 |
| 6. Did the study report the number, training and expertise of the persons executing and reading the index tests and the reference standard? | 3 | 13 |
| 7. Was there blinding to results of the index test and the reference standard? | 4 | 12 |
| Statistical methods | ||
| 8. Describe the statistical methods used to quantify uncertainty (i.e., 95% confidence intervals) | 6 | 10 |
| 9. Describe methods for calculating test reproducibility | 4 | 12 |
| Results: participants and test results | ||
| 10A. Describe when the study was carried out, including beginning and ending dates of recruitment | 13 | 3 |
| 10B. Did the study report clinical and demographic (postnatal hours or days, gestational age, birth weight, gender) features in those with and without sepsis? | 15 | 1 |
| 10C. Did the study report distribution of illness severity scores in those with and without sepsis? | 0 | 16 |
| 11. Report the number of participants satisfying the criteria for inclusion that did or did not undergo the index tests and/or or the reference standard; describe why participants failed to receive either test | 4 | 12 |
| 12. Report a cross-tabulation of the results (including indeterminate and missing results) using the results of the reference standard; for continuous results, report the distribution of the test results using the results of the reference standard | 2 | 14 |
| Results: estimates | ||
| 13. Report measures of statistical uncertainty (i.e., 95% confidence intervals) | 6 | 10 |
| 14. Report how indeterminate results, missing responses and outliers of index tests were handled | 1 | 15 |
| 15. Report estimates of test reproducibility | 5 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eichberger, J.; Resch, E.; Resch, B. Reliability of IL-6 Alone and in Combination for Diagnosis of Late Onset Sepsis: A Systematic Review. Children 2024, 11, 486. https://doi.org/10.3390/children11040486
Eichberger J, Resch E, Resch B. Reliability of IL-6 Alone and in Combination for Diagnosis of Late Onset Sepsis: A Systematic Review. Children. 2024; 11(4):486. https://doi.org/10.3390/children11040486
Chicago/Turabian StyleEichberger, Julia, Elisabeth Resch, and Bernhard Resch. 2024. "Reliability of IL-6 Alone and in Combination for Diagnosis of Late Onset Sepsis: A Systematic Review" Children 11, no. 4: 486. https://doi.org/10.3390/children11040486