Balancing Resolution with Analysis Time for Biodiesel–Diesel Fuel Separations Using GC, PCA, and the Mahalanobis Distance
Abstract
:1. Introduction
2. Theory
2.1. Nomenclature and Terminology
2.2. Mahalanobis Distance
3. Materials and Methods
3.1. Chemicals
3.2. Transesterification
3.3. Instrumentation
3.4. Data Processing
4. Results and Discussion
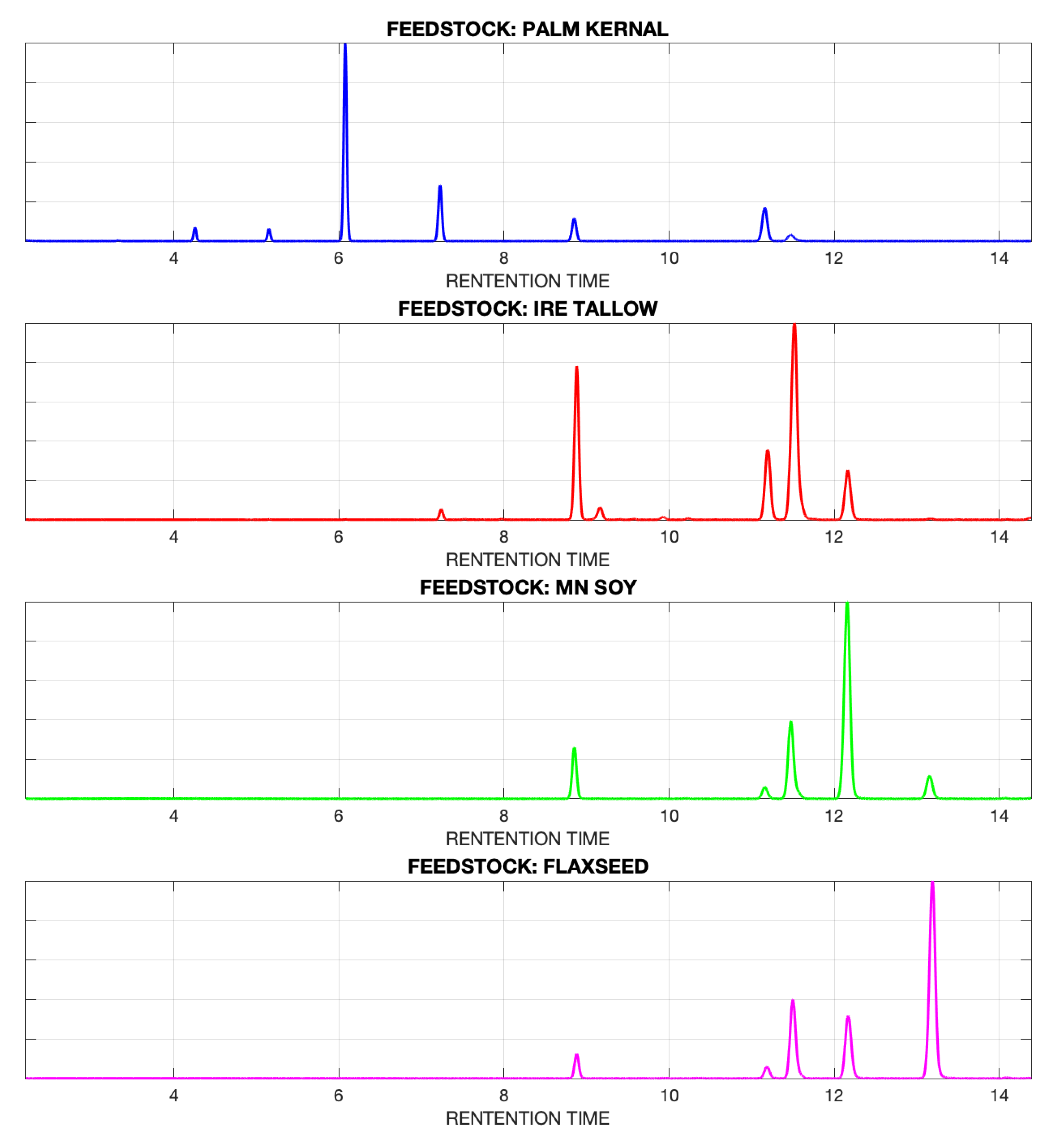
4.1. No Alignment within Each Feedstock Type
4.2. Alignment within Each Feedstock Type
4.3. Alignment across Feedstock Types
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| COW | correlated optimized warping |
| FAME | fatty acid methyl esters |
| GC | gas chromatography |
| MD | Mahalanobis distance |
| PCA | principal components analysis |
| SI | similarity index |
References
- Giddings, J.C. Dynamics of Chromatography Part I Principles and Theory; Marcel Dekker, Inc.: New York, NY, USA, 1965. [Google Scholar]
- Knothe, G.; Van Gerpen, J.; Krahl, J. (Eds.) The Biodiesel Handbook; AOCS Press: Urbana, IL, USA, 2005. [Google Scholar]
- Cruz-Hernandez, C.; Destaillats, F. Recent advances in fast Gas-Chromatography: Application to the separation of fatty acid methyl esters. J. Liq. Chrom. Rel. Technol. 2009, 32, 1672–1688. [Google Scholar] [CrossRef]
- Schale, S.P.; Le, T.M.; Pierce, K.M. Predicting feedstock and percent composition for blends of biodiesel with conventional diesel using chemometrics and gas chromatography-mass spectrometry. Talanta 2012, 94, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Hupp, A.M.; Marshall, L.J.; Campbell, D.I.; Waddell Smith, R.; McGuffin, V.L. Chemometric analysis of diesel fuel for forensic and environmental applications. Anal. Chim. Acta 2008, 606, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Fortunato de Carvalho Rocha, W.; Schantz, M.M.; Sheen, D.A.; Chu, P.M.; Lippa, K.A. Unsupervised classification of petroleum Certified Reference Materials and other fuels by chemometric analysis of gas chromatography-mass spectrometry data. Fuel 2017, 197, 248–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauls, R.E. Fast Gas Chromatographic Separation of Biodiesel. J. Chromatogr. Sci. 2011, 49, 370–374. [Google Scholar] [CrossRef]
- Turner, T.; Rolland, D.C.; Aldai, N.; Dugan, M.E.R. Rapid Separation of cis9, trans11- and trans7, cis9-18:2 (CLA) isomers from ruminant tissue using a 30 m SLB-IL111 ionic column. Can. J. Anim. Sci. 2011, 91, 711–713. [Google Scholar] [CrossRef]
- Goding, J.C.; Ragon, D.Y.; O’Connor, J.B.; Boehm, S.B.; Hupp, A.M. Comparison of GC stationary phases for the separation of fatty acid methyl esters in biodiesel fuels. Anal. Bioanal. Chem. 2013, 405, 6087–6094. [Google Scholar] [CrossRef]
- Masood, A.; Stark, K.D.; Salem, N., Jr. A simplified and efficient method for the analysis of fatty acid methyl esters suitable for large clinical studies. J. Lipid. Res. 2005, 46, 2299–2305. [Google Scholar] [CrossRef] [Green Version]
- Tauler, R. Multivariate curve resolution applied to second order data. Chemometr. Intell. Lab. 1995, 30, 133–146. [Google Scholar] [CrossRef]
- De Luca, S.; Ciotoli, E.; Biancolillo, A.; Bucci, R.; Magri, A.D.; Marini, F. Simultaneous quantification of caffeine and chlorogenic acid in coffee beans and varietal classification of the samples by HPLC-DAD coupled with chemometrics. Environ. Sci. Pollut. R 2018, 25, 28748–28759. [Google Scholar] [CrossRef]
- Tauler, R.; Smilde, A.; Kowalski, B. Selectivity, local rank, three-way data analysis and ambiguity in multivariate curve resolution. J. Chemometr. 1995, 9, 31–58. [Google Scholar] [CrossRef]
- Maeder, M. Evolving factor analysis for the resolution of overlapping chromatographic peaks. Anal. Chem. 1987, 59, 527–530. [Google Scholar] [CrossRef]
- Bahaghighat, H.D.; Freye, C.E.; Gough, D.V.; Synovec, R.E. Comprehensive two-dimensional gas chromatography and time-of-flight mass spectrometry detection with a 50ms modulation period. J. Chromatogr. A 2019, 1583, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Azimi, F.; Fatemi, M.H. Multivariate curve resolution-correlation optimized warping applied to the complex GC-MS signals: Toward comparative study of peel chemical variability of Citrus aurantium L. varieties. Microchem. J. 2018, 143, 99–109. [Google Scholar] [CrossRef]
- Aliakbarzadeh, G.; Sereshti, H.; Parastar, H. Fatty acids profiling of avocado seed and pulp using gas chromatography-mass spectrometry combined with multivariate chemometric techniques. J. Iran. Chem. Soc. 2016, 13, 1905–1913. [Google Scholar] [CrossRef]
- Rocha, W.F.C.; Vaz, B.G.; Sarmanho, G.F.; Leal, L.H.C.; Nogueira, R.; Silva, V.F.; Borges, C.N. Chemometric techniques applied for classification and quantification of binary biodiesel/diesel blends. Anal. Lett. 2012, 45, 2398–2411. [Google Scholar] [CrossRef]
- Flood, M.E.; Goding, J.C.; O’Connor, J.B.; Ragon, D.Y.; Hupp, A.M. Analysis of Biodiesel feedstock using GCMS and unsupervised chemometric methods. J. Am. Oil. Chem. Soc. 2014, 91, 1443–1452. [Google Scholar] [CrossRef]
- Soares, E.J.; Yalla, G.R.; O’Connor, J.B.; Walsh, K.A.; Hupp, A.M. Hotelling trace criterion as a figure of merit for optimization of chromatogram alignment. J. Chemom. 2014, 29, 200–212. [Google Scholar] [CrossRef]
- Flood, M.E.; Connolly, M.P.; Comiskey, M.C.; Hupp, A.M. Evaluation of single and multi-feedstock biodiesel-diesel blends using GC/MS and chemometric methods. Fuel 2016, 186, 58–67. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis; Springer: New York, NY, USA, 2002. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemometr. Intell. Lab. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Philos. Mag. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a Complex of Statistical Variable into Principal Components; Warwick and York: Baltimore, MD, USA, 1933. [Google Scholar]
- Malmquist, G.; Danielsson, R. Alignment of chromatographic profiles for principal component analysis: A prerequisite for fingerprinting methods. J. Chromatogr. A 1994, 687, 71–88. [Google Scholar] [CrossRef]
- Van Nederkassel, A.M.; Daszkowski, M.; Eilers, P.H.C.; Vander Heyden, Y. A comparison of three algorithms for chromatograms alignment. J. Chromatogr. A 2006, 1118, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Daszykowski, M.; Walczak, B. Target selection for alignment of chromatographic signals obtained using monochannel detectors. J. Chromatogr. A 2007, 1176, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vest Nielsen, N.; Carstensen, J.M.; Smedsgaard, J. Aligning of single and multiple wavelength chromatographic profiles for chemometric data analysis using correlation optimized warping. J. Chromatogr. A 1998, 805, 17–35. [Google Scholar] [CrossRef]
- Tomasi, G.; van den Berg, F.; Andersson, C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemom. 2004, 18, 231–241. [Google Scholar] [CrossRef]
- Andric, F.; Heberger, K. How to compare separation selectivity of high-performance liquid chromatographic columns properly? J. Chromatogr. A 2017, 1488, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowik, W.; Heron, S.; Bonose, M.; Tchapla, A. Separation system suitability (3S): A new criterion of chromatogram classification in HPLC based on cross-evaluation of separation capacity/peak symmetry and its application to complex mixtures of anthraquinones. Analyst 2013, 138, 5801–5810. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Re-evaluating the role of the Mahalanobis distance measure. J. Chemom. 2016, 30, 134–143. [Google Scholar] [CrossRef]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Inst. Sci. India Phys. Sci. 1936, 2, 49–55. [Google Scholar]
- Mardia, K.V.; Kent, J.T.; Bibby, J.M. Multivariate Analysis; Academic Press: New York, NY, USA, 1979. [Google Scholar]
- Skov, T.; van den Berg, F.; Bro, R. Automated alignment of chromatographic data. J. Chemom. 2006, 20, 484–497. [Google Scholar] [CrossRef]


| Feedstock | MD | p-Value | Seg. Length (ZB) | Max. Warp (ZB) | Seg. Length (DB) | Max. Warp (DB) |
|---|---|---|---|---|---|---|
| ADM Canola | 9.83 × 10 | 1 | 19 | 14 | 31 | 7 |
| Canola | 1.43 × 10 | 1 | 48 | 14 | 21 | 8 |
| Coconut | 8.04 × 10 | 1 | 35 | 6 | 24 | 7 |
| Flaxseed | 5.96 × 10 | 1 | 70 | 14 | 9 | 1 |
| IRE Tallow | 3.24 × 10 | 1 | 20 | 4 | 19 | 2 |
| MN Soy | 2.46 × 10 | 1 | 22 | 10 | 13 | 8 |
| Palm Kernal | 2.75 × 10 | 1 | 51 | 9 | 17 | 1 |
| Safflower | 6.29 × 10 | 1 | 43 | 4 | 21 | 6 |
| Soyabean | 2.15 × 10 | 1 | 14 | 9 | 17 | 2 |
| Sunflower | 3.07 × 10 | 1 | 46 | 9 | 25 | 8 |
| TexasTallow | 1.85 × 10 | 1 | 15 | 4 | 12 | 2 |
| Waste Grease | 1.59 × 10 | 1 | 50 | 2 | 27 | 8 |
| Feedstock | MD | p-Value | Seg. Length (ZB) | Max. Warp (ZB) | Seg. Length (DB) | Max. Warp (DB) |
|---|---|---|---|---|---|---|
| ADM Canola | 0.0015 | 0.9999 | 67 | 7 | 22 | 2 |
| Canola | 0.0004 | 1.0000 | 64 | 14 | 30 | 2 |
| Coconut | 0.0029 | 0.9998 | 43 | 9 | 27 | 3 |
| Flaxseed | 0.0010 | 1.0000 | 46 | 2 | 26 | 3 |
| IRE Tallow | 0.0018 | 0.9999 | 69 | 2 | 16 | 4 |
| MN Soy | 0.0030 | 0.9998 | 68 | 13 | 26 | 4 |
| Palm Kernal | 0.0187 | 0.9973 | 69 | 15 | 8 | 1 |
| Safflower | 0.0030 | 0.9998 | 44 | 1 | 20 | 7 |
| Soyabean | 0.0018 | 0.9999 | 48 | 14 | 20 | 5 |
| Sunflower | 0.0018 | 0.9999 | 23 | 5 | 23 | 3 |
| TexasTallow | 0.0025 | 0.9999 | 46 | 4 | 15 | 2 |
| Waste Grease | 0.0003 | 1.0000 | 56 | 2 | 14 | 1 |
| Feedstock | MD | p-Value | Seg. Length (ZB) | Max. Warp (ZB) | Seg. Length (DB) | Max. Warp (DB) |
|---|---|---|---|---|---|---|
| ADM Canola | 0.3294 | 0.9833 | 64 | 11 | 16 | 7 |
| Canola | 0.1119 | 0.9986 | 60 | 13 | 18 | 4 |
| Coconut | 0.1047 | 0.9988 | 52 | 6 | 10 | 2 |
| Flaxseed | 0.4001 | 0.9747 | 58 | 3 | 20 | 8 |
| IRE Tallow | 0.1356 | 0.9978 | 68 | 6 | 16 | 1 |
| MN Soy | 0.6257 | 0.9384 | 50 | 5 | 15 | 2 |
| Palm Kernal | 0.1048 | 0.9988 | 22 | 15 | 35 | 2 |
| Safflower | 0.2250 | 0.9928 | 48 | 2 | 25 | 1 |
| Soyabean | 0.1127 | 0.9986 | 39 | 4 | 21 | 2 |
| Sunflower | 0.0817 | 0.9993 | 57 | 5 | 13 | 2 |
| TexasTallow | 0.3184 | 0.9845 | 38 | 1 | 18 | 2 |
| Waste Grease | 0.0932 | 0.9991 | 35 | 1 | 14 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, E.J.; Clifford, A.J.; Brown, C.D.; Dean, R.R.; Hupp, A.M. Balancing Resolution with Analysis Time for Biodiesel–Diesel Fuel Separations Using GC, PCA, and the Mahalanobis Distance. Separations 2019, 6, 28. https://doi.org/10.3390/separations6020028
Soares EJ, Clifford AJ, Brown CD, Dean RR, Hupp AM. Balancing Resolution with Analysis Time for Biodiesel–Diesel Fuel Separations Using GC, PCA, and the Mahalanobis Distance. Separations. 2019; 6(2):28. https://doi.org/10.3390/separations6020028
Chicago/Turabian StyleSoares, Edward J., Alexandra J. Clifford, Carolyn D. Brown, Ryan R. Dean, and Amber M. Hupp. 2019. "Balancing Resolution with Analysis Time for Biodiesel–Diesel Fuel Separations Using GC, PCA, and the Mahalanobis Distance" Separations 6, no. 2: 28. https://doi.org/10.3390/separations6020028





