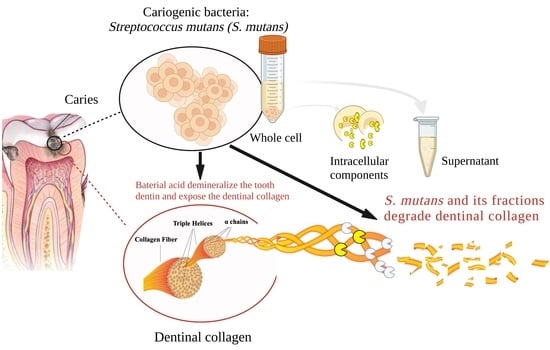
Streptococcus mutans Proteases Degrade Dentinal Collagen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Type I Collagen Degradation by S. mutans UA159 and Its Discrete Fractions
- (1)
- 125 CDU/mL Clostridium histolyticum (C. histolyticum) collagenase (0.2 mg) (positive control to validate the assay)
- (2)
- ¼ THYE medium (negative control to exclude degradative effect from medium)
- (3)
- Overnight (O/N) culture of S. mutans UA159 (OD600 = 0.8) in ¼ THYE
- (4)
- 1:100 fresh inoculated culture (NEW) of S. mutans UA159 (concentrated to OD600 = 0.8) in ¼ THYE
- (5)
- Intracellular protein fraction of lysed O/N S. mutans UA159 (see further details in the Appendix A)
- (6)
- Supernatant (cell-free fraction) from O/N of S. mutans UA159 culture
- (7)
- Bacterial membrane fraction of lysed O/N S. mutans UA159 (see further details in the Appendix A)
2.2. Dentinal Collagen Degradation by S. mutans UA159 and Its Discrete Fractions
2.3. Statistical Analysis
2.4. Verification of Dentinal Collagen Degradation by Intracellular Proteins of S. mutans Using SDS-PAGE and Mass Spectrometry
- (1)
- C. histolyticum collagenase (positive control)
- (2)
- PBS (negative control)
- (3)
- 75 μg of protein (experimental groups)
3. Results
3.1. Type I Collagen Degradation by S. mutans UA159 and Its Discrete Fractions
3.2. Dentinal Collagen Degradation by S. mutans UA159 and Its Discrete Fractions
3.3. Verification of Dentinal Collagen Degradation by Intracellular Proteins of S. mutans Using SDS-PAGE and Mass Spectrometry
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Chaussain, C.; Boukpessi, T.; Khaddam, M.; Tjaderhane, L.; George, A.; Menashi, S. Dentin matrix degradation by host matrix metalloproteinases: Inhibition and clinical perspectives toward regeneration. Front. Physiol. 2013, 4, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzoni, A.; Tjäderhane, L.; Checchi, V.; Di Lenarda, R.; Salo, T.; Tay, F.; Pashley, D.; Breschi, L. Role of Dentin MMPs in Caries Progression and Bond Stability. J. Dent. Res. 2015, 94, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.J.; Lim, D.V.; Dao, M.L. Identification and analysis of a collagenolytic activity in Streptococcus mutans. Curr. Microbiol. 1997, 34, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Marashdeh, M.; Gitalis, R.; Lévesque, C.; Finer, Y. Endodontic pathogens possess collagenolytic properties that degrade human dentine collagen matrix. Int. Endod. J. 2019, 52, 416–423. [Google Scholar] [CrossRef]
- Gitalis, R.; Zhou, L.; Marashdeh, M.Q.; Sun, C.; Glogauer, M.; Finer, Y. Human neutrophils degrade methacrylate resin composites and tooth dentin. Acta Biomater. 2019, 88, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Delaviz, Y.; Finer, Y.; Santerre, J.P. Biodegradation of resin composites and adhesives by oral bacteria and saliva: A rationale for new material designs that consider the clinical environment and treatment challenges. Dent. Mater. Off. Publ. Acad. Dent. Mater. 2014, 30, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Breschi, L.; Maravic, T.; Cunha, S.R.; Comba, A.; Cadenaro, M.; Tjäderhane, L.; Pashley, D.H.; Tay, F.R.; Mazzoni, A. Dentin bonding systems: From dentin collagen structure to bond preservation and clinical applications. Dent. Mater. 2018, 34, 78–96. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, A.; Nascimento, F.; Carrilho, M.; Tersariol, I.; Papa, V.; Tjäderhane, L.; Di Lenarda, R.; Tay, F.; Pashley, D.H.; Breschi, L. MMP activity in the hybrid layer detected with in situ zymography. J. Dent. Res. 2012, 91, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Pashley, D.H.; Tay, F.; Yiu, C.; Hashimoto, M.; Breschi, L.; Carvalho, R.M.D.; Ito, S. Collagen degradation by host-derived enzymes during aging. J. Dent. Res. 2004, 83, 216–221. [Google Scholar] [CrossRef]
- Sorsa, T.; Tjäderhane, L.; Salo, T. Matrix metalloproteinases (MMPs) in oral diseases. Oral Dis. 2004, 10, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Dung, S.Z. Effects of mutans streptococci, Actinomyces species and Porphyromonas gingivalis on collagen degradation. Zhonghua Yi Xue Za Zhi 1999, 62, 764–774. [Google Scholar] [PubMed]
- Rosengren, L.; Winblad, B. Proteolytic activity of Streptococcus mutans (GS-5). Oral Surg. Oral Med. Oral Pathol. 1976, 42, 801–809. [Google Scholar] [CrossRef]
- Harrington, D.J. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. Infect. Immun. 1996, 64, 1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damé-Teixeira, N.; Parolo, C.C.F.; Maltz, M.; Rup, A.G.; Devine, D.A.; Do, T. Gene expression of bacterial collagenolytic proteases in root caries. J. Oral Microbiol. 2018, 10, 1424475. [Google Scholar] [CrossRef] [Green Version]
- Aso, H.; Maeda, H.; Nambu, T.; Okinaga, T.; Shida, M. Identification of MMP-like protein from Streptococcus mitis. World J. Adv. Res. Rev. 2020, 8, 154–161. [Google Scholar]
- Arora, P.D.; McCulloch, C.A. Dependence of collagen remodelling on α-smooth muscle actin expression by fibroblasts. J. Cell. Physiol. 1994, 159, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Bidlingmeyer, B.A.; Cohen, S.A.; Tarvin, T.L. Rapid analysis of amino acids using pre-column derivatization. J. Chromatogr. 1984, 336, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.D.; Narani, N.; McCulloch, C.A. The compliance of collagen gels regulates transforming growth factor-β induction of α-smooth muscle actin in fibroblasts. Am. J. Pathol. 1999, 154, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Sauro, S.; Mannocci, F.; Tay, F.R.; Pashley, D.H.; Cook, R.; Carpenter, G.H.; Watson, T.F. Deproteinization effects of NaOCl on acid-etched dentin in clinically-relevant vs. prolonged periods of application. A confocal and environmental scanning electron microscopy study. Oper. Dent. 2009, 34, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, W.L.; Bakkal, M.; Xiao, Y.; Sutton, J.N.; Mendes, F.M. Quantitative proteomic analysis of the effect of fluoride on the acquired enamel pellicle. PLoS ONE 2012, 7, e42204. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kato, T.; Kuramitsu, H.K. Isolation and preliminary characterization of the Porphyromonas gingivalis prtC gene expressing collagenase activity. FEMS Microbiol. Lett. 1991, 84, 135–138. [Google Scholar] [CrossRef]
- Krieger, J.R.; Taylor, P.; Moran, M.F.; McGlade, C.J. Comprehensive identification of phosphorylation sites on the Numb endocytic adaptor protein. Proteomics 2015, 15, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Travis, J.; Potempa, J. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1477, 35–50. [Google Scholar] [CrossRef]
- Maeda, H. Role of microbial proteases in pathogenesis. Microbiol. Immunol. 1996, 40, 685–699. [Google Scholar] [CrossRef]
- Loesche, W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Mol. Biol. Rev. 1986, 50, 353. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Siqueira, W.L.; Cvitkovitch, D.G.; Finer, Y. Esterase from a cariogenic bacterium hydrolyzes dental resins. Acta Biomater. 2018, 71, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Tjäderhane, L.; Buzalaf, M.A.R.; Carrilho, M.; Chaussain, C. Matrix metalloproteinases and other matrix proteinases in relation to cariology: The era of ‘dentin degradomics’. Caries Res. 2015, 49, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Takahashi, N.; Kuramitsu, H.K. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J. Bacteriol. 1992, 174, 3889–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makinen, P.L.; Clewell, D.B.; An, F.; Makinen, K.K. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 1989, 264, 3325–3334. [Google Scholar] [CrossRef]
- Persadmehr, A.; Torneck, C.D.; Cvitkovitch, D.G.; Pinto, V.; Talior, I.; Kazembe, M.; Shrestha, S.; McCulloch, C.A.; Kishen, A. Bioactive chitosan nanoparticles and photodynamic therapy inhibit collagen degradation in vitro. J. Endod. 2014, 40, 703–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.A.; Hong, J.H.; Hatton, B.D.; Finer, Y. Antimicrobial antidegradative dental adhesive preserves restoration-tooth bond. Dent. Mater. Off. Publ. Acad. Dent. Mater. 2020, 36, 1666–1679. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.G.; Spatafora, G.A. Gene Regulation in S. mutans: Complex Control in a Complex Environment. J. Dent. Res. 2012, 91, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; MacDonald, J.B. Degradation of collagenous substrates by Bacteroides melaninogenicus. J. Bacteriol. 1961, 81, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertassoni, L.E. Dentin on the nanoscale: Hierarchical organization, mechanical behavior and bioinspired engineering. Dent. Mater. 2017, 33, 637–649. [Google Scholar] [CrossRef]
- Veis, A.; Schlueter, R.J. The macromolecular organization of dentine matrix collagen. I. Characterization of dentine collagen. Biochemistry 1964, 3, 1650–1657. [Google Scholar] [CrossRef]
- Almahdy, A.; Koller, G.; Sauro, S.; Bartsch, J.W.; Sherriff, M.; Watson, T.F.; Banerjee, A. Effects of MMP inhibitors incorporated within dental adhesives. J. Dent. Res. 2012, 91, 605–611. [Google Scholar] [CrossRef] [Green Version]
- Niu, L.N.; Zhang, L.; Jiao, K.; Li, F.; Ding, Y.X.; Wang, D.Y.; Wang, M.Q.; Tay, F.R.; Chen, J.H. Localization of MMP-2, MMP-9, TIMP-1, and TIMP-2 in human coronal dentine. J. Dent. 2011, 39, 536–542. [Google Scholar] [CrossRef]
- Juarez, Z.E.; Stinson, M.W. An extracellular protease of Streptococcus gordonii hydrolyzes type IV collagen and collagen analogues. Infect. Immun. 1999, 67, 271–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K. Collagenolytic proteases from bacteria. Appl. Microbiol. Biotechnol. 2004, 63, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Dufour, D.; Lévesque, C.M. Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J. Bacteriol. 2013, 195, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Burne, R.A. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J. Bacteriol. 2011, 193, 2826–2837. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Cvitkovitch, D.G.; Lévesque, C.M. Cell death in Streptococcus mutans biofilms: A link between CSP and extracellular DNA. FEMS Microbiol. Lett. 2009, 299, 261–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, A.J.; Woessner, J.F.; Rawlings, N.D. Handbook of Proteolytic Enzymes, Volume 1; Elsevier: Amsterdam, The Netherlands, 2012; Volume 1. [Google Scholar]
- Van Wart, H.E. Clostridium collagenases. In Handbook of Proteolytic Enzymes; Elsevier: Amsterdam, The Netherlands, 2004; pp. 416–419. [Google Scholar]
- Sulkala, M.; Tervahartiala, T.; Sorsa, T.; Larmas, M.; Salo, T.; Tjäderhane, L. Matrix metalloproteinase-8 (MMP-8) is the major collagenase in human dentin. Arch. Oral Biol. 2007, 52, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Yonejima, Y.; Tsujimoto, Y.; Suzuki, Y.; Watanabe, K. A thermostable collagenolytic protease with a very large molecular mass produced by thermophilic Bacillus sp. strain MO-1. Appl. Microbiol. Biotechnol. 2001, 57, 103–108. [Google Scholar] [PubMed]
- Tsuruoka, N.; Nakayama, T.; Ashida, M.; Hemmi, H.; Nakao, M.; Minakata, H.; Oyama, H.; Oda, K.; Nishino, T. Collagenolytic serine-carboxyl proteinase from Alicyclobacillus sendaiensis strain NTAP-1: Purification, characterization, gene cloning, and heterologous expression. Appl. Environ. Microbiol. 2003, 69, 162–169. [Google Scholar] [CrossRef] [Green Version]
- Svensater, G.; Welin, J.; Wilkins, J.C.; Beighton, D.; Hamilton, I.R. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 2001, 205, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.J.; Moore, J.C.; Lane, J.D.; Wilson, R.; Pribul, P.K.; Younes, Z.N.; Dobson, R.J.; Everest, P.; Reason, A.J.; Redfern, J.M.; et al. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 2002, 70, 1254–1259. [Google Scholar] [CrossRef] [Green Version]
- Kling, D.E.; Madoff, L.C.; Michel, J.L. Subcellular fractionation of group B Streptococcus. Biotechniques 1999, 27, 24–28. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, B.; Stewart, C.A.; McCulloch, C.A.; Santerre, J.P.; Cvitkovitch, D.G.; Finer, Y. Streptococcus mutans Proteases Degrade Dentinal Collagen. Dent. J. 2022, 10, 223. https://doi.org/10.3390/dj10120223
Huang B, Stewart CA, McCulloch CA, Santerre JP, Cvitkovitch DG, Finer Y. Streptococcus mutans Proteases Degrade Dentinal Collagen. Dentistry Journal. 2022; 10(12):223. https://doi.org/10.3390/dj10120223
Chicago/Turabian StyleHuang, Bo, Cameron A. Stewart, Christopher A. McCulloch, J. Paul Santerre, Dennis G. Cvitkovitch, and Yoav Finer. 2022. "Streptococcus mutans Proteases Degrade Dentinal Collagen" Dentistry Journal 10, no. 12: 223. https://doi.org/10.3390/dj10120223