Kombucha: Production and Microbiological Research †
Abstract
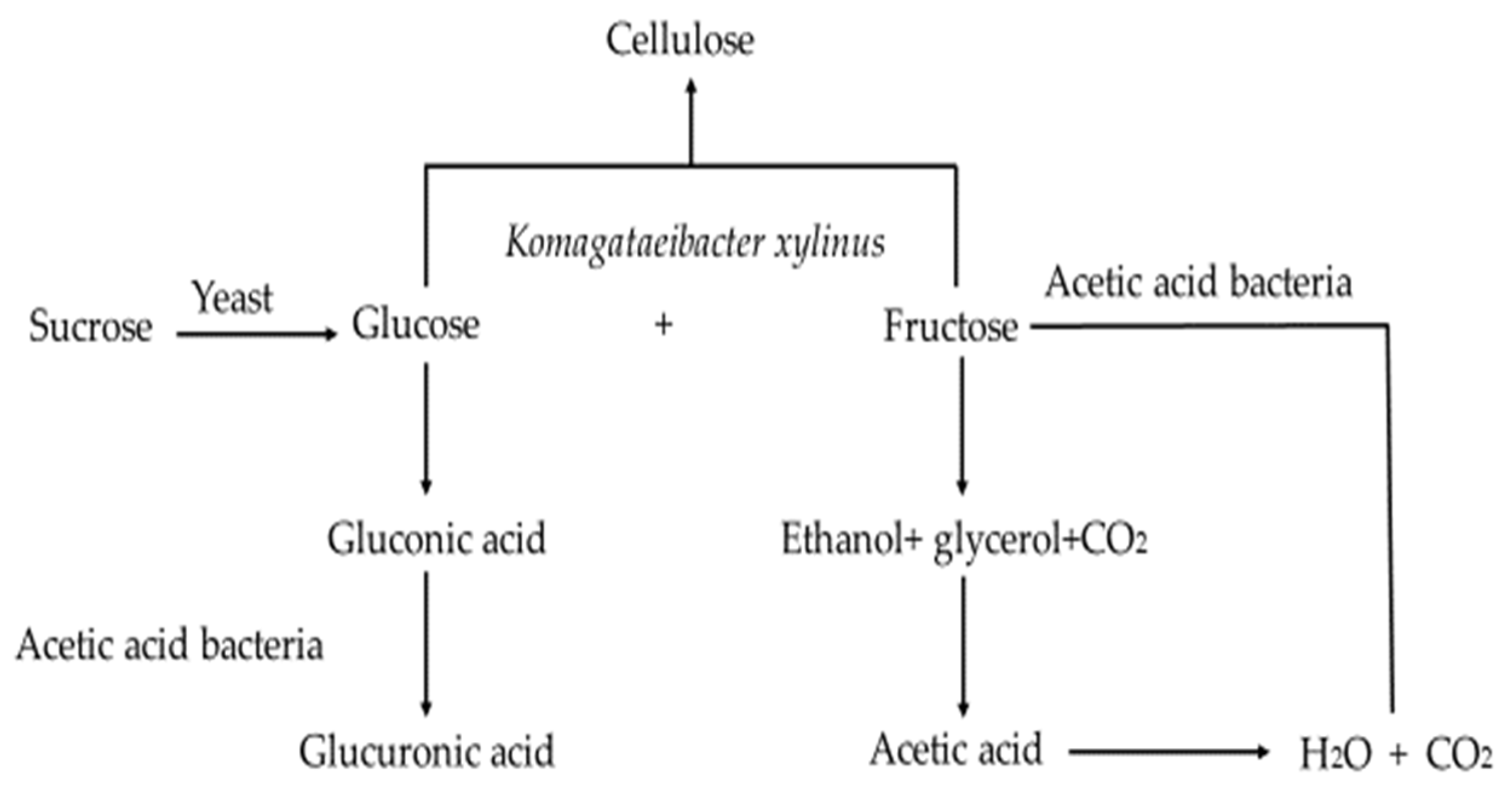
:1. Introduction
2. Microbiological Characteristics of Kombucha
2.1. Presence of Acetic Acid Bacteria in Fermented Foods
2.1.1. AAB Used in the Production of Fermented Products
2.1.2. Dominant Acetic Acid Bacteria Found in Kombucha
2.2. Lactic Acid Bacteria Isolated from Kombucha
2.3. Yeast Isolated from Kombucha
2.3.1. General Characteristics of Yeast
2.3.2. Dominant Yeast Present in Kombucha
3. Isolation of AAB and Yeast from Kombucha
3.1. Isolation, Enumeration and Preservation of AAB
3.2. Isolation and Enumeration of Yeast
4. Phenotypic Characterisation and Identification of Acetic Acid Bacteria and Yeast from Kombucha
4.1. Phenotypic Characterisation of AAB
4.2. Phenotypic Characterisation and Identification of Yeast from Kombucha
Identification of Yeast Using Commercial Kits
5. Genotypic Identification of AAB and Yeast from Kombucha
5.1. Genotypic Identification of AAB from Kombucha
5.2. Genotypic Identification of Yeast
6. Conclusions and Future Research on Kombucha
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dufresne, C.; Farnworth, E. Tea, Kombucha, and health: A review. Food Res. Int. 2000, 33, 409–421. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbasa, R.V.; Loncar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha Tea-Microbiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr. Rev. Food. Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, C.J.; Steinkraus, K.H.; Ledford, R.A. Kombucha, the fermented tea: Microbiology, composition, and claimed health effects. J. Food Prot. 2000, 63, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Joshi, V. Kombucha: Technology, microbiology, production, composition and therapeutic value. Int. J. Food Ferment. Technol. 2016, 6, 13–24. [Google Scholar] [CrossRef]
- Kim, J.; Adhikari, K. Current trends in kombucha: Marketing perspectives and the need for improved sensory research. Beverages 2020, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbial. 2014, 38, 171–178. [Google Scholar] [CrossRef]
- Coelho, R.M.D.; de Almeida, A.L.; do Amaral, R.Q.G.; da Mota, R.N.; de Sousa, P.H.M. Kombucha. Int. J. Gastron. Food Sci. 2020, 22, 100272. [Google Scholar] [CrossRef]
- Vargas, B.K.; Fabricio, M.F.; Ayub, M.A.Z. Health effects and probiotic and prebiotic potential of Kombucha: A bibliometric and systematic review. Food Biosci. 2021, 44, 101332. [Google Scholar] [CrossRef]
- Velićanski, A.; Cvetković, D.; Markov, S. Characteristics of Kombucha fermentation on medicinal herbs from Lamiaceae family. Rom. Biotechnol. Lett. 2013, 18, 8034–8042. [Google Scholar]
- Reiss, J. Influence of Different Sugars on the Metabolism of the Tea Fungus. Z. Für Lebensm.-Unters. Und-Forsch. 1994, 198, 258–261. [Google Scholar] [CrossRef]
- Steinkraus, K.H.; Shapiro, K.B.; Hotchkiss, J.H.; Mortlock, R.P. Investigations into the antibiotic activity of tea Fungus/Kombucha beverage. Acta Biotechnol. 1996, 16, 199–205. [Google Scholar] [CrossRef]
- Vijayaraghavan, R.; Singh, M.; Rao, P.V.; Bhattacharya, R.; Kumar, P.; Sugendran, K.; Kumar, O.; Pant, S.C.; Singh, R. Subacute (90 days) oral toxicity studies of Kombucha tea. Biomed. Environ. Sci. 2000, 13, 293–299. Available online: https://www.researchgate.net/profile/Pravin-Kumar-23/publication/11985545_Subacute_90_Days_Oral_Toxicity_Studies_of_Kombucha_Tea/links/5719be5608aed8a339e7057f (accessed on 20 September 2022).
- Kapp, J.M.; Sumner, W. Kombucha: A systematic review of the empirical evidence of human health benefits. Ann. Epidemiol. 2019, 30, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Dutta, H.; Paul, S.K. Kombucha drink: Production, quality, and safety aspects. In Production and Management of Beverages; Woodhead Publishing: Sawston, UK, 2020; pp. 259–288. [Google Scholar] [CrossRef]
- Laureys, D.; Britton, S.J.; De Clippeleer, J. Kombucha tea fermentation: A review. J. Am. Soc. Brew. Chem. 2020, 78, 165–174. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.P.; Taillandier, P. Understanding Kombucha Tea Fermentation: A Review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha tea fermentation: Microbial and biochemical dynamics. Int. J. Food Microbiol. 2016, 220, 63–72. [Google Scholar] [CrossRef]
- Markov, S.L.; Jerinić, V.M.; Cvetković, D.D.; Lončar, E.S.; Malbaša, R.V. Kombucha-functional beverage: Composition, characteristics and process of biotransformation. Hem. Ind. 2003, 57, 456–462. [Google Scholar] [CrossRef] [Green Version]
- Sievers, M.; Lanini, C.; Weber, A.; SchulerSchmid, U.; Teuber, M. Microbiology and fermentation balance in a kombucha beverage obtained from a tea fungus fermentation. Syst. Appl. Microbiol. 1996, 18, 590–594. [Google Scholar] [CrossRef]
- Ray, R.C. Acetic Acid Bacteria: Prospective Applications in Food Biotechnology. In Fermented Foods; Part I; CRC Press: Boca Raton, FL, USA, 2016; pp. 107–121. [Google Scholar]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of acetic acid bacteria and their role in vinegar and fermented beveragesCompr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef] [Green Version]
- Karabiyikli, S.; Sengun, I. Beneficial effects of acetic acid bacteria and their food products. In Acetic Acid Bacteria; CRC Press: Boca Raton, FL, USA, 2017; pp. 321–342. [Google Scholar]
- Jarrell, J.; Cal, T.; Bennett, J. The Kombucha Consortia of Yeasts and Bacteria. Mycologist 2000, 14, 166–170. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaśa, R.V.; Sathishkumar, M. Kombucha Tea: Metabolites. In Fungal Metabolites; Springer: Cham, Switzerland, 2017; pp. 965–978. [Google Scholar] [CrossRef]
- Fontana, J.D.; Franco, V.C.; De Souza, S.J.; Lyra, I.N.; De Souza, A.M. Nature of Plant Stimulators in the Production of Acetobacter xylinum (“Tea Fungus”) Biofilm Used in Skin Therapy. Appl. Biochem. Biotechnol 1991, 28, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, R.; Chen, P.-N.; Hsieh, Y.-S.; Prabhakaran, K.; Pitchai, P.; Marimuthu, S.; Thangaraj, P.; Swaminathan, K.; Yun, S.E. Effect of solvent fractions of kombucha tea on viability and invasiveness of cancer cells—Characterization of dimethyl 2-(2-hydroxy-2-methoxypropylidine) malonate and vitexin. Indian J. Biotechnol. 2011, 10, 62–75. [Google Scholar]
- Liu, C.-H.; Hsu, W.-H.; Lee, F.-L.; Liao, C.-C. The isolation and identification of microbes from a fermented tea beverage, Haipao, and their interactions during Haipao fermentation. Food Microbiol. 1996, 13, 407–415. [Google Scholar] [CrossRef]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Isolation and characterisation of dominant acetic acid bacteria and yeast isolated from Kombucha samples at point of sale in New Zealand. Curr. Res. Food Sci. 2022, 5, 835–844. [Google Scholar] [CrossRef]
- Dutta, D.; Gachhui, R. Nitrogen-fixing and cellulose-producing Gluconacetobacter kombuchae sp. nov., isolated from Kombucha tea. Int. J. Syst. Evol. Microbiol. 2007, 57, 353–357. [Google Scholar] [CrossRef]
- Mukadam, T.A.; Punjabi, K.; Deshpande, S.D.; Vaidya, S.P.; Chowdhary, A.S. Isolation and characterization of bacteria and yeast from Kombucha tea. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 32–41. [Google Scholar] [CrossRef]
- Jacek, P.; da Silva, F.A.S.; Dourado, F.; Bielecki, S.; Gama, M. Optimization and characterization of bacterial nanocellulose produced by Komagataeibacter rhaeticus K3. Carbohydr. Polym. Technol. Appl. 2021, 2, 100022. [Google Scholar] [CrossRef]
- Trovatti, E.; Serafim, L.S.; Freire, C.S.; Silvestre, A.J.; Neto, C.P. Gluconacetobacter sacchari: An Efficient Bacterial Cellulose Cell-factory. Carbohydr. Polym. 2011, 86, 1417–1420. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, F.; Ji, B.; Li, B.; Luo, Y.; Yang, L.; Li, T. Symbiosis between microorganisms from kombucha and kefir: Potential significance to the enhancement of kombucha function. Appl. Biochem. Biotechnol. 2010, 160, 446–455. [Google Scholar] [CrossRef]
- Dutta, D.; Gachhui, R. Novel nitrogen-fixing Acetobacter nitrogenifigens sp. nov., isolated from Kombucha tea. Int. J. Syst. Evol. Microbiol. 2006, 56, 1899–1903. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.K.; Dong, N.T.; Nguyen, H.T.; Le, P.H. Lactic acid bacteria: Promising supplements for enhancing the biological activities of kombucha. Springerplus 2015, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Diguta, C.F.; Nitoi, G.D.; Matei, F.; Luta, G.; Cornea, C.P. The Biotechnological Potential of Pediococcus spp. Isolated from Kombucha Microbial Consortium. Foods 2020, 9, 1780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, B.; Wu, W.; Wang, R.; Yang, Z.; Zhang, D.; Tian, W. Hepatoprotective effects of kombucha tea: Identification of functional strains and quantification of functional components. J. Sci. Food Agric. 2014, 94, 265–272. [Google Scholar] [CrossRef]
- Pei, J.; Jin, W.; Abd El-Aty, A.; Baranenko, D.A.; Gou, X.; Zhang, H.; Geng, J.; Jiang, L.; Chen, D.; Yue, T. Isolation, purification, and structural identification of a new bacteriocin made by Lactobacillus plantarum found in conventional kombucha. Food Control 2020, 110, 106923. [Google Scholar] [CrossRef]
- Yang, J.; Lagishetty, V.; Kurnia, P.; Henning, S.M.; Ahdoot, A.I.; Jacobs, J.P. Microbial and chemical profiles of commercial Kombucha products. Nutrients 2022, 14, 670. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, M.; Justine, S.; Filofteia, D.C.; Petruta, C.C.; Gabriela, L.; Roxana, U.E.; Florentina, M.; Camelia Filofteia, D.; Călina Petruța, C.; Gabriela, L. Lactic acid bacteria strains isolated from Kombucha with potential probiotic effect. Roman. Biotechnol. Lett. 2018, 23, 13592–13598. [Google Scholar]
- Deak, T.; Beuchat, L.R. Comparison of the SIM, API 20C, and ID 32C Systems for Identification of Yeasts Isolated from Fruit Juice Concentrates and Beverages. J. Food Prot. 1993, 56, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Lachance, M.-A.; Yurkov, A. Yeasts in Natural Ecosystems: Diversity; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Bekatorou, A.; Psarianos, C.; Koutinas, A.A. Production of food grade yeasts. Food Technol. Biotechnol. 2006, 44, 407–415. [Google Scholar]
- Fleet, G. The commercial and community significance of yeasts in food and beverage production. In Yeasts in Food and Beverages; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1–12. [Google Scholar] [CrossRef]
- Lachance, M.-A.; Starmer, W.T.; Rosa, C.A.; Bowles, J.M.; Barker, J.S.F.; Janzen, D.H. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001, 1, 1–8. [Google Scholar] [CrossRef]
- Teoh, A.L.; Heard, G.; Cox, J. Yeast ecology of Kombucha fermentation. Int. J. Food Microbiol. 2004, 95, 119–126. [Google Scholar] [CrossRef]
- Matei, B.; Diguță, C.F.; Popa, O.; Cornea, C.P.; Matei, F. Molecular identification of yeast isolated from different kombucha sources. Ann. Univ. Dunarea Jos Galati. Fascicle VI-Food Technol. 2018, 42, 17–25. [Google Scholar]
- Thomas, D.; Davenport, R. Zygosaccharomyces bailii—A profile of characteristics and spoilage activities. Food Microbiol. 1985, 2, 157–169. [Google Scholar] [CrossRef]
- Escott, C.; Del Fresno, J.M.; Loira, I.; Morata, A.; Suárez-Lepe, J.A. Zygosaccharomyces rouxii: Control strategies and applications in food and winemaking. Fermentation 2018, 4, 69. [Google Scholar] [CrossRef] [Green Version]
- Loira, I.; Morata, A.; Palomero, F.; González, C.; Suárez-Lepe, J.A. Schizosaccharomyces pombe: A promising biotechnology for modulating wine composition. Fermentation 2018, 4, 70. [Google Scholar] [CrossRef] [Green Version]
- Vejarano, R. Saccharomycodes ludwigii, control and potential uses in winemaking processes. Fermentation 2018, 4, 71. [Google Scholar] [CrossRef] [Green Version]
- Choonut, A.; Saejong, M.; Sangkharak, K. The production of ethanol and hydrogen from pineapple peel by Saccharomyces cerevisiae and Enterobacter aerogenes. Energy Procedia 2014, 52, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Agnolucci, M.; Tirelli, A.; Cocolin, L.; Toffanin, A. Brettanomyces bruxellensis yeasts: Impact on wine and winemaking. World J. Microbiol. Biotechnol. 2017, 33, 180. [Google Scholar] [CrossRef]
- Mayser, P.; Fromme, S.; Leitzmann, C.; Grunder, K. The yeast spectrum of the ‘tea fungus Kombucha’. Mycoses 1995, 38, 289–295. [Google Scholar] [CrossRef]
- Ramadani, A.; Abulreesh, H. Isolation and Identification of Yeast Flora in Local Kombucha Sample: Al Nabtah. Umm Al Qura Univ. J. App. Sci. 2010, 2, 42–51. [Google Scholar]
- Kozaki, M.; Kakuo, K.; Koizumi, A. Microorganisms of zoogloeal mats formed in tea decoction. Food Hyg. Saf. Sci. (Shokuhin Eiseigaku Zasshi) 1972, 13, 89–96. [Google Scholar] [CrossRef]
- Urbahillah, A.; Jayus, J.; Nurhayati, N. Improving SCOBY starter using co-culture of tapai and bakery yeast. Biodivers. J. Biol. Divers. 2021, 22, 4617–4624. [Google Scholar] [CrossRef]
- Bellut, K.; Krogerus, K.; Arendt, E.K. Lachancea fermentati strains isolated from kombucha: Fundamental insights, and practical application in low alcohol beer brewing. Front. Microbiol. 2020, 11, 764. [Google Scholar] [CrossRef] [Green Version]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar-Powers, E. Zygosaccharomyces kombuchaensis, a new ascosporogenous yeast from ‘Kombucha tea’. FEMS Yeast Res. 2001, 1, 133–138. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P. Genera and Species in Acetic Acid Bacteria. Int. J. Food Microbiol. 2008, 125, 15–24. [Google Scholar] [CrossRef]
- Gomes, R.J.; Borges, M.F.; Rosa, M.F.; Castro-Gomez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef]
- Sengun, I.Y.; Karabiyikli, S. Importance of acetic acid bacteria in food industry. Food Control 2011, 22, 647–656. [Google Scholar] [CrossRef]
- Sievers, M. Family II. Acetobacteraceae. Bergey’s Man. Systmatic Bacteriol. 2005, 2, 41–50. [Google Scholar]
- Gullo, M.; Giudici, P. Acetic acid bacteria in traditional balsamic vinegar: Phenotypic traits relevant for starter cultures selection. Int. J. Food Microbiol. 2008, 125, 46–53. [Google Scholar] [CrossRef]
- Cleenwerck, I.; Camu, N.; Engelbeen, K.; De Winter, T.; Vandemeulebroecke, K.; De Vos, P.; De Vuyst, L. Acetobacter ghanensis sp. nov., a novel acetic acid bacterium isolated from traditional heap fermentations of Ghanaian cocoa beans. Int. J. Syst. Evolut. Microbiol. 2007, 57, 1647–1652. [Google Scholar] [CrossRef]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Carr, J.G.; Passmore, S.M. Methods for identifying acetic acid bacteria. Identif. Methods Microbiol. 1979, 14, 33–47. [Google Scholar]
- Cleenwerck, I.; De Wachter, M.; Gonzalez, A.; De Vuyst, L.; De Vos, P. Differentiation of species of the family Acetobacteraceae by AFLP DNA fingerprinting: Gluconacetobacter kombuchae is a later heterotypic synonym of Gluconacetobacter hansenii. Int. J. Syst. Evol. Microbiol. 2009, 59, 1771–1786. [Google Scholar] [CrossRef]
- Wu, J.J.; Ma, Y.K.; Zhang, F.F.; Chen, F.S. Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “Shanxi aged vinegar”, a traditional Chinese vinegar. Food Microbiol. 2012, 30, 289–297. [Google Scholar] [CrossRef]
- Entani, E.; Ohmori, S.; Masai, H.; Suzuki, K.-I. Acetobacter polyoxogenes sp. nov., a new species of an acetic acid bacterium useful for producing vinegar with high acidity. J. Gen. Appl. Microbiol. 1985, 31, 475–490. [Google Scholar] [CrossRef]
- Hestrin, S.; Schramm, M. Synthesis of cellulose by Acetobacter xylinum. 2. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem. J. 1954, 58, 345. [Google Scholar] [CrossRef]
- Ohmori, S.; Masai, H.; Arima, K.; Beppu, T. Isolation and identification of acetic acid bacteria for submerged acetic acid fermentation at high temperature. Agric. Biol. Chem. 1980, 44, 2901–2906. [Google Scholar] [CrossRef] [Green Version]
- Fugelsang, K.C. Wine Microbiology, 2nd ed.; Ripol Classic: Moscow, Russia, 2007. [Google Scholar]
- Bartowsky, E.J.; Henschke, P.A. Acetic acid bacteria spoilage of bottled red wine—A review. Int. J. Food Microbiol. 2008, 125, 60–70. [Google Scholar] [CrossRef]
- Wiegand, C.; Klemm, D. Influence of protective agents for preservation of Gluconacetobacter xylinus on its cellulose production. Cellulose 2006, 13, 485–492. [Google Scholar] [CrossRef]
- Da Silva, N.; Taniwaki, M.H.; Junqueira, V.C.A.; de Arruda Silveira, N.F.; Okazaki, M.M.; Gomes, R.A.R. Microbiological Examination Methods of Food and Water: A Laboratory Manual; CRC Press: Boca Raton, FL, USA, 2018; p. 25. [Google Scholar]
- Deak, T. Handbook of Food Spoilage Yeasts, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2007; pp. 204–205. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts; Elsevier: Amsterdam, The Netherlands, 2011; pp. 87–110. [Google Scholar] [CrossRef]
- Schifferdecker, A.J.; Dashko, S.; Ishchuk, O.P.; Piskur, J. The Wine and Beer Yeast Dekkera bruxellensis. Yeast 2014, 31, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Malimas, T.; Vu, H.T.L.; Muramatsu, Y.; Yukphan, P.; Tanasupawat, S. Systematics of acetic acid bacteria. In Acetic Acid Bacteria, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 3–43. [Google Scholar]
- Frateur, J. Essai Sur La Systematique Des Acetobacters. In Extrait de La CeUuZe 63; fascicule 3; Joseph Van In & Cie: Lierre, Belgium, 1950. [Google Scholar]
- Yamada, Y.; Okada, Y.; Kondo, K. Isolation and characterization of “polarly flagellated intermediate strains” in acetic acid bacteria. J. Gen. Appl. Microbiol. 1976, 22, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frebort, I.; Frebortova, J. Acetic acid bacteria: A group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Fung, D. Conventional and rapid methods for yeast identification. Crit. Rev. Microbiol. 1987, 14, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Robert, V. Yeasts in Food, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Sangeetha, J.; Thangadurai, D. Staining techniques and biochemical methods for the identification of fungi. In Laboratory Protocols in Fungal Biology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 237–257. [Google Scholar] [CrossRef]
- St Germain, G.; Beauchesne, D. Evaluation of the MicroScan rapid yeast identification panel. J. Clin. Microbiol. 1991, 29, 2296–2299. [Google Scholar] [CrossRef] [Green Version]
- Fricker-Hidalgo, H.; Lebeau, B.; Kervroedan, P.; Faure, O.; Ambroise-Thomas, P.; Grillot, R. Auxacolor, a new commercial system for yeast identification: Evaluation of 182 strains comparatively with ID 32C. Proc. Ann. Biol. Clin. 1995, 53, 221–225. [Google Scholar]
- Ramani, R.; Gromadzki, S.; Pincus, D.H.; Salkin, I.F.; Chaturvedi, V. Efficacy of API 20C and ID 32C Systems for Identification of Common and Rare Clinical Yeast Isolates. J. Clin. Microbiol. 1998, 36, 3396–3398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milan, E.P.; Malheiros, E.S.; Fischman, O.; Colombo, A.L. Evaluation of the AUXACOLOR system for the identification of clinical yeast isolates. Mycopathologia 1997, 137, 153–157. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Stockman, L.; Roberts, G.; Pincus, D.; Pollack, J.; Marler, J. Comparison of RapID yeast plus system with API 20C system for identification of common, new, and emerging yeast pathogens. J. Clin. Microbiol. 1998, 36, 883–886. [Google Scholar] [CrossRef] [Green Version]
- Bowman, P.; Ahearn, D. Evaluation of the Uni-Yeast-Tek kit for the identification of medically important yeasts. J. Clin. Microbiol. 1975, 2, 354–358. [Google Scholar] [CrossRef]
- Land, G.; Vinton, E.; Adcock, G.; Hopkins, J. Improved auxanographic method for yeast assimilations: A comparison with other approaches. J Clin. Microbiol. 1975, 2, 206–217. [Google Scholar] [CrossRef]
- Aubertine, C.; Rivera, M.; Rohan, S.; Larone, D. Comparative study of the new colorimetric VITEK 2 yeast identification card versus the older fluorometric card and of CHROMagar Candida as a source medium with the new card. J. Clin. Microbiol. 2006, 44, 227–228. [Google Scholar] [CrossRef] [Green Version]
- Abd El-Salam, S.S. 16S rRNA gene sequence detection of acetic acid bacteria isolated from tea Kombucha. N. Y. Sci. J. 2012, 5, 55–61. [Google Scholar]
- Andrés-Barrao, C.; Barja, F.; Pérez, R.O.; Chappuis, M.-L.; Braito, S. Identification Techniques of Acetic Acid Bacteria: Comparison between MALDI-TOF MS and Molecular Biology Techniques. In Acetic Acid Bacteria; CRC Press: Boca Raton, FL, USA, 2017; pp. 162–192. [Google Scholar]
- Rasmussen, H.B. Restriction fragment length polymorphism analysis of PCR-amplified fragments (PCR-RFLP) and gel electrophoresis-valuable tool for genotyping and genetic fingerprinting. In Gel Electrophoresis-Principles and Basics; InTechopen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef] [Green Version]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.; Curtin, C. Microbial composition of SCOBY starter cultures used by commercial kombucha brewers in North America. Microorganisms 2021, 9, 1060. [Google Scholar] [CrossRef]
- Reva, O.N.; Zaets, I.E.; Ovcharenko, L.P.; Kukharenko, O.E.; Shpylova, S.P.; Podolich, O.V.; de Vera, J.P.; Kozyrovska, N.O. Metabarcoding of the kombucha microbial community grown in different microenvironments. AMB Express 2015, 5, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93, fix048. [Google Scholar] [CrossRef]
- Savary, O.; Mounier, J.; Thierry, A.; Poirier, E.; Jourdren, J.; Maillard, M.-B.; Penland, M.; Decamps, C.; Coton, E.; Coton, M. Tailor-made microbial consortium for Kombucha fermentation: Microbiota-induced biochemical changes and biofilm formation. Food Res. Int. 2021, 147, 110549. [Google Scholar] [CrossRef]
- Pradhan, S.; Prabhakar, M.R.; Karthika Parvathy, K.; Dey, B.; Jayaraman, S.; Behera, B.; Paramasivan, B. Metagenomic and physicochemical analysis of Kombucha beverage produced from tea waste. J. Food Sci. Technol. 2022, 59, 1–9. [Google Scholar] [CrossRef]
- Arikan, M.; Mitchell, A.L.; Finn, R.D.; Gurel, F. Microbial composition of Kombucha determined using amplicon sequencing and shotgun metagenomics. J. Food Sci. 2020, 85, 455–464. [Google Scholar] [CrossRef]
- Kumar, N.S.; Gurusubramanian, G. Random amplified polymorphic DNA (RAPD) markers and its applications. Sci. Vis. 2011, 11, 116–124. [Google Scholar]
- Gaggìa, F.; Baffoni, L.; Galiano, M.; Nielsen, D.S.; Jakobsen, R.R.; Castro-Mejía, J.L.; Bosi, S.; Truzzi, F.; Musumeci, F.; Dinelli, G. Kombucha beverage from green, black and rooibos teas: A comparative study looking at microbiology, chemistry and antioxidant activity. Nutrients 2018, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Hierro, N.; Poblet, M.; Rozes, N.; Mas, A.; Guillamón, J. Application of molecular methods for the differentiation of acetic acid bacteria in a red wine fermentation. J. Appl. Microbiol. 2004, 96, 853–860. [Google Scholar] [CrossRef] [PubMed]
- La China, S.; Vero, L.D.; Anguluri, K.; Brugnoli, M.; Mamlouk, D.; Gullo, M. Kombucha tea as a reservoir of cellulose producing bacteria: Assessing diversity among Komagataeibacter isolates. Appl. Sci. 2021, 11, 1595. [Google Scholar] [CrossRef]
- Semjonovs, P.; Ruklisha, M.; Paegle, L.; Saka, M.; Treimane, R.; Skute, M.; Rozenberga, L.; Vikele, L.; Sabovics, M.; Cleenwerck, I. Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Appl. Microbiol. Biotechnol. 2017, 101, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and Phylogeny of Ascomycetous Yeasts from Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial Sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Pincus, D.; Orenga, S.; Chatellier, S. Yeast identification—Past, present, and future methods. Med. Mycol. 2007, 45, 97–121. [Google Scholar] [CrossRef] [Green Version]
- Mackay, I.M. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef]
| Brand | Origin | Packaging | Flavour(s) | Storage Conditions |
|---|---|---|---|---|
| Remedy | Australia | 330 mL (glass bottles) 250 mL (cans) 1.25 L (plastic bottles) | Ginger lemon, Raspberry Lemonade, Cola, Wildberry, Mango Passion, Passionfruit, Apple Crisp, Peach | Chilled/ambient temperature |
| Batchwell | New Zealand | 375 mL (glass bottles) | Braeburn apple, Pineapple and Ginger, Motueka hops, Ginger and Turmeric, Early grey | Chilled |
| Daily Organics | New Zealand | 200 mL (glass bottles) 1 L (glass bottles) | Original Kombucha, Chai spices and ginger, Lemon and ginger | Chilled |
| LO BROS | New Zealand | 330 mL (glass bottles) 250 mL (cans) 750 mL (glass bottles) | Feijoa, Raspberry and lemon, Orange and mango, Ginger and lemon, Mango, Ginger and turmeric, Blueberry, Passionfruit, Cola, Ginger beer, Lemon lime and bitters, Pineapple and lime | Chilled/ ambient temperature |
| MAMA’S Brew Shop | New Zealand | 375 mL (glass bottles) 330 mL (cans) | Lemongrass and ginger, Lavender and hibiscus | Chilled |
| Karma Drinks | New Zealand | 330 mL (glass bottles) | Lemon and ginger, Raspberry and lemon, Mango and passionfruit, Cherryand berry | Chilled |
| Good Buzz | New Zealand | 250 mL (cans) 328 mL (glass bottles) 375 mL (glass bottles) 888 mL (glass bottles) | Passionfruit and guava, Blueberry and peach, Pineapple and mango, Feijoa, Raspberry and lemon, Mango, Gisborne lemon and Manuka leaf, Hawkes Bay, peach and kawakawa | Chilled or ambient temperature |
| Species | Region | Reference |
|---|---|---|
| Lacticaseibacillus casei | NS | [35] |
| Lactiplantibacillus plantarum | China | [38] |
| Lactobacillus nagelii | United State of America (USA) | [39] |
| Lactobacillus rhamnosus | NS | [5] |
| Lactobacillus mali | USA | [39] |
| Pediococcus (P.) pentosaceus | Romania | [40] |
| P.acidiliactici | Romania | [36] |
| Species | Morphology | Characteristics |
|---|---|---|
| Zygosaccharomyces (Z.) bailii [1] | White to cream colonies with brownish top, cylindrical or ellipsoidal shape, (3.5–6.0) × (4.5–11.5) μm in size | Tolerant to organic acids, Forms acetic acid, heat tolerance < 75 °C Growth pH > 2 and < 7 [48] |
| Zycosaccharomyces (Z.) rouxii | White to cream smooth colonies, round or oval shape | High osmotic stress and salt/sugar tolerant, grows under low oxygen and low water activity [49] |
| Schizosaccharomyces (S.) pombe | Cream to tan, butyrous colonies, rod-shaped | Can convert malic acid to ethanol, high resistance to low water activity, low pH and wide range of temperature environments, highly sugar content tolerant [50] |
| Saccharomycodes (S.) ludwigi [10] | Cream, butyrous colonies, elongated shape, and swelling in the centre | Resistant to pressurised carbon dioxide, high sugar tolerant [51] |
| S. cerevisiae [27] | White to cream, butyrous colonies, spherical or ovoid shape, 2.5–10.0 µm (diameter) | Can convert glucose to ethanol, high ethanol tolerance, rapid fermentation rate [52] |
| Brettanomyces (B.) bruxellensis [24,47] | Distinctive elongated shape, 2.5–10.0 µm (diameter) | Can produce high amounts of acetic acid and ethanol under aerobic conditions, high ethanol concentration (up to 15%), able to grow under low pH and oxygen environment, high efficiency to utilise nitrogen sources [53] |
| Species | Country | References |
|---|---|---|
| Brettanomyces (B.) anamalus | New Zealand | [28] |
| B. lambicus | Germany | [54] |
| B. custerisii | Germany | [54] |
| B. intermedius | NS | [24] |
| B. claussenii | NS | [24] |
| Candida (C.) albican C. colleculosa C. kefir C. krusei | Japan Saudi Arabia Saudi Arabia Saudi Arabia | [24] [55] [55] [55] |
| C. guilliermondii C. obtuse C. stellata | Japan/Saudi Arabia Formosa Australia | [56] [46] [46] |
| C. famata | Indonesia | [57] |
| C. magnoliae | Indonesia | [57] |
| Dekkera (D.) bruxelensis | New Zealand | [28] |
| Hanseniaspora (H.) valbyensis | New Zealand | [28] |
| Lachancea (L.) fermentati | USA | [58] |
| Kloeckera (K.) apiculata Kluyceromyces (K.) africanus | NS NS | [24] [24] |
| Pichia (P.) fermentans | NS | [4] |
| P. membranefaciens | NS | [26] |
| P. kudriavzevii | New Zealand | [28] |
| Torulaspora (T.) delbrueckii | Australia | [46] |
| Torulopsis (T.)famata | Japan | [56] |
| Zygosaccharomyces (Z.) kombucahensis | Russia | [59] |
| Characteristics | Acetobacter | Gluconacetobacter | Gluconobacter | Komagataeibacter |
|---|---|---|---|---|
| Cell shape | Ellipsoidal to rods | Ellipsoidal to rods | Ellipsoidal to Rods | Coccoid to rods |
| Cell size (μm) | 0.4–1.0 × 1.0–3.0 | 0.5–0.9 × 1.0–2.0 | 0.6–1.0 × 1.0–3.0 | 0.6–0.8 × 1.0–3.0 |
| Colony appearance | Creamy to brown | Light brown to brownish | Smooth, entire, shiny, white, pink or brown | Raised, convex to umbonate, smooth to rough, entire to irregular |
| Catalase | + | + | + | + |
| Gram staining | Gram-negative | Gram-negative | Gram-negative | Gram-negative |
| Oxidase | − | − | − | − |
| Motility | Motile or non-motile | Motile or non-motile | Non-motile | No |
| Flagellation | Peritrichous | peritrichous | polar | No |
| Oxidation of ethanol to acetic acid | + | + | + | + |
| Oxidation of acetic acid to CO2 and water | + | + | − | + |
| Oxidation of lactate/acetate | + | + | − | + |
| Production of cellulose | − | ± | − | ± |
| Growth on 0.35% acetic acid | + | + | + | + |
| Growth in the presence of 1% KNO3 | − | − | − | ND |
| Growth on methanol | ± | − | − | ND |
| Growth in 30% D-glucose | − | ± | ± | ND |
| Ketogenesis (dihydroxyacetone) from glycerol | + | + | + | + |
| Acid production from: Glycerol D-Mannitol Raffinose Sorbitol | ± − − | + − − − | + + − + | ND − ND − |
| Production of DHA from glycerol | ± | ± | + | + |
| Production of levan-like polysaccharide from sucrose | ± | − | − | − |
| Ubiquinone type | Q9 | Q10 | Q10 | Q10 |
| G + C content (mol %) | 50.5–60.3 | 55–67 | 52–64 | 56–64 |
| Sources | Flowers, fruits, palm wine, vinegar, kefir | Rhizosphere of coffee plants, roots and stem of sugar cane | Strawberry, grape and spoiled jackfruit and sugar-rich environments | Kombucha, vinegar, wine vinegar |
| Different Reproduction Mode of Yeast | Morphology Characteristics of Yeast |
|---|---|
| Vegetative or asexual reproduction | Budding: new cell is produced on the surface of parent cell and then separate Fission: an asexual cell is produced by a septum grown inward from cell wall to halve the long axis of the cell. Blastoconidiation: a mother cell of stalk-like tubular sterigmata produce a terminal conidium |
| Sexual reproduction in ascomycetous yeast | Parent cell-bud conjugation Gametangial conjugation Heterothallism conjugation Conjugation between hyphae |
| Sexual reproduction in basidiomycetous yeast | Budding haplophase Dikaryotic hyphal phase or self-spore forming diplophase |
| Physiological Test | Biochemical Test |
|---|---|
| Assimilation of carbon and nitrogen sources | Diazonium Blue B reaction |
| Fermentation of carbohydrates | Urease test |
| Growth at different temperature | |
| Growth in vitamin-free medium | |
| Growth in high osmotic pressure condition | |
| Starch hydrolysis activity |
| Commercial System | Description of Tests and Controls Included in the Kits | Incubation Condition | Accuracy (%) | Reference |
|---|---|---|---|---|
| API 20 C | 19 carbon assimilation test and 1 control test in 20 strips | 30 °C for 72 h | 98.9 | [88] |
| API Candida | 5 carbohydrate and 7 enzymes colorimetric test in 10 strips | 35 °C for 18–24 h | 97.4 | [89] |
| API 32 C | 29 assimilation tests (carbohydrate, organic acids, and amino acids); 1 negative control, 1 susceptibility test (cycloheximide) and 1 colorimetric test (esculin) in 32 wells. Includes 63 different species in database | 30 °C for 48 h | 92 | [90] |
| Auxacolor system | 13 carbohydrate tests with bromocresol purple, test for cycloheximide resistance and phenoloxidase production in 16 wells. | 37 °C for 48 h | 79.4 | [91] |
| RapID Yeast Plus system | 5 carbon assimilation tests and 13 enzymatic hydrolysis substrate tests | 30 °C for 4 h | 96 | [92] |
| The Uni-Yeast-Tek (UYT) system | 7 carbon assimilation tests, urease, Nitrate and corn meal with Tween 80 agar | 22–26 °C for 2–10 days | 99.8 | [93] |
| MicroScan yeast identification panel | 13 aminopeptidase, 3 carbohydrates, 9 glycosidase, phosphatase and urease tests. | 37 °C for 4 h | 86.9 | [94] |
| VITEK 2 YST | 4 aminopeptidase, 25 carbohydrate, esculin, 3 glycosidase, nitrate, 2 nitrogen, 9 organic acid, and urea tests | 35 °C for 18 h | 94.8 | [95] |
| Technique | Level | Advantage | Disadvantage |
|---|---|---|---|
| PCR-RFLP | Species | Rapid, cheap, and easy to set up; suitable for genotyping of AAB isolates | Difficult to identify small insertions and expensive, unable to discriminate closely related species |
| DGGE | Species | Rapid and cost-effective; suitable for estimation of AAB diversity | Cannot discriminate closely related species. |
| Real-time PCR | Species | Rapid, reliable and quantitative; suitable for comparing microbial abundance. | Complex and expensive |
| RAPD | Strain | Quick and simple | Low reproducibility, as the quality and concentration of template DNA influence the results |
| ALFP | Strain | Can be used for any DNA samples of any origin, and reveal multiple polymorphic bands in one lane. | Complex and sensitive |
| Method | Microorganism | Primer Sequence (5′–3′) | Reference |
|---|---|---|---|
| 16S rRNA, Sanger sequencing | Gluconacetobacter; | 27F, AGAGTTTGATCMTGGCTCAG | [107] |
| Komagataeibacter; | 1494R, TGACTGACTGAGGYTACCTTGTTACGACTT | ||
| Gluconacetobacter; | fD1, CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG | [29] | |
| kombuchae sp. nov.; | rD1, CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC | ||
| Acetobacter aceti; | 16S d, GCTGGCGGCATGCTTAACACA | [96] | |
| 16S r, GCAGGTGATCCAGCCGCA | |||
| 16S rRNA V1-V3 region, Illumina MiSeq | Bacterial communities | Forward, TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG | [108] |
| Reverse, GTVTVGTGGGCTCGGAGATGTGTATAAGAGACAG | |||
| 16S rRNA V3-V4 region, Illumina MiSeq | Bacterial communities | Forward, CCTACGGGNGGCWGCAG | [102] [109] |
| Reverse, GACTACHVGGGTATCTAATCC | |||
| 16S rRNA, Real-time PCR | Bacterial abundance | 926f, AAACTCAAKGAATTGACGG | [100] |
| 1062r, CTCACRRCACGAGCTGAC | |||
| 16S rRNA, T-RFLP | Bacterial communities | 27F, AGAGTTTGATCMTGGCTCAG | [17] |
| 1525R, AAGGAGGTGATCCAGCC | |||
| RAPD | Komagataeibacter spp. | M13, GAGGGTGGCGGTTCT | [104] |
| AFLP | Komagataeibacter rhaeticus | A03, GACTGCGTACAGGCCCCG | [110] |
| T03, CGATGAGTCCTGACCGAG | |||
| REP-PCR | G. oxydans; A. aceti | REPIR-I, IIICGICGICATCIGGC | [105] |
| REP2-I, ICGICTTATCIGGCCTAC | |||
| ERIC-PCR | G. oxydans; A. aceti | ERIC1R, ATGTAAGCTCCTGGGGATTCAC | [105] |
| ERIC2, AAGTAAGTGACTGGGGTGAGCG | |||
| Shotgun metagenomic sequencing | Bacterial and fungal communities | [101] [102] |
| Method | Microorganism | Primer Sequence (5′–3′) | Reference |
|---|---|---|---|
| 26S rDNA, D1/D2 domain | Brettanomyces/Dekkera; Pichia; B. bruxellensis; | NL1, GCATATCAATAAGCGGAGGAAAAG | [107] [104] |
| D. bruxellensis; Hanseniaspora (H.) uvarum | NL4, GGTCCGTGTTTCAAGACGG | [109] | |
| 26S rDNA, D1/D2 domain | D. bruxellensis; D. anaomala; Z. bailii; H. valbyensis | NL1, GCATATCAATAAGCGGAGGAAAAG | [108] |
| NL4, GGTCCGTGTTTCAAGACGG | |||
| 18S rDNA, D1/D2 domain | Z. kombuchaensis sp. | NS-1, GTAGTCATATGCTTGTCTC | [59] |
| NS-8A, CCTTCCGCAGGTTCACCTACGGAAACC | |||
| ITS, Illumina MiSeq | Z. bailii | ITS1F, CTTGGTCATTTAGAGGAAGTAA | [102] |
| ITS2R, GCTGCGTTCTTCATCGATGC | |||
| Real-time PCR | Brettanomyces | Yeast-F, GAGTCGAGTTGTTTGGGAATGC | [100] |
| Yeast-R, TCTCTTTCCAAAGTTCTTTTCATCTTT | |||
| PCR-ITS RFLP | D. bruxellensis; | ITS1, TCCGTAGGTGAACCTGCGG | [47] |
| ITS4, TCCTCCGCTTATTGATATGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Kombucha: Production and Microbiological Research. Foods 2022, 11, 3456. https://doi.org/10.3390/foods11213456
Wang B, Rutherfurd-Markwick K, Zhang X-X, Mutukumira AN. Kombucha: Production and Microbiological Research. Foods. 2022; 11(21):3456. https://doi.org/10.3390/foods11213456
Chicago/Turabian StyleWang, Boying, Kay Rutherfurd-Markwick, Xue-Xian Zhang, and Anthony N. Mutukumira. 2022. "Kombucha: Production and Microbiological Research" Foods 11, no. 21: 3456. https://doi.org/10.3390/foods11213456







