Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Bacterial Strains
2.3. Antimicrobial Susceptibility Testing
2.4. Detection of Genes Encoding Virulence Factors
2.5. PCR Detection of Genes Encoding AR
2.6. Statistical Analysis
2.7. Network Generation
2.8. Predictive Analysis Using Machine Learning Random Forest Algorithm
3. Results
3.1. Virulotypes and Phenotypic Profiling of AR
3.2. Molecular Detection of AR Genes
3.3. Statistical Analysis of Phenotypic AR with Virulence Genes
3.4. Network Analysis of Resistance, Virulence Genes, and Phenotypic AR
3.5. Predictive Analysis of AR/virulence Genes Links Using the Machine Learning Random Forest Algorithm
4. Discussion
4.1. Antimicrobial Resistance and Corresponding Genotypes
4.2. Virulence Power of Campylobacter Isolates
4.3. Relationship between Virulence Genes and Phenotypic and Genotypic Antimicrobial Resistance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Function | Gene | Primers (5-3′) | References |
|---|---|---|---|
| Motility | flaA | F: AATAAAAATGCTCATAAAAACAGGTG R: TACCGAACCAATGTCTGCTCTGATT | [72] |
| Adhesion | cadF | F: TTGAAGGTAATTTAGATATG R: CTAATACCTAAAGTTGAAAC | [73] |
| racR | F: GATGATCCTGACTTTG R: TCTCCTATTTTTACCC | [74] | |
| dnaJ | F: AAGGCTTTGGCTCATC R: CTTTTTGTTCATCGTT | [75] | |
| Cytolethal distending toxin | cdtA | F: CCTTGTGATGCAAGCAATC R: ACACTCCATTTGCTTTCTG | [76] |
| cdtB | F: CAGAAAGCAAATGGAGTGTT R: AGCTAAAAGCGGTGGAGTAT | [76] | |
| cdtC | F: CGATGAGTTAAAACAAAAAGATA R: TTGGCATTATAGAAAATACAGTT | [76] | |
| Invasion | virB11 | F: TCTTGTGAGTTGCCTTACCCCTTTT R: CCTGCGTGTCCTGTGTTATTTACCC | [72] |
| pldA | F: AAGCTTATGCGTTTTT R: TATAAGGCTTTCTCCA | [75] | |
| ciaB | F: TTTTTATCAGTCCTTA R: TTTCGGTATCATTAGC | [75] | |
| Neurological complications | WlaN | F: TTAAGAGCAAGATATGAAGGTG R: CCATTTGAATTGATATTTTTG | [77] |
| CgtB | F: TAAGAGCAAGATATGAAGGTG R: GCACATAGAGAACGCTACAA | ||
| CeuE, lipoprotein involved in iron acquisition | ceuE-Cj | F: CCTGCTACGGTGAAAGTTTTGC R: GATCTTTTTGTTTTGTGCTGC | [78] |
| ceuE-Cc | F: ATGAAAAAATA TTTAGTTTTTGCA R: ATTTTATTATTTG TAGCAGCG |
| Antibiotic/(MDR) | Target Gene | Primer Sequence (5′-3′) | References |
|---|---|---|---|
| Tetracycline | tet(O) | F: GCGTTTTGTTTATGTGCG R: ATGGACAACCCGACAGAAG | [79] |
| Multidrug CmeABC efflux system | cmeB | F: AGGCGGTTTTGAAATGTATGTT R: TGTGCCGCTGGGAAAAG | [80] |
| Ampicillin/Amoxicillin | blaOXA-61 | F: AGAGTATAATACAAGCG R: TAGTGAGTTGTCAAGCC | [81] |
| Gentamicin | aphA-3 | F: TGCGTAAAAGATACGGAAG R: CAATCAGGCTTGATCCCC | [81] |
| Erythromycin | 23S rRNA | 23S rRNA. F: TTAGCTAATGTTGCCCGTACCG ERY2075: TAGTAAAGGTCCACGGGGTCGC ERY2074: AGTAAAGGTCCACGGGGTCTGG | [39] |
| ermB | F: TGAAAAAGTACTCAACCAAAT R: TCCTCCCGTTAAATAATAGAT | [28] | |
| Ciprofloxacin/Nalidixic acid | gyrA-Cj | gryA1: TTTTTAGCAAAGATTCTGAT gyrA5: AAAGCATCATAAACTGCAA gyrA4: CAAAGCATCATAAACTGCAG | [38] |
| gyrA-Cc | gyrA3: TATGAGCGTTATTATCGGTC gyrA8: TAAGGCATCGTAAACAGCCA gyrA4: GTCCATCTACAAGCTCGTTA | [37] |
References
- Alaboudi, A.R.; Malkawi, I.M.; Osaili, T.M.; Abu-Basha, E.A.; Guitian, J. Prevalence, antibiotic Rrsistance and genotypes of Campylobacter Jejuni and Campylobacter coli isolated from chickens in Irbid governorate, Jordan. Int. J. Food Microbiol. 2020, 327, 108656. [Google Scholar] [CrossRef] [PubMed]
- Sifré, E.; Salha, B.A.; Ducournau, A.; Floch, P.; Chardon, H.; Mégraud, F.; Lehours, P. EUCAST Recommendations for Antimicrobial Susceptibility Testing Applied to the Three Main Campylobacter Species Isolated in Humans. J. Microbiol. Methods 2015, 119, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Food, E.; Authority, S. The European Union One Health 2019 zoonoses report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Igwaran, A.; Okoh, A.I. Human Campylobacteriosis: A public health concern of global Importance. Heliyon 2019, 5, e02814. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Zhou, Q.; Zhang, X.; Zhou, S.; Zhang, J.; Tang, X.; Lu, J.; Gao, Y. Antibiotic Resistance Profiles and Molecular Mechanisms of Campylobacter from Chicken and Pig in China. Front. Microbiol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, Y.; Deng, F.; Liu, D.; Yao, H.; Zhang, Q.; Shen, J.; Liu, Z.; Gao, Y.; Wu, C.; et al. Species Shift and Multidrug Resistance of Campylobacter from Chicken and Swine, China, 2008–2014. J. Antimicrob. Chemother. 2016, 71, 666–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poehlsgaard, J.; Andersen, N.M.; Warrass, R.; Douthwaite, S. Visualizing the 16-membered ring macrolides tildipirosin and tilmicosin bound to their ribosomal site. ACS Chem. Biol. 2012, 7, 1351–1355. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A One Health perspective. Microbiol. Spectr. 2018, 6, 6-2. [Google Scholar] [CrossRef] [Green Version]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.W.N.; Tiam-Lee, D.C.; Paclibare, P.A.P.; Subejano, M.S.E.P.; Cabero-Palma, J.A.S.; Penuliar, G.M. High rates of contamination of poultry meat products with drug-resistant campylobacter in Metro Manila, Philippines. Jpn. J. Infect. Dis. 2017, 70, 311–313. [Google Scholar] [CrossRef]
- Hafez, H.M.; Attia, Y.A. Challenges to the poultry industry: Current perspectives and strategic future after the COVID-19 outbreak. Front. Vet. Sci. 2020, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Premarathne, J.M.K.J.K.; Anuar, A.S.; Thung, T.Y.; Satharasinghe, D.A.; Jambari, N.N.; Abdul-Mutalib, N.A.; Yew Huat, J.T.; Basri, D.F.; Rukayadi, Y.; Nakaguchi, Y.; et al. Prevalence and Antibiotic Resistance against Tetracycline in Campylobacter Jejuni and C. Coli in Cattle and Beef Meat from Selangor, Malaysia. Front. Microbiol. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Woźniak-Biel, A.; Bugla-Płoskońska, G.; Kielsznia, A.; Korzekwa, K.; Tobiasz, A.; Korzeniowska-Kowal, A.; Wieliczko, A. High Prevalence of Resistance to Fluoroquinolones and Tetracycline Campylobacter spp. Isolated from Poultry in Poland. Microb. Drug Resist. 2018, 24, 314–322. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, A.; Dionisi, A.M.; Arena, S.; Iglesias-Torrens, Y.; Carattoli, A.; Luzzi, I. Human Campylobacteriosis in Italy: Emergence of Multi-Drug Resistance to Ciprofloxacin, Tetracycline, and Erythromycin. Front. Microbiol. 2018, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Feizabadi, M.M.; Dolatabadi, S.; Zali, M.R. Isolation and Drug-Resistant Patterns of Campylobacter Strains Cultured from Diarrheic Children in Tehran. Jpn J. Infect. Dis. 2007, 60, 217–219. [Google Scholar]
- Li, B.; Ma, L.; Li, Y.; Jia, H.; Wei, J.; Shao, D.; Liu, K.; Shi, Y.; Qiu, Y.; Ma, Z. Antimicrobial Resistance of Campylobacter Species Isolated from Broilers in Live Bird Markets in Shanghai, China. Foodborne Pathog. Dis. 2017, 14, 96–102. [Google Scholar] [CrossRef]
- Neogi, S.B.; Islam, M.M.; Islam, S.K.S.; Akhter, A.H.M.T.; Sikder, M.M.H.; Yamasaki, S.; Kabir, S.M.L. Risk of Multi-Drug Resistant Campylobacter spp. And Residual Antimicrobials at Poultry Farms and Live Bird Markets in Bangladesh. BMC Infect. Dis. 2020, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Iovine, N.M. Resistance mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.C. Update on Acquired Tetracycline Resistance Genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, M.C. Genetic mobility and distribution of tetracycline resistance determinants. In Ciba Foundation Symposium 207-Antibiotic Resistance: Origins, Evolution, Selection and Spread: Antibiotic Resistance: Origins, Evolution, Selection and Spread: Ciba Foundation Symposium 207; John Wiley & Sons, Ltd.: Chichester, UK, 2007; pp. 206–222. [Google Scholar]
- Hormeño, L.; Campos, M.J.; Vadillo, S.; Quesada, A. Occurrence of tet(O/M/O) mosaic gene in tetracycline-resistant Campylobacter. Microorganisms 2020, 8, 1710. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.; Hawkins, K.; Lynch, H.; Egan, J.; Bolton, D.; Coffey, A.; Lucey, B. Investigation of Molecular Mechanisms Underlying Tetracycline Resistance in Thermophilic Campylobacter spp. Suggests That Previous Reports of Tet(A)-Mediated Resistance in These Bacteria Are Premature. Gut Pathog. 2019, 11, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jiang, Q.; Tang, H.; Wang, Z.; Yin, Y.; Ren, F.; Kong, L.; Jiao, X.; Huang, J. Characterization and Prevalence of Campylobacter Spp. From Broiler Chicken Rearing Period to the Slaughtering Process in Eastern China. Front. Vet. Sci. 2020, 7, 1–9. [Google Scholar] [CrossRef]
- Gibreel, A.; Tracz, D.M.; Nonaka, L.; Ngo, T.M.; Connell, S.R.; Taylor, D.R. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 2004, 48, 3442–3450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic Resistance in Campylobacter: Emergence, Transmission and Persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Overbye Michel, L.; Zhang, Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Wang, Y.; Zhang, Q.; Zhang, M.; Deng, F.; Shen, Z.; Wu, C.; Wang, S.; Zhang, J.; Shen, J. Report of Ribosomal RNA Methylase Gene Erm(B) in Multidrug-Resistant Campylobacter coli. J. Antimicrob. Chemother. 2014, 69, 964–968. [Google Scholar] [CrossRef] [Green Version]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [Green Version]
- Roux, D.; Aubier, B.; Cochard, H.; Quentin, R.; Van Der Mee-Marquet, N. Contaminated Sinks in Intensive Care Units: An Underestimated Source of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in the Patient Environment. J. Hosp. Infect. 2013, 85, 106–111. [Google Scholar] [CrossRef]
- Wesche, A.M.; Gurtler, J.B.; Marks, B.P.; Ryser, E.T. Stress, Sublethal Injury, Resuscitation, and Virulence of Bacterial Foodborne Pathogens. J. Food Prot. 2009, 72, 1121–1138. [Google Scholar] [CrossRef] [Green Version]
- Guerin, É.; Cambray, G.; Sanchez-Alberola, N.; Campoy, S.; Erill, I.; Re, S.D.; Gonzalez-Zorn, B.; Barbé, J.; Ploy, M.C.; Mazel, D. The SOS Response Controls Integron Recombination. Science 2009, 324, 1034. [Google Scholar] [CrossRef] [PubMed]
- ISO 10272-1 (Annex E); Microbiology of Food and Animal Feeding Stuffs: Horizontal Method for Detection and Enumera-tion of Campylobacter spp. I. Detection Method. International Organization for Standardization: Geneva, Switzerland, 2006.
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Alaya, N.; Hamrouni, S.; Bessoussa, G.; Ghram, A.; Maaroufi, A. Campylobacter spp. in Eggs and Laying Hens in the North-East of Tunisia: High Prevalence and Multidrug-Resistance Phenotypes. Vet. Sci. 2022, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Jouini, A.; Ghedira, K.; Zrelli, C.; Hamrouni, S.; Aouadhi, C.; Bessoussa, G.; Ghram, A.; et al. Prevalence and Antibiotic Resistance Patterns of Campylobacter spp. Isolated from Broiler Chickens in the North of Tunisia. BioMed Res. Int. 2018, 2018, 7943786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Ghedira, K.; Ghram, A.; Maaroufi, A. Distribution of Virulence and Antibiotic Resistance Genes in Campylobacter jejuni and Campylobacter coli Isolated from Broiler Chickens in Tunisia. J. Microbiol. Immunol. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zirnstein, G.; Helsel, L.; Li, Y.; Swaminathan, B.; Besser, J. Characterization of GyrA Mutations Associated with Fluoroquinolone Resistance in Campylobacter coli by DNA Sequence Analysis and MAMA PCR. FEMS Microbiol. Lett. 2000, 190, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zirnstein, G.; Li, Y.; Swaminathan, B.; Angulo, F. Ciprofloxacin Resistance in Campylobacter jejuni Isolates: Detection of GyrA Resistance Mutations by Mismatch Amplification Mutation Assay PCR and DNA Sequence Analysis. J. Clin. Microbiol. 1999, 37, 3276–3280. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Mateo, E.; Churruca, E.; Martinez, I.; Girbau, C.; Fernández-Astorga, A. MAMA-PCR Assay for the Detection of Point Mutations Associated with High-Level Erythromycin Resistance in Campylobacter jejuni and Campylobacter coli Strains. J. Microbiol. Methods 2005, 63, 99–103. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Martinez-Taboada, F.; Redondo, J.I. The SIESTA (SEAAV Integrated Evaluation Sedation Tool for Anaesthesia) Project: Initial Development of a Multifactorial Sedation Assessment Tool for Dogs. PLoS ONE 2020, 15, e0230799. [Google Scholar] [CrossRef] [Green Version]
- Kugelberg, E.; Löfmark, S.; Wretlind, B.; Andersson, D.I. Reduction of the Fitness Burden of Quinolone Resistance in Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2005, 55, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Skurnik, D.; Roux, D.; Cattoir, V.; Danilchanka, O.; Lu, X.; Yoder-Himes, D.R.; Han, K.; Guillard, T.; Jiang, D.; Gaultier, C.; et al. Enhanced in Vivo Fitness of Carbapenem-Resistant OprD Mutants of Pseudomonas Aeruginosa Revealed through High-Throughput Sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 20747–20752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Choi, Y.; He, H.; Dodd, M.C. Degradation Kinetics of Antibiotic Resistance Gene Meca of Methicillin-Resistant Staphylococcus Aureus (Mrsa) during Water Disinfection with Chlorine, Ozone, and Ultraviolet Light. Environ. Sci. Technol. 2021, 55, 2541–2552. [Google Scholar] [CrossRef]
- Elhadidy, M.; Miller, W.G.; Arguello, H.; Álvarez-Ordóñez, A.; Duarte, A.; Dierick, K.; Botteldoorn, N. Genetic basis and clonal population structure of antibiotic resistance in Campylobacter jejuni isolated from broiler carcasses in Belgium. Front. Microbiol. 2018, 9, 1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, C.T.; Lynch, H.; Burke, S.; Hawkins, K.; Buttimer, C.; Mc Carthy, C.; Egan, J.; Whyte, P.; Bolton, D.; Coffey, A.; et al. Antimicrobial Resistance Determinants Circulating among Thermophilic Campylobacter Isolates Recovered from Broilers in Ireland Over a One-Year Period. Antibiotics 2020, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Song, D.G.; Yoon, K.Y.; Mboera, L.E.G.; Matee, M.I.; Mutangana, D.; Komba, E.V.G.; Pan, C.H.; Amachawadi, R.G. Genomic Characterization of Fluoroquinolone-Resistant Thermophilic Campylobacter Strains Isolated from Layer Chicken Feces in Gangneung, South Korea by Whole-Genome Sequencing. Genes 2021, 12, 1131. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Mughini-Gras, L.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Mevius, D.J.; Hordijk, J. Successful Host Adaptation of IncK2 Plasmids. Front. Microbiol. 2019, 10, 2384. [Google Scholar] [CrossRef] [Green Version]
- Fraqueza, M.J.; Martins, A.; Borges, A.C.; Fernandes, M.H.; Fernandes, M.J.; Vaz, Y.; Bessa, R.J.B.; Barreto, A.S. Antimicrobial Resistance among Campylobacter spp. Strains Isolated from Different Poultry Production Systems at Slaughterhouse Level. Poult. Sci. 2014, 93, 1578–1586. [Google Scholar] [CrossRef]
- Frazão, M.R.; Cao, G.; Medeiros, M.I.C.; Duque, S.D.S.; Allard, M.W.; Falcão, J.P. Antimicrobial Resistance Profiles and Phylogenetic Analysis of Campylobacter jejuni Strains Isolated in Brazil by Whole Genome Sequencing. Microb. Drug Resist. 2021, 27, 660–669. [Google Scholar] [CrossRef]
- Kim, J.; Park, H.; Kim, J.; Kim, J.H.; Jung, J.I.; Cho, S.; Ryu, S.; Jeon, B. Comparative Analysis of Aerotolerance, Antibiotic Resistance, and Virulence Gene Prevalence in Campylobacter jejuni Isolates from Retail Raw Chicken and Duck Meat in South Korea. Microorganisms 2019, 7, 433. [Google Scholar] [CrossRef] [Green Version]
- Hlashwayo, D.F.; Sigaúque, B.; Bila, C.G. Epidemiology and Antimicrobial Resistance of Campylobacter spp. in Animals in Sub-Saharan Africa: A Systematic Review. Heliyon 2020, 6, e03537. [Google Scholar] [CrossRef]
- Bolinger, H.K.; Zhang, Q.; Miller, W.G.; Kathariou, S. Lack of Evidence for Erm (B) Infiltration into Erythromycin-Resistant Campylobacter coli and Campylobacter jejuni from Commercial Turkey Production in Eastern North Carolina: A Major Turkey-Growing Region in the United States. Foodborne Pathog. Dis. 2018, 15, 698–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.C.; Tagg, K.A.; Joung, Y.J.; Bennett, C.; Francois Watkins, L.; Eikmeier, D.; Folster, J.P. Report of Erm (B)+ Campylobacter jejuni in the United States. Antimicrob. Agents Chemother. 2018, 62, e02615-17. [Google Scholar] [CrossRef] [PubMed]
- Kashoma, I.P.; Kassem, I.I.; John, J.; Kessy, B.M.; Gebreyes, W.; Kazwala, R.R.; Rajashekara, G. Prevalence and Antimicrobial Resistance of Campylobacter Isolated from Dressed Beef Carcasses and Raw Milk in Tanzania. Microb. drug Resist. 2016, 22, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, S.P.W.; Vurayai, M.; Holmes, M.; Gupta, S.; Bateman, M.; Goldfarb, D.; Maskell, D.J.; Matsheka, M.I.; Grant, A.J. Phylogenetic Analyses and Antimicrobial Resistance Profiles of Campylobacter spp. from Diarrhoeal Patients and Chickens in Botswana. PLoS ONE 2018, 13, e0194481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäesaar, M.; Kramarenko, T.; Meremäe, K.; Sõgel, J.; Lillenberg, M.; Häkkinen, L.; Ivanova, M.; Kovalenko, K.; Hörman, A.; Hänninen, M. Antimicrobial Resistance Profiles of Campylobacter spp. Isolated from Broiler Chicken Meat of Estonian, Latvian and Lithuanian Origin at Estonian Retail Level and from Patients with Severe Enteric Infections in Estonia. Zoonoses Public Health 2016, 63, 89–96. [Google Scholar] [CrossRef]
- Cantero, G.; Correa-Fiz, F.; Ronco, T.; Strube, M.; Cerdà-Cuéllar, M.; Pedersen, K. Characterization of Campylobacter jejuni and Campylobacter coli Broiler Isolates by Whole-Genome Sequencing. Foodborne Pathog. Dis. 2018, 15, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Pilay, S.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Shobo, C.O.; Perrett, K.; Bester, L.A.; Essack, S.Y. Characterisation of Campylobacter spp. Isolated from Poultry in KwaZulu-Natal, South Africa. Antibiotics 2020, 9, 42. [Google Scholar] [CrossRef] [Green Version]
- Elhadidy, M.; Ali, M.M.; El-Shibiny, A.; Miller, W.G.; Elkhatib, W.F.; Botteldoorn, N.; Dierick, K. Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS ONE 2021, 15, e0227833. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Guan, X.; Zeng, H.; Li, J.; Huang, X.; Wen, Y.; Zhao, Q.; Huang, X.; Yan, Q.; Huang, Y. Prevalence, Antimicrobial Resistance Profiles and Virulence-Associated Genes of Thermophilic Campylobacter spp. Isolated from Ducks in a Chinese Slaughterhouse. Food Control. 2019, 104, 157–166. [Google Scholar] [CrossRef]
- Oh, J.-Y.; Kwon, Y.-K.; Wei, B.; Jang, H.-K.; Lim, S.-K.; Kim, C.-H.; Jung, S.-C.; Kang, M.-S. Epidemiological Relationships of Campylobacter Jejuni Strains Isolated from Humans and Chickens in South Korea. J. Microbiol. 2017, 55, 13–20. [Google Scholar] [CrossRef]
- Wysok, B.; Wojtacka, J. Detection of Virulence Genes Determining the Ability to Adhere and Invade in Campylobacter spp. from Cattle and Swine in Poland. Microb. Pathog. 2018, 115, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Riso, R.; Avventuroso, E.; Visalli, G.; Delia, S.A.; Laganà, P. Campylobacter: From Microbiology to Prevention. J. Prev. Med. Hyg. 2017, 58, E79. [Google Scholar] [PubMed]
- Otigbu, A.C.; Clarke, A.M.; Fri, J.; Akanbi, E.O.; Njom, H.A. Antibiotic Sensitivity Profiling and Virulence Potential of Campylobacter jejuni Isolates from Estuarine Water in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 2018, 15, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Hein, G.; García, P.; Foerster, C.; Troncoso, M.; Figueroa, G. Campylobacter jejuni Isolated from Human Cases in Chile Showed Indistinguishable Pulsed Field Gel Electrophoresis Profiles with Strains Isolated from Poultry and Bovine Sources. CyTA-J. Food 2013, 11, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Ramires, T.; de Oliveira, M.G.; Kleinubing, N.R.; de Fátima Rauber Würfel, S.; Mata, M.M.; Iglesias, M.A.; Lopes, G.V.; Dellagostin, O.A.; da Silva, W.P. Genetic Diversity, Antimicrobial Resistance, and Virulence Genes of Thermophilic Campylobacter Isolated from Broiler Production Chain. Braz. J. Microbiol. 2020, 51, 2021–2032. [Google Scholar] [CrossRef]
- Datta, S.; Niwa, H.; Itoh, K. Age-Dependent Variation of Virulence-Associated Genes Retained in Campylobacter jejuni Isolated from Chickens in a Poultry Farm. J. Vet. Med. Sci. 2009, 71, 1247–1249. [Google Scholar] [CrossRef] [Green Version]
- Khoshbakht, R.; Tabatabaei, M.; Hosseinzadeh, S.; Shekarforoush, S.S.; Aski, H.S. Distribution of Nine Virulence-Associated Genes in Campylobacter Jejuni and C. Coli Isolated from Broiler Feces in Shiraz, Southern Iran. Foodborne Pathog. Dis. 2013, 10, 764–770. [Google Scholar] [CrossRef]
- Gilbert, M.; Brisson, J.-R.; Karwaski, M.-F.; Michniewicz, J.; Cunningham, A.-M.; Wu, Y.; Young, N.M.; Wakarchuk, W.W. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384: Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-MHz 1H and 13C NMR analysis. J. Biol. Chem. 2000, 275, 3896–3906. [Google Scholar] [CrossRef] [Green Version]
- Linton, D.; Gilbert, M.; Hitchen, P.G.; Dell, A.; Morris, H.R.; Wakarchuk, W.W.; Gregson, N.A.; Wren, B.W. Phase variation of a β-1, 3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 2000, 37, 501–514. [Google Scholar] [CrossRef]
- Datta, S.; Niwa, H.; Itoh, K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003, 52, 345–348. [Google Scholar] [CrossRef] [Green Version]
- Konkel, M.E.; Gray, S.A.; Kim, B.J.; Garvis, S.G.; Yoon, J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J. Clin. Microbiol. 1999, 37, 510–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermans, D.; Van Deun, K.; Martel, A.; Van Immerseel, F.; Messens, W.; Heyndrickx, M.; Haesebrouck, F.; Pasmans, F. Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 2011, 42, 82. [Google Scholar] [CrossRef] [PubMed]
- Ziprin, R.L.; Young, C.R.; Byrd, J.A.S.; tanker, L.H.; Hume, M.E.; Gray, S.A.; Kim, B.J.; Konkel, M.E. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 2001, 45, 549–557. [Google Scholar] [CrossRef]
- Hickey, T.E.; McVeigh, A.L.; Scott, D.A.; Michielutti, R.E.; Bixby, A.; Carroll, S.A.; Bourgeois, A.L.; Guerry, P. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin- from intestinal epithelial cells. Infect. Immun. 2000, 68, 6535–6541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linton, D.; Owen, R.J.; Stanley, J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 1996, 147, 707–718. [Google Scholar] [CrossRef]
- Gonzalez, I.; Grant, K.A.; Richardson, P.T.; Park, S.F.; Collins, M.D. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 1997, 35, 759–763. [Google Scholar] [CrossRef] [Green Version]
- Pratt, A.; Korolik, V. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 2005, 55, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Olah, P.A.; Doetkott, C.; Fakhr, M.K.; Logue, C.M. Prevalence of the Campylobacter multi-drug efflux pump (CmeABC) in Campylobacter spp. Isolated from freshly processed turkeys. Food Microbiol. 2006, 23, 453–460. [Google Scholar] [CrossRef]
- Obeng, A.S.; Rickard, H.; Sexton, M.; Pang, Y.; Peng, H.; Barton, M. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J. Appl. Microbiol. 2012, 113, 294–307. [Google Scholar] [CrossRef]

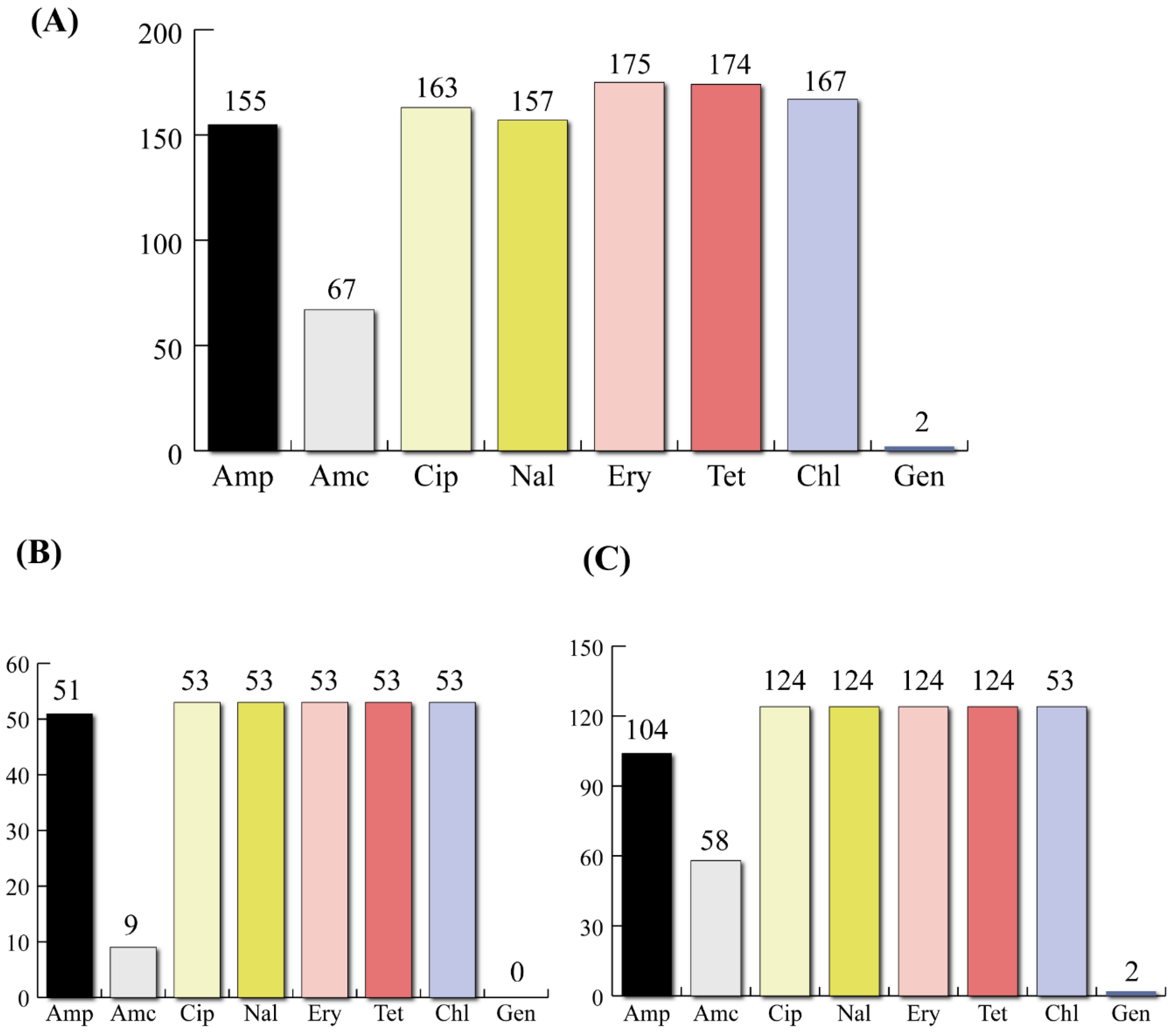
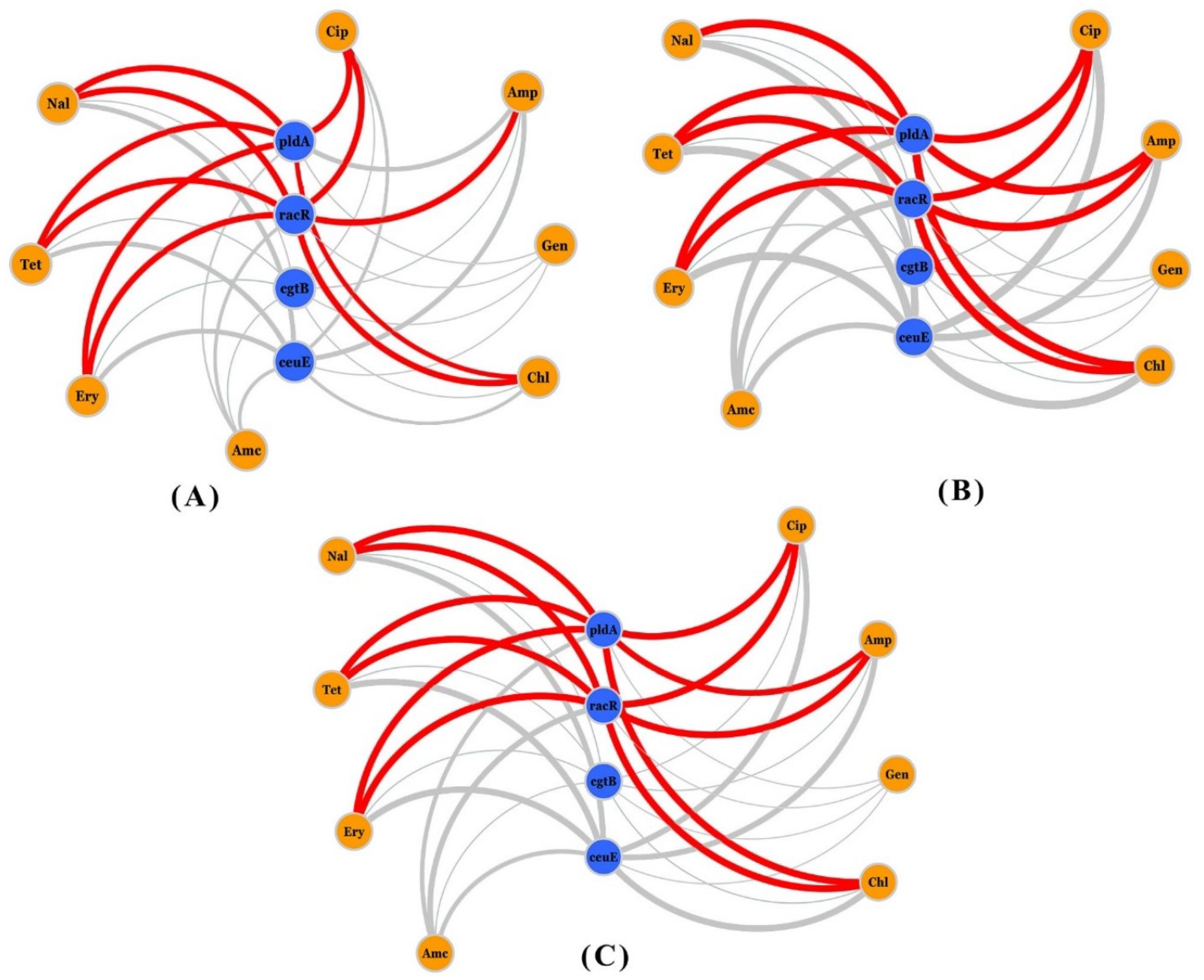
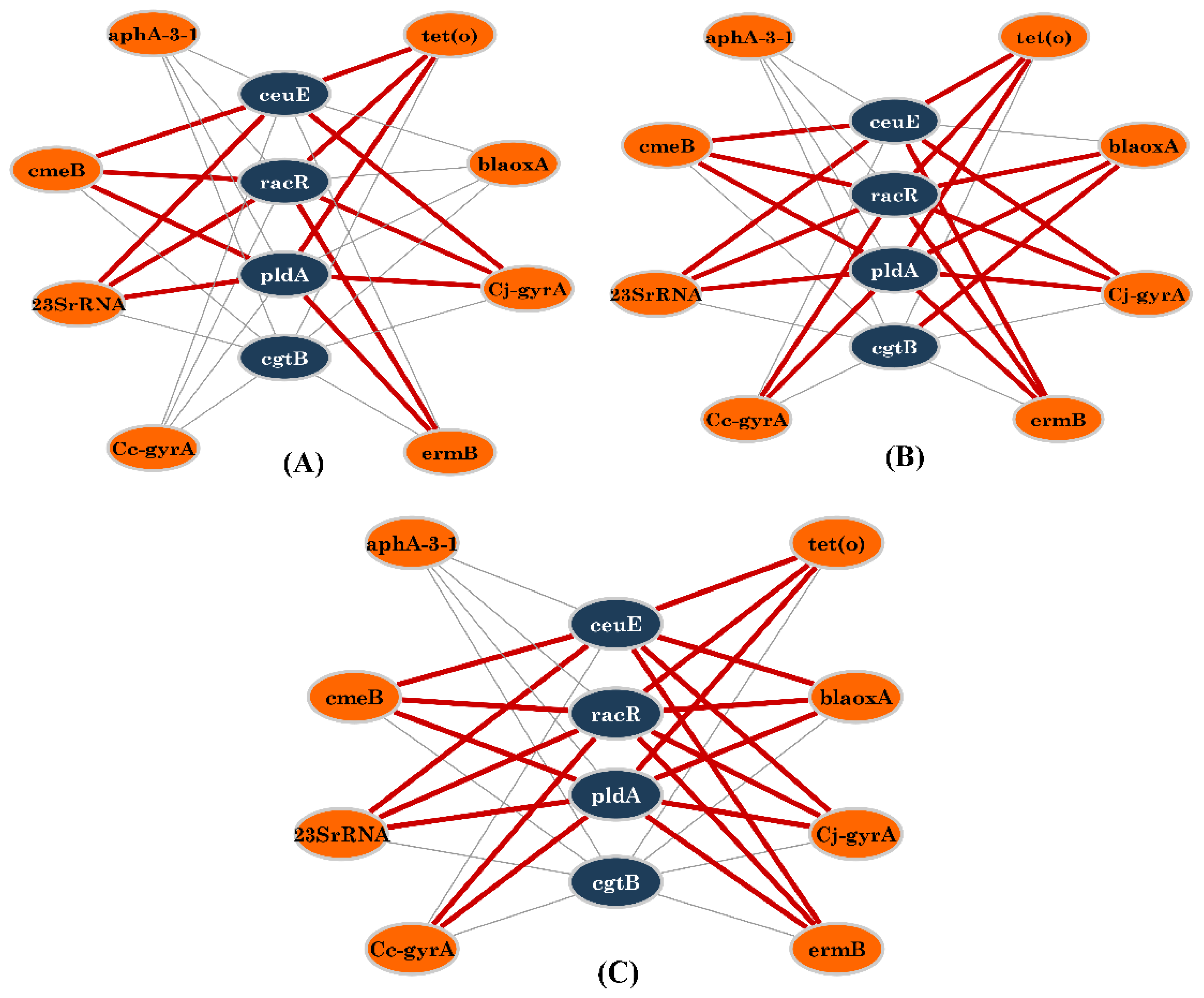


| Virulence Gene | Absence/Presence * | 4–6 Drug Resistance n (%) | >6 Drug Resistance n (%) | Chi-sq Value | p Value (Chi-sq/Fischer) |
|---|---|---|---|---|---|
| flaA | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 111 (62.71) | 66 (37.28) | |||
| cadF | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 111 (62.71) | 66 (37.28) | |||
| racR | 0 | 0 (0) | 16 (9.03) | 20.379 | 6.353 × 10−6 |
| 1 | 95 (53.67) | 66 (37.28) | |||
| dnaJ | 0 | 13 (7.34) | 11 (6.21) | 0.64529 | 0.4218 |
| 1 | 96 (54.23) | 57 (32.20) | |||
| virB11 | 0 | 18 (10.16) | 12 (6.8) | 1.3439 | 0.2463 |
| 1 | 104 (58.75) | 43 (24.3) | |||
| ciaB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 110 (62.14%) | 67 (37.85%) | |||
| pldA | 0 | 62(35.03) | 9(5.085) | 30.712 | 2.99 × 10−8 |
| 1 | 49(27.68) | 57(32.20) | |||
| cdtA | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 111 (62.71) | 66 (37.28) | |||
| cdtB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 112 (63.27) | 65 (36.72) | |||
| cdtC | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 111 (62.71) | 66 (37.28) | |||
| wlaN | 0 | 111 (62.71) | 66 (37.28) | NaN | 1 |
| 1 | 0 (0) | 0 (0) | |||
| ceuE(c,j) | 0 | 54 (30.50) | 8 (4.51) | 24.265 | 8.393 × 10−7 |
| 1 | 57 (32.20) | 58 (32.76) | |||
| cgtB | 0 | 108 (61.02) | 60 (33.9) | 6.4249 | 0.02778 |
| 1 | 2 (1.13) | 7 (3.95) |
| Virulence Gene | Absence/Presence * | 4–6 Drug Resistance n (%) | >6 Drug Resistance n (%) | Chi-sq Value | p Value (Chi-sq/Fischer) |
|---|---|---|---|---|---|
| flaA | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77%) | |||
| cadF | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77) | |||
| racR | 0 | 16 (12.9) | 0 (0) | 16.144 | 5.871 × 10−5 |
| 1 | 50 (40.32) | 58 (46.77) | |||
| DnaJ | 0 | 11 (8.87) | 4 (3.22) | 2.9925 | 0.1025 |
| 1 | 54 (43.54) | 55 (44.35) | |||
| virB11 | 0 | 17 (13.70) | 4 (3.22) | 8.2523 | 0.004213 |
| 1 | 48 (38.70) | 55 (44.35) | |||
| ciaB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77) | |||
| pldA | 0 | 18 (14.5) | 3 (2.41) | 10.718 | 0.001369 |
| 1 | 48 (38.70) | 55 (44.35) | |||
| cdtA | 0 | 0 (0) | 0 (0%) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77) | |||
| cdtB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77) | |||
| cdtC | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 66 (53.22) | 58 (46.77) | |||
| wlaN | 0 | 66 (53.22) | 58 (46.77) | NaN | 1 |
| 1 | 0 (0) | 0 (0) | |||
| ceuE(c,j) | 0 | 9 (7.25) | 0 (0) | NaN | 1 |
| 1 | 57 (45.96) | 58 (46.77) | |||
| cgtB | 0 | 66 (53.22) | 52 (41.93) | 3.5443 | 0.0933 |
| 1 | 1 (0.80) | 5 (4.03) |
| Virulence Gene | Absence/Presence * | 4–6 Drug Resistance n (%) | >6 Drug Resistance n (%) | Chi-sq Value | p Value (Chi-sq/Fischer) |
|---|---|---|---|---|---|
| flaA | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| cadF | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| racR | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| dnaJ | 0 | 0 (0) | 8 (15.09) | NaN | 1 |
| 1 | 45 (84.9) | 0 (0) | |||
| virB11 | 0 | 0 (0) | 8 (15.09) | NaN | 1 |
| 1 | 45 (84.9) | 0 (0) | |||
| ciaB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| pldA | 0 | 0 (0) | 1 (1.88) | 5.7332 | 0.1509 |
| 1 | 45 (84.9) | 7 (13.20) | |||
| cdtA | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| cdtB | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| cdtC | 0 | 0 (0) | 0 (0) | NaN | 1 |
| 1 | 45 (84.9) | 8 (15.09) | |||
| wlaN | 0 | 45 (84.9) | 8 (15.09) | NaN | 1 |
| 1 | 0 (0) | 0 (0) | |||
| ceuE(c,j) | 0 | 45 (84.9) | 8 (15.09) | NaN | 1 |
| 1 | 0 (0) | 0 (0) | |||
| cgtB | 0 | 44 (83.09) | 6 (11.32) | 6.5995 | 0.05618 |
| 1 | 1 ((1.88) | 2 (3.77) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharbi, M.; Kamoun, S.; Hkimi, C.; Ghedira, K.; Béjaoui, A.; Maaroufi, A. Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights. Foods 2022, 11, 3554. https://doi.org/10.3390/foods11223554
Gharbi M, Kamoun S, Hkimi C, Ghedira K, Béjaoui A, Maaroufi A. Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights. Foods. 2022; 11(22):3554. https://doi.org/10.3390/foods11223554
Chicago/Turabian StyleGharbi, Manel, Selim Kamoun, Chaima Hkimi, Kais Ghedira, Awatef Béjaoui, and Abderrazak Maaroufi. 2022. "Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights" Foods 11, no. 22: 3554. https://doi.org/10.3390/foods11223554





