Latest Trends in Industrial Vinegar Production and the Role of Acetic Acid Bacteria: Classification, Metabolism, and Applications—A Comprehensive Review
Abstract
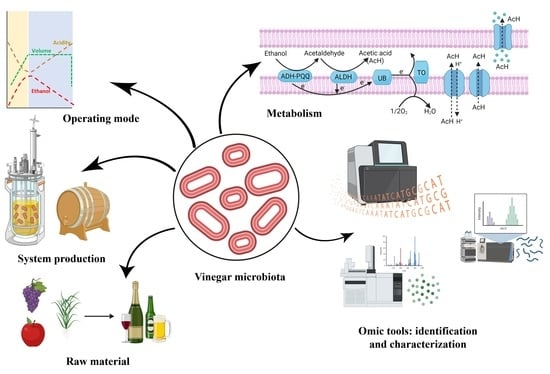
:1. Introduction
2. The Historical Context of Vinegar: Origin and Uses
3. Varieties of Vinegar
3.1. Mediterranean Vinegar: Wine and Balsamic Vinegar
3.2. Spirit Vinegar
3.3. Cereal Vinegar
3.4. Fruit Vinegar
4. Systems of Vinegar Production
4.1. Traditional Systems: Solid-State Fermentation and Surface Culture
4.2. Submerged Culture System
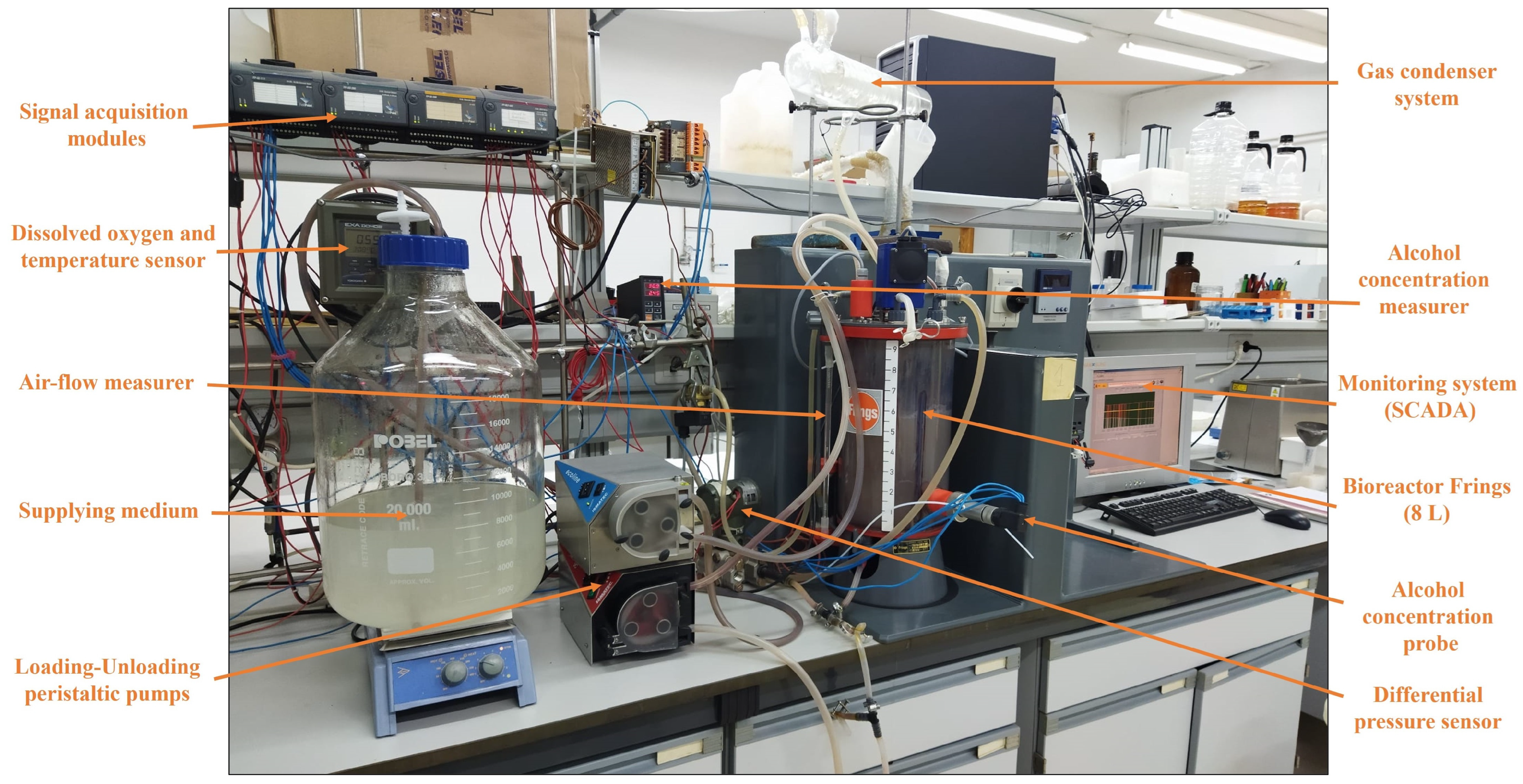
4.2.1. The Bioreactor: Acetator Frings
4.2.2. Operating Modes
4.2.3. Automation Systems
5. General Characteristics of Acetic Acid Bacteria
6. Current Taxonomy of Acetic Acid Bacteria
7. Metabolism of Acetic Acid Bacteria
7.1. Biotransformation of Ethanol to Acetic Acid
7.2. Carbohydrates Oxidation
8. Biotechnological Applications of Acetic Acid Bacteria
9. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ho, C.W.; Lazim, A.M.; Fazry, S.; Zaki, U.K.H.H.; Lim, S.J. Varieties, Production, Composition and Health Benefits of Vinegars: A Review. Food Chem. 2017, 221, 1621–1630. [Google Scholar] [CrossRef]
- Luzón-Quintana, L.M.; Castro, R.; Durán-Guerrero, E. Biotechnological Processes in Fruit Vinegar Production. Foods 2021, 10, 945. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, P.; Feng, F.; Luo, L.X. Microbial Diversity and Their Roles in the Vinegar Fermentation Process. Appl. Microbiol. Biotechnol. 2015, 99, 4997–5024. [Google Scholar] [CrossRef] [PubMed]
- Mas, A.; Torija, M.J.; García-Parrilla, M.D.C.; Troncoso, A.M. Acetic Acid Bacteria and the Production and Quality of Wine Vinegar. Sci. World J. 2014, 2014, 394671. [Google Scholar] [CrossRef] [PubMed]
- García-García, I.; Santos-Dueñas, I.M.; Jiménez-Ot, C.; Jiménez-Hornero, J.E.; Bonilla-Venceslada, J.L. Vinegar Engineering. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 97–120. ISBN 978-88-470-0865-6. [Google Scholar]
- Román-Camacho, J.J.; Mauricio, J.C.; Santos-Dueñas, I.M.; García-Martínez, T.; García-García, I. Unraveling the Role of Acetic Acid Bacteria Comparing Two Acetification Profiles From Natural Raw Materials: A Quantitative Approach in Komagataeibacter Europaeus. Front. Microbiol. 2022, 13, 840119. [Google Scholar] [CrossRef]
- Gullo, M.; Verzelloni, E.; Canonico, M. Aerobic Submerged Fermentation by Acetic Acid Bacteria for Vinegar Production: Process and Biotechnological Aspects. Process Biochem. 2014, 49, 1571–1579. [Google Scholar] [CrossRef]
- Trček, J.; Mahnič, A.; Rupnik, M. Diversity of the Microbiota Involved in Wine and Organic Apple Cider Submerged Vinegar Production as Revealed by DHPLC Analysis and Next-Generation Sequencing. Int. J. Food Microbiol. 2016, 223, 57–62. [Google Scholar] [CrossRef]
- Álvarez-Cáliz, C.M.; Santos-Dueñas, I.M.; Jiménez-Hornero, J.E.; García-García, I. Optimization of the Acetification Stage in the Production of Wine Vinegar by Use of Two Serial Bioreactors. Appl. Sci. 2021, 11, 1217. [Google Scholar] [CrossRef]
- Qi, Z.; Yang, H.; Xia, X.; Quan, W.; Wang, W.; Yu, X. Achieving High Strength Vinegar Fermentation via Regulating Cellular Growth Status and Aeration Strategy. Process Biochem. 2014, 49, 1063–1070. [Google Scholar] [CrossRef]
- Garcia-Garcia, I.; Gullo, M.; Chen, F.; Garcia-Martinez, T. Editorial: Acetic Acid Bacteria. Front. Microbiol. 2023, 14, 1142659. [Google Scholar] [CrossRef]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frébort, I.; Frebortova, J. Acetic Acid Bacteria: A Group of Bacteria with Versatile Biotechnological Applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Trček, J.; Mira, N.P.; Jarboe, L.R. Adaptation and Tolerance of Bacteria against Acetic Acid. Appl. Microbiol. Biotechnol. 2015, 99, 6215–6229. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xie, Z.; Zhang, H.; Liebl, W.; Toyama, H.; Chen, F. Oxidative Fermentation of Acetic Acid Bacteria and Its Products. Front. Microbiol. 2022, 13, 879246. [Google Scholar] [CrossRef]
- Anguluri, K.; La China, S.; Brugnoli, M.; De Vero, L.; Pulvirenti, A.; Cassanelli, S.; Gullo, M. Candidate Acetic Acid Bacteria Strains for Levan Production. Polymers 2022, 14, 2000. [Google Scholar] [CrossRef]
- La China, S.; Zanichelli, G.; De Vero, L.; Gullo, M. Oxidative Fermentations and Exopolysaccharides Production by Acetic Acid Bacteria: A Mini Review. Biotechnol. Lett. 2018, 40, 1289–1302. [Google Scholar] [CrossRef]
- Kuo, C.H.; Chen, J.H.; Liou, B.K.; Lee, C.K. Utilization of Acetate Buffer to Improve Bacterial Cellulose Production by Gluconacetobacter xylinus. Food Hydrocoll. 2016, 53, 98–103. [Google Scholar] [CrossRef]
- Rizo, J.; Guillén, D.; Farrés, A.; Díaz-Ruiz, G.; Sánchez, S.; Wacher, C.; Rodríguez-Sanoja, R. Omics in Traditional Vegetable Fermented Foods and Beverages. Crit. Rev. Food Sci. Nutr. 2020, 60, 791–809. [Google Scholar] [CrossRef]
- Román-Camacho, J.J.; Santos-Dueñas, I.M.; García-García, I.; Moreno-García, J.; García-Martínez, T.; Mauricio, J.C. Metaproteomics of Microbiota Involved in Submerged Culture Production of Alcohol Wine Vinegar: A First Approach. Int. J. Food Microbiol. 2020, 333, 108797. [Google Scholar] [CrossRef]
- Román-Camacho, J.J.; Mauricio, J.C.; Santos-Dueñas, I.M.; García-Martínez, T.; García-García, I. Functional Metaproteomic Analysis of Alcohol Vinegar Microbiota during an Acetification Process: A Quantitative Proteomic Approach. Food Microbiol. 2021, 98, 103799. [Google Scholar] [CrossRef]
- Román-Camacho, J.J.; García-García, I.; Santos-Dueñas, I.M.; Ehrenreich, A.; Liebl, W.; García-Martínez, T.; Mauricio, J.C. Combining Omics Tools for the Characterization of the Microbiota of Diverse Vinegars Obtained by Submerged Culture: 16S RRNA Amplicon Sequencing and MALDI-TOF MS. Front. Microbiol. 2022, 13, 1055010. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Claesson, M.J.; Cotter, P.D. Translating Omics to Food Microbiology. Annu. Rev. Food Sci. Technol. 2017, 8, 113–134. [Google Scholar] [CrossRef]
- Andrés-Barrao, C.; Barja, F. Acetic Acid Bacteria Strategies Contributing to Acetic Acid Resistance during Oxidative Fermentation. In Acetic Acid Bacteria: Fundamentals and Food Applications; Sengun, I.Y., Ed.; Food Biology Series “A Science Publishers Book”; CRC Press: Boca Raton, FL, USA, 2017; pp. 92–119. ISBN 978-1-315-15349-0. [Google Scholar]
- Johnston, C.S.; Gaas, C.A. Vinegar: Medicinal Uses and Antiglycemic Effect. Med. Gen. Med. 2006, 8, 61. [Google Scholar]
- Budak, N.H.; Aykin, E.; Seydim, A.C.; Greene, A.K.; Guzel-Seydim, Z.B. Functional Properties of Vinegar. J. Food Sci. 2014, 79, R757–R764. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Wang, Z.; Amir, R.M.; Younas, S.; Wali, A.; Adowa, N.; Ayim, I. Potential Uses of Vinegar as a Medicine and Related in Vivo Mechanisms. Int. J. Vitam. Nutr. Res. 2016, 86, 140–151. [Google Scholar] [CrossRef]
- Budak, N.H.; Kumbul Doguc, D.; Savas, C.M.; Seydim, A.C.; Kok Tas, T.; Ciris, M.I.; Guzel-Seydim, Z.B. Effects of Apple Cider Vinegars Produced with Different Techniques on Blood Lipids in High-Cholesterol-Fed Rats. J. Agric. Food Chem. 2011, 59, 6638–6644. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.N.; Guzel-Seydim, Z.B. Antioxidant Activity and Phenolic Content of Wine Vinegars Produced by Two Different Techniques. J. Sci. Food Agric. 2010, 90, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Fang, T. Survival of Escherichia coli O157:H7 and Salmonella enterica Serovars Typhimurium in Iceberg Lettuce and the Antimicrobial Effect of Rice Vinegar against E. coli O157:H7. Food Microbiol. 2007, 24, 745–751. [Google Scholar] [CrossRef]
- Chou, C.H.; Liu, C.W.; Yang, D.J.; Wu, Y.H.S.; Chen, Y.C. Amino Acid, Mineral, and Polyphenolic Profiles of Black Vinegar, and Its Lipid Lowering and Antioxidant Effects in Vivo. Food Chem. 2015, 168, 63–69. [Google Scholar] [CrossRef]
- Honsho, S.; Sugiyama, A.; Takahara, A.; Satoh, Y.; Nakamura, Y.; Hashimoto, K. A Red Wine Vinegar Beverage Can Inhibit the Renin-Angiotensin System: Experimental Evidence in Vivo. Biol. Pharm. Bull. 2005, 28, 1208–1210. [Google Scholar] [CrossRef]
- Yamashita, H.; Fujisawa, K.; Ito, E.; Idei, S.; Kawaguchi, N.; Kimoto, M.; Hiemori, M.; Tsuji, H. Improvement of Obesity and Glucose Tolerance by Acetate in Type 2 Diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) Rats. Biosci. Biotechnol. Biochem. 2007, 71, 1236–1243. [Google Scholar] [CrossRef]
- Lazim, A.M.; Lim, S.J.; Ho, C.W.; Fazry, S. Types of Vinegars. In Advances in Vinegar Production; Bekatorou, A., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 19–28. ISBN 978-1-351-20847-5. [Google Scholar]
- Parrondo, J.; Garcia, L.A.; Diaz, M. Whey Vinegar. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 273–288. ISBN 978-88-470-0865-6. [Google Scholar]
- Maestre, O.; Santos-Dueñas, I.M.; Peinado, R.; Jiménez-Ot, C.; García-García, I.; Mauricio, J.C. Changes in Amino Acid Composition during Wine Vinegar Production in a Fully Automatic Pilot Acetator. Process Biochem. 2008, 43, 803–807. [Google Scholar] [CrossRef]
- Sellmer-Wilsberg, S. Wine and Grape Vinegars. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 145–156. ISBN 978-88-470-0865-6. [Google Scholar]
- Durán-Guerrero, E.; Castro, R.; García-Moreno, M.D.V.; Rodríguez-Dodero, M.D.C.; Schwarz, M.; Guillén-Sánchez, D. Aroma of Sherry Products: A Review. Foods 2021, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Giudici, P.; Gullo, M.; Solieri, L. Traditional Balsamic Vinegar. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 157–177. ISBN 978-88-470-0865-6. [Google Scholar]
- Gullo, M.; Giudici, P. Acetic Acid Bacteria in Traditional Balsamic Vinegar: Phenotypic Traits Relevant for Starter Cultures Selection. Int. J. Food Microbiol. 2008, 125, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Grierson, B. Malt and Distilled Malt Vinegar. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 135–143. ISBN 978-88-470-0865-6. [Google Scholar]
- Andrés-Barrao, C.; Saad, M.M.; Cabello Ferrete, E.; Bravo, D.; Chappuis, M.-L.; Ortega Pérez, R.; Junier, P.; Perret, X.; Barja, F. Metaproteomics and Ultrastructure Characterization of Komagataeibacter Spp. Involved in High-Acid Spirit Vinegar Production. Food Microbiol. 2016, 55, 112–122. [Google Scholar] [CrossRef]
- Giudici, P.; Corradini, G.; Bonciani, T.; Wu, J.; Chen, F.; Lemmetti, F. The Flavor and Taste of Cereal Chinese Vinegars. Acetic Acid Bact. 2017, 6, 6370. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, P.; Xu, D.; Wang, W.; Zhao, Y. Aroma Patterns of Beijing Rice Vinegar and Their Potential Biomarker for Traditional Chinese Cereal Vinegars. Food Res. Int. 2019, 119, 398–410. [Google Scholar] [CrossRef]
- Murooka, Y.; Nanda, K.; Yamashita, M. Rice Vinegars. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 121–133. ISBN 978-88-470-0865-6. [Google Scholar]
- Chen, F.; Li, L.; Qu, J.; Chen, C. Cereal Vinegars Made by Solid-State Fermentation in China. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 243–259. ISBN 978-88-470-0865-6. [Google Scholar]
- Jiang, Y.; Lv, X.; Zhang, C.; Zheng, Y.; Zheng, B.; Duan, X.; Tian, Y. Microbial Dynamics and Flavor Formation during the Traditional Brewing of Monascus Vinegar. Food Res. Int. 2019, 125, 108531. [Google Scholar] [CrossRef]
- Xu, W.; Huang, Z.; Zhang, X.; Li, Q.; Lu, Z.; Shi, J.; Xu, Z.; Ma, Y. Monitoring the Microbial Community during Solid-State Acetic Acid Fermentation of Zhenjiang Aromatic Vinegar. Food Microbiol. 2011, 28, 1175–1181. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, F.; Zhang, C.; Yang, L.; Fan, G.; Xu, Y.; Sun, B.; Li, X. Dynamic Microbial Succession of Shanxi Aged Vinegar and Its Correlation with Flavor Metabolites during Different Stages of Acetic Acid Fermentation. Sci. Rep. 2018, 8, 8612. [Google Scholar] [CrossRef]
- Wu, L.-H.; Lu, Z.-M.; Zhang, X.-J.; Wang, Z.-M.; Yu, Y.-J.; Shi, J.-S.; Xu, Z.-H. Metagenomics Reveals Flavour Metabolic Network of Cereal Vinegar Microbiota. Food Microbiol. 2017, 62, 23–31. [Google Scholar] [CrossRef]
- Joshi, V.K.; Sharma, S. Cider Vinegar: Microbiology, Technology and Quality. In Vinegars of the World; Solieri, L., Giudici, P., Eds.; Springer: Milan, Italy, 2009; pp. 197–207. ISBN 978-88-470-0865-6. [Google Scholar]
- Ousaaid, D.; Mechchate, H.; Laaroussi, H.; Hano, C.; Bakour, M.; El Ghouizi, A.; Conte, R.; Lyoussi, B.; El Arabi, I. Fruits Vinegar: Quality Characteristics, Phytochemistry, and Functionality. Molecules 2021, 27, 222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J.-A.; Park, G.-G.; Jang, J.-K.; Park, Y.-S. Semi-Continuous Fermentation of Onion Vinegar and Its Functional Properties. Molecules 2017, 22, 1313. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gullo, M.; Chen, F.; Giudici, P. Diversity of Acetobacter pasteurianus Strains Isolated From Solid-State Fermentation of Cereal Vinegars. Curr. Microbiol. 2010, 60, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Zheng, Y.; Zhang, X.; Xie, S.; Wu, Y.; Xia, T.; Wang, M. Solid-State Fermentation Systems for Vinegar Production. In Advances in Vinegar Production; Bekatorou, A., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 141–154. ISBN 978-1-351-20847-5. [Google Scholar]
- Bekatorou, A. Advances in Immobilized Biocatalyst Technologies for Vinegar Production. In Advances in Vinegar Production; Bekatorou, A., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 117–139. ISBN 978-1-351-20847-5. [Google Scholar]
- Tesfaye, W.; Morales, M.L.; García-Parrilla, M.C.; Troncoso, A.M. Wine Vinegar: Technology, Authenticity and Quality Evaluation. Trends Food Sci. 2002, 13, 12–21. [Google Scholar] [CrossRef]
- Andres-Barrao, C.; Weber, A.; Chappuis, M.L. Acetic Acid Bacteria Population Dynamics and Natural Imposition of Gluconacetobacter europaeus during Submerged Vinegar Production. Arch. Sci. 2011, 64, 99–114. [Google Scholar] [CrossRef]
- Raspor, P.; Goranovič, D. Biotechnological Applications of Acetic Acid Bacteria. Crit. Rev. Biotechnol. 2008, 28, 101–124. [Google Scholar] [CrossRef]
- El Vinagre de Vino; Llaguno Marchena, C.; Polo, M.C. (Eds.) Consejo Superior de Investigaciones Científicas: Madrid, Spain, 1991; ISBN 978-84-00-07205-6. [Google Scholar]
- García-García, I.; Jiménez-Hornero, J.E.; Santos-Dueñas, I.M.; González-Granados, Z.; Cañete-Rodríguez, A.M. Modeling and Optimization of Acetic Acid Fermentation. In Advances in Vinegar Production; Bekatorou, A., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 299–325. ISBN 978-1-351-20847-5. [Google Scholar]
- Hromatka, O.; Ebner, H. Vinegar by Submerged Oxidative Fermentation. Ind. Eng. Chem. 1959, 51, 1279–1280. [Google Scholar] [CrossRef]
- De Ory, I.; Romero, L.E.; Cantero, D. Operation in Semi-Continuous with a Closed Pilot Plant Scale Acetifier for Vinegar Production. J. Food Eng. 2004, 63, 39–45. [Google Scholar] [CrossRef]
- Fernández-Pérez, R.; Torres, C.; Sanz, S.; Ruiz-Larrea, F. Strain Typing of Acetic Acid Bacteria Responsible for Vinegar Production by the Submerged Elaboration Method. Food Microbiol. 2010, 27, 973–978. [Google Scholar] [CrossRef]
- Baena-Ruano, S.; Santos-Dueñas, I.M.; Mauricio, J.C.; García-García, I. Relationship between Changes in the Total Concentration of Acetic Acid Bacteria and Major Volatile Compounds during the Acetic Acid Fermentation of White Wine. J. Sci. Food Agric. 2010, 90, 2675–2681. [Google Scholar] [CrossRef]
- Baena-Ruano, S.; Jiménez-Ot, C.; Santos-Dueñas, I.M.; Jiménez-Hornero, J.E.; Bonilla-Venceslada, J.L.; Álvarez-Cáliz, C.; García-García, I. Influence of the Final Ethanol Concentration on the Acetification and Production Rate in the Wine Vinegar Process. J. Chem. Technol. Biotechnol. 2010, 85, 908–912. [Google Scholar] [CrossRef]
- García-García, I.; Cantero-Moreno, D.; Jiménez-Ot, C.; Baena-Ruano, S.; Jiménez-Hornero, J.; Santos-Dueñas, I.; Bonilla-Venceslada, J.; Barja, F. Estimating the Mean Acetification Rate via On-Line Monitored Changes in Ethanol during a Semi-Continuous Vinegar Production Cycle. J. Food Eng. 2007, 80, 460–464. [Google Scholar] [CrossRef]
- Jiménez-Hornero, J.E.; Santos-Dueñas, I.M.; García-García, I. Modelling Acetification with Artificial Neural Networks and Comparison with Alternative Procedures. Processes 2020, 8, 749. [Google Scholar] [CrossRef]
- Santos-Dueñas, I.M.; Jimenez-Hornero, J.E.; Cañete-Rodriguez, A.M.; Garcia-Garcia, I. Modeling and Optimization of Acetic Acid Fermentation: A Polynomial-Based Approach. Biochem. Eng. J. 2015, 99, 35–43. [Google Scholar] [CrossRef]
- Kalogianni, D. Rapid Detection Methods for Online Monitoring of Vinegar Fermentations. In Advances in Vinegar Production; Bekatorou, A., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 421–440. ISBN 978-1-351-20847-5. [Google Scholar]
- Wang, B.; Shao, Y.; Chen, F. Overview on Mechanisms of Acetic Acid Resistance in Acetic Acid Bacteria. World J. Microbiol. Biotechnol. 2015, 31, 255–263. [Google Scholar] [CrossRef]
- Gomes, R.J.; Borges, M.D.F.; Rosa, M.D.F.; Castro-Gómez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139. [Google Scholar] [CrossRef]
- Malimas, T.; Thi Lan Vu, H.; Muramatsu, Y.; Yukphan, P.; Tanasupawat, S.; Yamada, Y. Systematics of Acetic Acid Bacteria. In Acetic Acid Bacteria; Sengun, I.Y., Ed.; Food Biology Series “A Science Publishers Book”; CRC Press: Boca Raton, FL, USA, 2017; pp. 3–43. ISBN 978-1-315-15349-0. [Google Scholar]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, Y.; Hong, H. Classification of Acetic Acid Bacteria and Their Acid Resistant Mechanism. AMB Expr. 2021, 11, 29. [Google Scholar] [CrossRef]
- Guzman, J.; Vilcinskas, A. Genome Analysis Suggests the Bacterial Family Acetobacteraceae Is a Source of Undiscovered Specialized Metabolites. Antonie Leeuwenhoek 2022, 115, 41–58. [Google Scholar] [CrossRef]
- Komagata, K.; Iino, T.; Yamada, Y. The Family Acetobacteraceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–78. ISBN 978-3-642-30196-4. [Google Scholar]
- Skerman, V.B.D.; McGowan, V.; Sneath, P.H.A. Approved Lists of Bacterial Names. Int. J. Syst. Bacteriol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Gillis, M.; De Ley, J. Intra- and Intergeneric Similarities of the Ribosomal Ribonucleic Acid Cistrons of Acetobacter and Gluconobacter. Int. J. Syst. Bacteriol. 1980, 30, 7–27. [Google Scholar] [CrossRef]
- Yamada, Y. Acetobacter xylinus Sp. nov., nom. rev., for the Cellulose-Forming and Cellulose-Less, Acetate-Oxidizing Acetic Acid Bacteria with the Q-10 System. J. Gen. Appl. Microbiol. 1983, 29, 417–420. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Lan Vu, H.T.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter gen. nov., with Proposals of New Combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Cleenwerck, I.; De Vos, P. Polyphasic Taxonomy of Acetic Acid Bacteria: An Overview of the Currently Applied Methodology. Int. J. Food Microbiol. 2008, 125, 2–14. [Google Scholar] [CrossRef]
- Trček, J.; Jernejc, K.; Matsushita, K. The Highly Tolerant Acetic Acid Bacterium Gluconacetobacter europaeus Adapts to the Presence of Acetic Acid by Changes in Lipid Composition, Morphological Properties and PQQ-Dependent ADH Expression. Extremophiles 2007, 11, 627–635. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Bacteriol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Cleenwerck, I.; Vandemeulebroecke, K.; Janssens, D.; Swings, J. Re-Examination of the Genus Acetobacter, with Descriptions of Acetobacter cerevisiae Sp. nov. and Acetobacter malorum Sp. nov. Int. J. Syst. Bacteriol. 2002, 52, 1551–1558. [Google Scholar] [CrossRef]
- Lisdiyanti, P.; Kawasaki, H.; Seki, T.; Yamada, Y.; Uchimura, T.; Komagata, K. Identification of Acetobacter Strains Isolated from Indonesian Sources, and Proposals of Acetobacter syzygii Sp. nov., Acetobacter cibinongensis Sp. nov., and Acetobacter orientalis Sp. nov. J. Gen. Appl. Microbiol. 2001, 47, 119–131. [Google Scholar] [CrossRef]
- Sombolestani, A.S.; Cleenwerck, I.; Cnockaert, M.; Borremans, W.; Wieme, A.D.; De Vuyst, L.; Vandamme, P. novel Acetic Acid Bacteria from Cider Fermentations: Acetobacter conturbans Sp. nov. and Acetobacter fallax Sp. nov. Int. J. Syst. Bacteriol. 2020, 70, 6163–6171. [Google Scholar] [CrossRef]
- Lisdiyanti, P.; Kawasaki, H.; Seki, T.; Yamada, Y.; Uchimura, T.; Komagata, K. Systematic Study of the Genus Acetobacter with Descriptions of Acetobacter indonesiensis Sp. nov., Acetobacter tropicalis Sp. nov., Acetobacter orleanensis (Henneberg 1906) Comb. nov., Acetobacter lovaniensis (Frateur 1950) Comb. nov., and Acetobacter estunensis (Carr 1958) Comb. nov. J. Gen. Appl. Microbiol. 2000, 46, 147–165. [Google Scholar] [CrossRef] [PubMed]
- Cleenwerck, I.; Gonzalez, A.; Camu, N.; Engelbeen, K.; De Vos, P.; De Vuyst, L. Acetobacter fabarum Sp. nov., an Acetic Acid Bacterium from a Ghanaian Cocoa Bean Heap Fermentation. Int. J. Syst. Bacteriol. 2008, 58, 2180–2185. [Google Scholar] [CrossRef] [PubMed]
- Tanasupawat, S.; Kommanee, J.; Yukphan, P.; Muramatsu, Y.; Nakagawa, Y.; Yamada, Y. Acetobacter Farinalis Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2011, 57, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Yukphan, P.; Charoenyingcharoen, P.; Kingcha, Y.; Likhitrattanapisal, S.; Muangham, S.; Tanasupawat, S.; Yamada, Y. Acetobacter garciniae Sp. nov., an Acetic Acid Bacterium Isolated from Fermented Mangosteen Peel in Thailand. Int. J. Syst. Bacteriol. 2021, 71, 005052. [Google Scholar] [CrossRef] [PubMed]
- Cleenwerck, I.; Camu, N.; Engelbeen, K.; De Winter, T.; Vandemeulebroecke, K.; De Vos, P.; De Vuyst, L. Acetobacter ghanensis Sp. nov., a novel Acetic Acid Bacterium Isolated from Traditional Heap Fermentations of Ghanaian Cocoa Beans. Int. J. Syst. Bacteriol. 2007, 57, 1647–1652. [Google Scholar] [CrossRef] [PubMed]
- Spitaels, F.; Li, L.; Wieme, A.; Balzarini, T.; Cleenwerck, I.; Van Landschoot, A.; De Vuyst, L.; Vandamme, P. Acetobacter lambici Sp. nov., Isolated from Fermenting Lambic Beer. Int. J. Syst. Bacteriol. 2014, 64, 1083–1089. [Google Scholar] [CrossRef]
- Ferrer, S.; Mañes-Lázaro, R.; Benavent-Gil, Y.; Yépez, A.; Pardo, I. Acetobacter musti Sp. nov., Isolated from Bobal Grape Must. Int. J. Syst. Bacteriol. 2016, 66, 957–961. [Google Scholar] [CrossRef]
- Dutta, D.; Gachhui, R. novel Nitrogen-Fixing Acetobacter nitrogenifigens Sp. nov., Isolated from Kombucha Tea. Int. J. Syst. Bacteriol. 2006, 56, 1899–1903. [Google Scholar] [CrossRef]
- Silva, L.R. Acetobacter oeni Sp. nov., Isolated from Spoiled Red Wine. Iny. J. Syst. Bacteriol. 2006, 56, 21–24. [Google Scholar] [CrossRef]
- Lino, T.; Suzuki, R.; Kosako, Y.; Ohkuma, M.; Komagata, K.; Uchimura, T. Acetobacter okinawensis Sp. nov., Acetobacter papayae Sp. nov., and Acetobacter persicus Sp. nov.; novel Acetic Acid Bacteria Isolated from Stems of Sugarcane, Fruits, and a Flower in Japan. J. Gen. Appl. Microbiol. 2012, 58, 235–243. [Google Scholar] [CrossRef]
- Baek, J.H.; Kim, K.H.; Moon, J.Y.; Yeo, S.-H.; Jeon, C.O. Acetobacter oryzoeni Sp. nov., Isolated from Korean Rice Wine Vinegar. Int. J. Syst. Bacteriol. 2020, 70, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Cho, G.Y.; Chun, B.H.; Weckx, S.; Moon, J.Y.; Yeo, S.H.; Jeon, C.O. Acetobacter oryzifermentans sp. nov., Isolated from Korean Traditional Vinegar and Reclassification of the Type Strains of Acetobacter pasteurianus subsp. ascendens (Henneberg 1898) and Acetobacter pasteurianus subsp. paradoxus (Frateur 1950) as Acetobacter ascendens sp. nov., comb. nov. Syst. Appl. Microbiol. 2018, 41, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Sokollek, S.J.; Hertel, C.; Hammes, W.P. Description of Acetobacter oboediens Sp. nov. and Acetobacter pomorum Sp. nov., Two New Species Isolated from Industrial Vinegar Fermentations. Int. J. Syst. Bacteriol. 1998, 48, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.L.; Yukphan, P.; Bui, V.T.T.; Charoenyingcharoen, P.; Malimas, S.; Nguyen, L.K.; Muramatsu, Y.; Tanaka, N.; Tanasupawat, S.; Le, B.T.; et al. Acetobacter sacchari Sp. nov., for a Plant Growth-Promoting Acetic Acid Bacterium Isolated in Vietnam. Ann. Microbiol. 2019, 69, 1155–1163. [Google Scholar] [CrossRef]
- Ndoye, B.; Cleenwerck, I.; Engelbeen, K.; Dubois-Dauphin, R.; Guiro, A.T.; Van Trappen, S.; Willems, A.; Thonart, P. Acetobacter senegalensis Sp. nov., a Thermotolerant Acetic Acid Bacterium Isolated in Senegal (Sub-Saharan Africa) from Mango Fruit (Mangifera indica L.). Int. J. Syst. Bacteriol. 2007, 57, 1576–1581. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wieme, A.; Spitaels, F.; Balzarini, T.; Nunes, O.C.; Manaia, C.M.; Van Landschoot, A.; De Vuyst, L.; Cleenwerck, I.; Vandamme, P. Acetobacter sicerae Sp. nov., Isolated from Cider and Kefir, and Identification of Species of the Genus Acetobacter by DnaK, GroEL and RpoB Sequence Analysis. Int. J. Syst. Bacteriol. 2014, 64, 2407–2415. [Google Scholar] [CrossRef]
- Pitiwittayakul, N.; Theeragool, G.; Yukphan, P.; Chaipitakchonlatarn, W.; Malimas, T.; Muramatsu, Y.; Tanasupawat, S.; Nakagawa, Y.; Yamada, Y. Acetobacter suratthanensis Sp. nov., an Acetic Acid Bacterium Isolated in Thailand. Ann. Microbiol. 2016, 66, 1157–1166. [Google Scholar] [CrossRef]
- Pitiwittayakul, N.; Yukphan, P.; Chaipitakchonlatarn, W.; Yamada, Y.; Theeragool, G. Acetobacter thailandicus Sp. nov., for a Strain Isolated in Thailand. Ann. Microbiol. 2015, 65, 1855–1863. [Google Scholar] [CrossRef]
- Urakami, T.; Tamaoka, J.; Suzuki, K.-I.; Komagata, K. Acidomonas Gen. nov., Incorporating Acetobacter methanolicus as Acidomonas methanolica comb. nov. Int. J. Syst. Bacteriol. 1989, 39, 50–55. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Muramatsu, Y.; Takahashi, M.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Hamana, K.; Tahara, Y.; et al. Ameyamaea chiangmaiensis gen. nov., Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. Biosci. Biotechnol. Biochem. 2009, 73, 2156–2162. [Google Scholar] [CrossRef]
- Suzuki, R.; Zhang, Y.; Iino, T.; Kosako, Y.; Komagata, K.; Uchimura, T. Asaia astilbes Sp. nov., Asaia platycodi Sp. nov., and Asaia prunellae Sp. nov., novel Acetic Acid Bacteria Isolated from Flowers in Japan. J. Gen. Appl. Microbiol. 2010, 56, 339–346. [Google Scholar] [CrossRef]
- Yamada, Y.; Katsura, K.; Kawasaki, H.; Widyastuti, Y.; Saono, S.; Seki, T.; Uchimura, T.; Komagata, K. Asaia bogorensis gen. nov., Sp. nov., an Unusual Acetic Acid Bacterium in the α-Proteobacteria. Int. J. Syst. Bacteriol. 2000, 50, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Yukphan, P.; Potacharoen, W.; Tanasupawat, S.; Tanticharoen, M.; Yamada, Y. Asaia krungthepensis Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. Int. J. Syst. Bacteriol. 2004, 54, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Asaia lannaensis Sp. nov., a New Acetic Acid Bacterium in the Alphaproteobacteria. Biosci. Biotechnol. Biochem. 2008, 72, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Katsura, K.; Kawasaki, H.; Potacharoen, W.; Saono, S.; Seki, T.; Yamada, Y.; Uchimura, T.; Komagata, K. Asaia siamensis Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. Int. J. Syst. Bacteriol. 2001, 51, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Kommanee, J.; Tanasupawat, S.; Yukphan, P.; Malimas, T.; Muramatsu, Y.; Nakagawa, Y.; Yamada, Y. Asaia spathodeae Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2010, 56, 81–87. [Google Scholar] [CrossRef]
- Yun, J.-H.; Lee, J.-Y.; Hyun, D.-W.; Jung, M.-J.; Bae, J.-W. Bombella apis Sp. nov., an Acetic Acid Bacterium Isolated from the Midgut of a Honey Bee. Int. J. Syst. Bacteriol. 2017, 67, 2184–2188. [Google Scholar] [CrossRef]
- Hilgarth, M.; Redwitz, J.; Ehrmann, M.A.; Vogel, R.F.; Jakob, F. Bombella favorum Sp. nov. and Bombella mellum Sp. nov., Two novel Species Isolated from the Honeycombs of Apis mellifera. Int. J. Syst. Bacteriol. 2021, 71, 004633. [Google Scholar] [CrossRef]
- Li, L.; Praet, J.; Borremans, W.; Nunes, O.C.; Manaia, C.M.; Cleenwerck, I.; Meeus, I.; Smagghe, G.; De Vuyst, L.; Vandamme, P. Bombella intestini Gen. nov., Sp. nov., an Acetic Acid Bacterium Isolated from Bumble Bee Crop. Int. J. Syst. Bacteriol. 2015, 65, 267–273. [Google Scholar] [CrossRef]
- Roh, S.W.; Nam, Y.-D.; Chang, H.-W.; Kim, K.-H.; Kim, M.-S.; Ryu, J.-H.; Kim, S.-H.; Lee, W.-J.; Bae, J.-W. Phylogenetic Characterization of Two novel Commensal Bacteria Involved with Innate Immune Homeostasis in Drosophila melanogaster. Appl. Environ. Microbiol. 2008, 74, 6171–6177. [Google Scholar] [CrossRef]
- Ramírez-Bahena, M.H.; Tejedor, C.; Martín, I.; Velázquez, E.; Peix, A. Endobacter medicaginis gen. nov., Sp. nov., Isolated from Alfalfa Nodules in an Acidic Soil. Int. J. Syst. Bacteriol. 2013, 63, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, M.; Tazato, N.; Handa, Y.; Tomita, J.; Kigawa, R.; Sano, C.; Sugiyama, J. Gluconacetobacter tumulisoli Sp. nov., Gluconacetobacter takamatsuzukensis Sp. nov. and Gluconacetobacter aggeris Sp. nov., Isolated from Takamatsuzuka Tumulus Samples before and during the Dismantling Work in 2007. Int. J. Syst. Bacteriol. 2013, 63, 3981–3988. [Google Scholar] [CrossRef] [PubMed]
- Tazato, N.; Nishijima, M.; Handa, Y.; Kigawa, R.; Sano, C.; Sugiyama, J. Gluconacetobacter tumulicola Sp. nov. and Gluconacetobacter asukensis Sp. nov., Isolated from the Stone Chamber Interior of the Kitora Tumulus. Int. J. Syst. Bacteriol. 2012, 62, 2032–2038. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Ramírez, L.E.; Bustillos-Cristales, R.; Tapia-Hernández, A.; Jiménez-Salgado, T.; Wang, E.T.; Martínez-Romero, E.; Caballero-Mellado, J. novel Nitrogen-Fixing Acetic Acid Bacteria, Gluconacetobacter johannae Sp. nov. and Gluconacetobacter azotocaptans Sp. nov., Associated with Coffee Plants. Int. J. Syst. Bacteriol. 2001, 51, 1305–1314. [Google Scholar] [CrossRef]
- Yamada, Y.; Hoshino, K.; Ishikawa, T. The Phylogeny of Acetic Acid Bacteria Based on the Partial Sequences of 16S Ribosomal RNA: The Elevation of the Subgenus Gluconoacetobacter to the Generic Level. Biosci. Biotechnol. Biochem. 1997, 61, 1244–1251. [Google Scholar] [CrossRef]
- Sombolestani, A.S.; Cleenwerck, I.; Cnockaert, M.; Borremans, W.; Wieme, A.D.; Moutia, Y.; Spaepen, S.; De Vuyst, L.; Vandamme, P. Gluconacetobacter dulcium Sp. nov., a novel Gluconacetobacter Species from Sugar-Rich Environments. Int. J. Syst. Bacteriol. 2021, 71, 004569. [Google Scholar] [CrossRef]
- Schüller, G.; Hertel, C.; Hammes, W.P. Gluconacetobacter entanii Sp. nov., Isolated from Submerged High-Acid Industrial Vinegar Fermentations. Int. J. Syst. Bacteriol. 2000, 50, 2013–2020. [Google Scholar] [CrossRef]
- Franke, I.H.; Fegan, M.; Hayward, C.; Leonard, G.; Stackebrandt, E.; Sly, L.I. Description of Gluconacetobacter sacchari Sp. nov., a New Species of Acetic Acid Bacterium Isolated from the Leaf Sheath of Sugar Cane and from the Pink Sugar-Cane Mealy Bug. Int. J. Syst. Bacteriol. 1999, 49, 1681–1693. [Google Scholar] [CrossRef]
- Yukphan, P.; Charoenyingcharoen, P.; Malimas, S.; Muramatsu, Y.; Nakagawa, Y.; Tanasupawat, S.; Yamada, Y. Gluconobacter aidae Sp. nov., an Acetic Acid Bacteria Isolated from Tropical Fruits in Thailand. Int. J. Syst. Bacteriol. 2020, 70, 4351–4357. [Google Scholar] [CrossRef]
- Yukphan, P.; Takahashi, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter albidus (Ex Kondo and Ameyama 1958) Sp. nov., nom. rev., an Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2004, 50, 235–242. [Google Scholar] [CrossRef]
- Sombolestani, A.S.; Cleenwerck, I.; Cnockaert, M.; Borremans, W.; Wieme, A.D.; De Vuyst, L.; Vandamme, P. Characterization of novel Gluconobacter Species from Fruits and Fermented Food Products: Gluconobacter cadivus Sp. nov., Gluconobacter vitians Sp. nov. and Gluconobacter potus Sp. nov. Int. J. Syst. Bacteriol. 2019, 71, 004751. [Google Scholar] [CrossRef] [PubMed]
- Spitaels, F.; Wieme, A.; Balzarini, T.; Cleenwerck, I.; Van Landschoot, A.; De Vuyst, L.; Vandamme, P. Gluconobacter cerevisiae Sp. nov., Isolated from the Brewery Environment. Int. J. Syst. Bacteriol. 2014, 64, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Akita, M. An Electrophoretic Comparison of Enzymes in Strains of Gluconobacter Species. J. Gen. Appl. Microbiol. 1984, 30, 115–126. [Google Scholar] [CrossRef]
- Mason, L.M.; Claus, G.W. Phenotypic Characteristics Correlated with Deoxyribonucleic Acid Sequence Similarities for Three Species of Gluconobacter: G. Oxydans (Henneberg 1897) De Ley 1961, G. frateurii Sp. nov., and G. asaii Sp. nov. Int. J. Syst. Bacteriol. 1989, 39, 174–184. [Google Scholar] [CrossRef]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Muramatsu, Y.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter japonicus Sp. nov., an Acetic Acid Bacterium in the Alphaproteobacteria. Int. J. Syst. Bacteriol. 2009, 59, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Malimas, T.; Yukphan, P.; Lundaa, T.; Muramatsu, Y.; Takahashi, M.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Suzuki, K.; et al. Gluconobacter kanchanaburiensis Sp. nov., a Brown Pigment-Producing Acetic Acid Bacterium for Thai Isolates in the Alphaproteobacteria. J. Gen. Appl. Microbiol. 2009, 55, 247–254. [Google Scholar] [CrossRef]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter kondonii Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2007, 53, 301–307. [Google Scholar] [CrossRef]
- De Ley, J. Comparative Carbohydrate Metabolism and a Proposal for a Phylogenetic Relationship of the Acetic Acid Bacteria. J. Gen. Microbiol. 1961, 24, 31–50. [Google Scholar] [CrossRef]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Muramatsu, Y.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter roseus (Ex Asai 1935) Sp. nov., nom. rev., a Pink-Colored Acetic Acid Bacterium in the Alphaproteobacteria. J. Gen. Appl. Microbiol. 2008, 54, 119–125. [Google Scholar] [CrossRef]
- Malimas, T.; Yukphan, P.; Takahashi, M.; Muramatsu, Y.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Gluconobacter sphaericus (Ameyama 1975) Comb. nov., a Brown Pigment-Producing Acetic Acid Bacterium in the Alphaproteobacteria. J. Gen. Appl. Microbiol. 2008, 54, 211–220. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Thawai, C.; Yukphan, P.; Moonmangmee, D.; Itoh, T.; Adachi, O.; Yamada, Y. Gluconobacter thailandicus Sp. nov., an Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2004, 50, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Yukphan, P.; Malimas, T.; Lundaa, T.; Muramatsu, Y.; Takahashi, M.; Kaneyasu, M.; Tanasupawat, S.; Nakagawa, Y.; Suzuki, K.; Tanticharoen, M.; et al. Gluconobacter wancherniae Sp. nov., an Acetic Acid Bacterium from Thai Isolates in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2010, 56, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, D.E.; Porcella, S.F.; Stock, F.; Wong, A.; Conville, P.S.; Murray, P.R.; Holland, S.M.; Zelazny, A.M. Granulibacter bethesdensis gen. nov., Sp. nov., a Distinctive Pathogenic Acetic Acid Bacterium in the Family Acetobacteraceae. Int. J. Syst. Evol. Microbiol. 2006, 56, 2609–2616. [Google Scholar] [CrossRef]
- Naloka, K.; Yukphan, P.; Matsutani, M.; Matsushita, K.; Theeragool, G. Komagataeibacter diospyri Sp. nov., a novel Species of Thermotolerant Bacterial Nanocellulose-Producing Bacterium. Int. J. Syst. Bacteriol. 2020, 70, 251–258. [Google Scholar] [CrossRef]
- Yamada, Y. Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the Genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov. and Komagataeibacter maltaceti comb. nov. Int. J. Syst. Bacteriol. 2014, 64, 1670–1672. [Google Scholar] [CrossRef]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti Sp. nov. and Komagataeibacter melomenusus Sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Lisdiyanti, P.; Kawasaki, H.; Widyastuti, Y.; Saono, S.; Seki, T.; Yamada, Y.; Uchimura, T.; Komagata, K. Kozakia baliensis Gen. nov., Sp. nov., a novel Acetic Acid Bacterium in the α-Proteobacteria. Int. J. Syst. Bacteriol. 2002, 52, 813–818. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Potacharoen, W.; Tanasupawat, S.; Tanticharoen, M.; Yamada, Y. Neoasaia chiangmaiensis gen. nov., Sp. nov., a novel Osmotolerant Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2005, 51, 301–311. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Muramatsu, Y.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Neokomagataea Gen. nov., with Descriptions of Neokomagataea thailandica Sp. nov. and Neokomagataea tanensis Sp. nov., Osmotolerant Acetic Acid Bacteria of the α-Proteobacteria. Biosci. Biotechnol. Biochem. 2011, 75, 419–426. [Google Scholar] [CrossRef]
- Thi Lan Vu, H.; Yukphan, P.; Chaipitakchonlatarn, W.; Malimas, T.; Muramatsu, Y.; Thi Tu Bui, U.; Tanasupawat, S.; Cong Duong, K.; Nakagawa, Y.; Thanh Pham, H.; et al. Nguyenibacter vanlangensis gen. nov., Sp. nov., an Unusual Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2013, 59, 153–166. [Google Scholar] [CrossRef]
- Brandão, P.R.; Crespo, M.T.B.; Nascimento, F.X. Phylogenomic and Comparative Analyses Support the Reclassification of Several Komagataeibacter Species as novel Members of the Novacetimonas gen. nov. and Bring New Insights into the Evolution of Cellulose Synthase Genes. Int. J. Syst. Bacteriol. 2022, 72, 005252. [Google Scholar] [CrossRef] [PubMed]
- Jojima, Y.; Mihara, Y.; Suzuki, S.; Yokozeki, K.; Yamanaka, S.; Fudou, R. Saccharibacter floricola gen. nov., Sp. nov., a novel Osmophilic Acetic Acid Bacterium Isolated from Pollen. Int. J. Syst. Bacteriol. 2004, 54, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, P.; Nair, S. Swaminathania salitolerans gen. nov., Sp. nov., a Salt-Tolerant, Nitrogen-Fixing and Phosphate-Solubilizing Bacterium from Wild Rice (Porteresia Coarctata Tateoka). Int. J. Syst. Bacteriol. 2004, 54, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Malimas, T.; Chaipitakchonlatarn, W.; Thi Lan Vu, H.; Yukphan, P.; Muramatsu, Y.; Tanasupawat, S.; Potacharoen, W.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Swingsia samuiensis gen. nov., Sp. nov., an Osmotolerant Acetic Acid Bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 2013, 59, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.L.; Malimas, T.; Chaipitakchonlatarn, W.; Bui, V.T.T.; Yukphan, P.; Bui, U.T.T.; Muramatsu, Y.; Sitdhipol, J.; Tanasupawat, S.; Duong, K.C.; et al. Tanticharoenia aidae Sp. nov., for Acetic Acid Bacteria Isolated in Vietnam. Ann. Microbiol. 2016, 66, 417–423. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Muramatsu, Y.; Takahashi, M.; Kaneyasu, M.; Tanasupawat, S.; Nakagawa, Y.; Suzuki, K.; Potacharoen, W.; Yamada, Y. Tanticharoenia sakaeratensis Gen. nov., Sp. nov., a New Osmotolerant Acetic Acid Bacterium in the α-Proteobacteria. Biosci. Biotechnol. Biochem. 2008, 72, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, P.; Lei, Q.; Li, B.; Sun, Y.; Li, S.; Lei, H.; Xie, N. Metabolic Adaptability Shifts of Cell Membrane Fatty Acids of Komagataeibacter hansenii HDM1-3 Improve Acid Stress Resistance and Survival in Acidic Environments. J. Ind. Microbiol. Biotechnol. 2019, 46, 1491–1503. [Google Scholar] [CrossRef]
- Sankuan, X.; Cuimei, Z.; Bingqian, F.; Yu, Z.; Menglei, X.; Linna, T.; Jia, S.; Xinyi, Z.; Min, W. Metabolic Network of Ammonium in Cereal Vinegar Solid-State Fermentation and Its Response to Acid Stress. Food Microbiol. 2021, 95, 103684. [Google Scholar] [CrossRef]
- Sriherfyna, F.H.; Matsutani, M.; Hirano, K.; Koike, H.; Kataoka, N.; Yamashita, T.; Nakamaru-Ogiso, E.; Matsushita, K.; Yakushi, T. The Auxiliary NADH Dehydrogenase Plays a Crucial Role in Redox Homeostasis of Nicotinamide Cofactors in the Absence of the Periplasmic Oxidation System in Gluconobacter oxydans NBRC3293. Appl. Environ. Microbiol. 2021, 87, e02155-20. [Google Scholar] [CrossRef]
- Wang, L.; Hong, H.; Zhang, C.; Huang, Z.; Guo, H. Transcriptome Analysis of Komagataeibacter europaeus CGMCC 20445 Responses to Different Acidity Levels During Acetic Acid Fermentation. Pol. J. Microbiol. 2021, 70, 305–313. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, R.; Yin, H.; Bai, X.; Chang, Y.; Xia, M.; Wang, M. Acetobacter pasteurianus Metabolic Change Induced by Initial Acetic Acid to Adapt to Acetic Acid Fermentation Conditions. Appl. Microbiol. Biotechnol. 2017, 101, 7007–7016. [Google Scholar] [CrossRef] [PubMed]
- Adachi, O.; Tayama, K.; Shinagawa, E.; Matsushita, K.; Ameyama, M. Purification and Characterization of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter suboxydans. Agric. Biol. Chem. 1980, 44, 503–515. [Google Scholar] [CrossRef]
- Ameyama, M.; Adachi, O. Alcohol Dehydrogenase from Acetic Acid Bacteria, Membrane-Bound. Meth. Enzymol. 1982, 89, 450–457. [Google Scholar]
- Qin, Z.; Yu, S.; Chen, J.; Zhou, J. Dehydrogenases of Acetic Acid Bacteria. Biotechnol. Adv. 2022, 54, 107863. [Google Scholar] [CrossRef]
- Mullins, E.A.; Francois, J.A.; Kappock, T.J. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA): Acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J. Bacteriol. 2008, 190, 4933–4940. [Google Scholar] [CrossRef]
- Arai, H.; Sakurai, K.; Ishii, M. Metabolic features of Acetobacter aceti. In Acetic Acid Bacteria; Matsushita, K., Toyama, H., Tonouchi, N., Okamoto-Kainuma, A., Eds.; Springer: Tokyo, Japan, 2016; pp. 255–271. ISBN 978-4-431-55931-3. [Google Scholar]
- Yakushi, T.; Matsushita, K. Alcohol Dehydrogenase of Acetic Acid Bacteria: Structure, Mode of Action, and Applications in Biotechnology. Appl. Microbiol. Biotechnol. 2010, 86, 1257–1265. [Google Scholar] [CrossRef]
- Adler, P.; Frey, L.J.; Berger, A.; Bolten, C.J.; Hansen, C.E.; Wittmann, C. The Key to Acetate: Metabolic Fluxes of Acetic Acid Bacteria under Cocoa Pulp Fermentation-Simulating Conditions. Appl. Environ. Microbiol. 2014, 80, 4702–4716. [Google Scholar] [CrossRef]
- García-García, I.; Cañete-Rodríguez, A.M.; Santos-Dueñas, I.M.; Jiménez-Hornero, J.E.; Ehrenreich, A.; Liebl, W.; García-Martínez, T.; Mauricio, J.C. Biotechnologically Relevant Features of Gluconic Acid Production by Acetic Acid Bacteria. Acetic Acid Bact. 2017, 6, 6458. [Google Scholar] [CrossRef]
- Yin, H.; Zhang, R.; Xia, M.; Bai, X.; Mou, J.; Zheng, Y.; Wang, M. Effect of Aspartic Acid and Glutamate on Metabolism and Acid Stress Resistance of Acetobacter pasteurianus. Microb Cell. Fact. 2017, 16, 109. [Google Scholar] [CrossRef]
- Bringer, S.; Bott, M. Central Carbon Metabolism and Respiration in Gluconobacter oxydans. In Acetic Acid Bacteria; Matsushita, K., Toyama, H., Tonouchi, N., Okamoto-Kainuma, A., Eds.; Springer: Tokyo, Japan, 2016; pp. 235–253. ISBN 978-4-431-55931-3. [Google Scholar]
- Kiefler, I.; Bringer, S.; Bott, M. Metabolic Engineering of Gluconobacter oxydans 621H for Increased Biomass Yield. Appl. Microbiol. Biotechnol. 2017, 101, 5453–5467. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.-P.; Taillandier, P. Understanding Kombucha Tea Fermentation: A Review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Ayed, L.; Ben Abid, S.; Hamdi, M. Development of a Beverage from Red Grape Juice Fermented with the Kombucha Consortium. Ann. Microbiol. 2017, 67, 111–121. [Google Scholar] [CrossRef]
- Mounir, M.; Shafiei, R.; Zarmehrkhorshid, R.; Hamouda, A.; Ismaili Alaoui, M.; Thonart, P. Simultaneous Production of Acetic and Gluconic Acids by a Thermotolerant Acetobacter Strain during Acetous Fermentation in a Bioreactor. J. Biosci. Bioeng. 2016, 121, 166–171. [Google Scholar] [CrossRef]
- Cañete-Rodríguez, A.M.; Santos-Dueñas, I.M.; Jiménez-Hornero, J.E.; Ehrenreich, A.; Liebl, W.; García-García, I. Gluconic Acid: Properties, Production Methods and Applications—An Excellent Opportunity for Agro-Industrial by-Products and Waste Bio-Valorization. Process Biochem. 2016, 51, 1891–1903. [Google Scholar] [CrossRef]
- Mohammadkazemi, F.; Azin, M.; Ashori, A. Production of Bacterial Cellulose Using Different Carbon Sources and Culture Media. Carbohydr. Polym. 2015, 117, 518–523. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, Y.; Phillips, G.O.; Yang, G. Utilization of Bacterial Cellulose in Food. Food Hydrocoll. 2014, 35, 539–545. [Google Scholar] [CrossRef]
- Ullah, H.; Wahid, F.; Santos, H.A.; Khan, T. Advances in Biomedical and Pharmaceutical Applications of Functional Bacterial Cellulose-Based Nanocomposites. Carbohydr. Polym. 2016, 150, 330–352. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Coughlan, L.M.; Briandet, R.; Cotter, P.D. Biofilms in Food Processing Environments: Challenges and Opportunities. Annu. Rev. Food Sci. Technol. 2019, 10, 173–195. [Google Scholar] [CrossRef]
- Peng, M.Y.; Zhang, X.J.; Huang, T.; Zhong, X.Z.; Chai, L.J.; Lu, Z.M.; Shi, J.S.; Xu, Z.H. Komagataeibacter europaeus improves community stability and function in solid-state cereal vinegar fermentation ecosystem: Non-abundant species plays important role. Food Res. Int. 2021, 150, 110815. [Google Scholar] [CrossRef]
- Yang, H.; He, Y.; Liao, J.; Li, X.; Zhang, J.; Liebl, W.; Chen, F. RNA-Seq Transcriptomic Analysis Reveals Gene Expression Profiles of Acetic Acid Bacteria under High-Acidity Submerged Industrial Fermentation Process. Front. Microbiol. 2022, 13, 956729. [Google Scholar] [CrossRef]





| Genera | Species | References | |
|---|---|---|---|
| Acetobacter | 1 | Acetobacter aceti | Skerman et al. [77] |
| 2 | Acetobacter cerevisiae | Cleenwerck et al. [85] | |
| 3 | Acetobacter cibinongensis | Lisdiyanti et al. [86] | |
| 4 | Acetobacter conturbans | Sombolestani et al. [87] | |
| 5 | Acetobacter estunensis | Lisdiyanti et al. [88] | |
| 6 | Acetobacter fabarum | Cleenwerck et al. [89] | |
| 7 | Acetobacter fallax | Sombolestani et al. [87] | |
| 8 | Acetobacter farinalis | Tanasupawat et al. [90] | |
| 9 | Acetobacter garciniae | Yukphan et al. [91] | |
| 10 | Acetobacter ghanensis | Cleenwerck et al. [92] | |
| 11 | Acetobacter indonesiensis | Lisdiyanti et al. [88] | |
| 12 | Acetobacter lambici | Spitaels et al. [93] | |
| 13 | Acetobacter lovaniensis | Lisdiyanti et al. [88] | |
| 14 | Acetobacter malorum | Cleenwerck et al. [85] | |
| 15 | Acetobacter musti | Ferrer et al. [94] | |
| 16 | Acetobacter nitrogenifiens | Dutta and Gachhui [95] | |
| 17 | Acetobacter oeni | Silva et al. [96] | |
| 18 | Acetobacter okinawensis | Lino et al. [97] | |
| 19 | Acetobacter orientalis | Lisdiyanti et al. [86] | |
| 20 | Acetobacter orleanensis | Lisdiyanti et al. [88] | |
| 21 | Acetobacter oryzoeni | Baek et al. [98] | |
| 22 | Acetobacter oryzifermentans | Kim et al. [99] | |
| 23 | Acetobacter papayae | Lino et al. [97] | |
| 24 | Acetobacter pasteurianus | Skerman et al. [77] | |
| 25 | Acetobacter persici | Lino et al. [97] | |
| 26 | Acetobacter pomorum | Sokollek et al. [100] | |
| 27 | Acetobacter sacchari | Vu et al. [101] | |
| 28 | Acetobacter senegalensis | Ndoye et al. [102] | |
| 29 | Acetobacter sicerae | Li et al. [103] | |
| 30 | Acetobacter suratthaniensis | Pitiwittayakul et al. [104] | |
| 31 | Acetobacter syzygii | Lisdiyanti et al. [86] | |
| 32 | Acetobacter thailandicus | Pitiwittayakul et al. [105] | |
| 33 | Acetobacter tropicalis | Lisdiyanti et al. [88] | |
| Acidomonas | 34 | Acidomonas methanolica | Urakami et al. [106] |
| Ameyamaea | 35 | Ameyamaea chiangmaiensis | Yukphan et al. [107] |
| Asaia | 36 | Asaia astilbis | Suzuki et al. [108] |
| 37 | Asaia bogorensis | Yamada et al. [109] | |
| 38 | Asaia krungthepensis | Yukphan et al. [110] | |
| 39 | Asaia lannensis | Malimas et al. [111] | |
| 40 | Asaia platycodi | Suzuki et al. [108] | |
| 41 | Asaia prunellae | Suzuki et al. [108] | |
| 42 | Asaia siamensis | Katsura et al. [112] | |
| 43 | Asaia spathodeae | Kommanee et al. [113] | |
| Bombella | 44 | Bombella apis | Yun et al. [114] |
| 45 | Bombella favorum | Hilgarth et al. [115] | |
| 46 | Bombella intestini | Li et al. [116] | |
| 47 | Bombella mellum | Hilgarth et al. [115] | |
| Commensalibacter | 48 | Commensalibacter intestini | Roh et al. [117] |
| Endobacter | 49 | Endobacter medicaginis | Ramírez-Bahena et al. [118] |
| Gluconacetobacter | 50 | Gluconacetobacter aggeris | Nishijima et al. [119] |
| 51 | Gluconacetobacter asukensis | Tazato et al. [120] | |
| 52 | Gluconacetobacter azotocaptans | Fuentes-Ramírez et al. [121] | |
| 53 | Gluconacetobacter diazotrophicus | Yamada et al. [122] | |
| 54 | Gluconacetobacter dulcium | Sombolestani et al. [123] | |
| 55 | Gluconacetobacter entanii | Schüller et al. [124] | |
| 56 | Gluconacetobacter johannae | Fuentes-Ramírez et al. [121] | |
| 57 | Gluconacetobacter liquefaciens | Yamada et al. [122] | |
| 58 | Gluconacetobacter sacchari | Franke et al. [125] | |
| 59 | Gluconacetobacter takamatsuzukensis | Nishijima et al. [119] | |
| 60 | Gluconacetobacter tumulicola | Tazato et al. [120] | |
| 61 | Gluconacetobacter tumulisoli | Nishijima et al. [119] | |
| Gluconobacter | 62 | Gluconobacter aidae | Yukphan et al. [126] |
| 63 | Gluconobacter albidus | Yukphan et al. [127] | |
| 64 | Gluconobacter cadivus | Sombolestani et al. [128] | |
| 65 | Gluconobacter cerevisiae | Spitaels et al. [129] | |
| 66 | Gluconobacter cerinus | Yamada and Akita [130] | |
| 67 | Gluconobacter frateurii | Mason and Claus [131] | |
| 68 | Gluconobacter japonicus | Malimas et al. [132] | |
| 69 | Gluconobacter kanchanaburiensis | Malimas et al. [133] | |
| 70 | Gluconobacter kondonii | Malimas et al. [134] | |
| 71 | Gluconobacter morbifer | Roh et al. [117] | |
| 72 | Gluconobacter oxydans | De Ley [135] | |
| 73 | Gluconobacter potus | Sombolestani et al. [128] | |
| 74 | Gluconobacter roseus | Malimas et al. [136] | |
| 75 | Gluconobacter sphaericus | Malimas et al. [137] | |
| 76 | Gluconobacter thailandicus | Tanasupawat et al. [138] | |
| 77 | Gluconobacter vitians | Sombolestani et al. [126] | |
| 78 | Gluconobacter wancherniae | Yukphan et al. [139] | |
| Granulibacter | 79 | Granulibacter bethesdensis | Greenberg et al. [140] |
| Komagataeibacter | 80 | Komagataeibacter diospyri | Naloka et al. [141] |
| 81 | Komagataeibacter europaeus | Yamada et al. [80] | |
| 82 | Komagataeibacter intermedius | Yamada et al. [80] | |
| 83 | Komagataeibacter kakiaceti | Yamada [142] | |
| 84 | Komagataeibacter kombuchae | Yamada et al. [80] | |
| 85 | Komagataeibacter medellinensis | Yamada [142] | |
| 86 | Komagataeibacter melaceti | Marič et al. [143] | |
| 87 | Komagataeibacter melomenusus | Marič et al. [143] | |
| 88 | Komagataeibacter nataicola | Yamada et al. [80] | |
| 89 | Komagataeibacter oboediens | Yamada et al. [80] | |
| 90 | Komagataeibacter rhaeticus | Yamada et al. [80] | |
| 91 | Komagataeibacter saccharivorans | Yamada et al. [80] | |
| 92 | Komagataeibacter sucrofermentans | Yamada et al. [80] | |
| 93 | Komagataeibacter swingsii | Yamada et al. [80] | |
| 94 | Komagataeibacter xylinus | Yamada et al. [80] | |
| Kozakia | 95 | Kozakia baliensis | Lisdiyanti et al. [144] |
| Neoasaia | 96 | Neoasaia chiangmaiensis | Yukphan et al. [145] |
| Neokomagataea | 97 | Neokomagataea tanensis | Yukphan et al. [146] |
| 98 | Neokomagataea thailandica | Yukphan et al. [146] | |
| Nguyenibacter | 99 | Nguyenibacter vanlangensis | Vu et al. [147] |
| Novacetimonas | 100 101 | Novacetimonas cocois Novacetimonas hansenii | Brandao et al. [148] Brandao et al. [148] |
| 102 | Novacetimonas maltaceti | Brandao et al. [148] | |
| 103 | Novacetimonas pomaceti | Brandao et al. [148] | |
| Saccharibacter | 104 | Saccharibacter floricola | Jojima et al. [149] |
| Swaminathania | 105 | Swaminathania saitolerans | Loganathan and Nair [150] |
| Swingsia | 106 | Swingsia samuiensis | Malimas et al. [151] |
| Tanticharoenia | 107 | Tanticharoenia aidae | Vu et al. [152] |
| 108 | Tanticharoenia sakaeratensis | Yukphan et al. [153] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Román-Camacho, J.J.; García-García, I.; Santos-Dueñas, I.M.; García-Martínez, T.; Mauricio, J.C. Latest Trends in Industrial Vinegar Production and the Role of Acetic Acid Bacteria: Classification, Metabolism, and Applications—A Comprehensive Review. Foods 2023, 12, 3705. https://doi.org/10.3390/foods12193705
Román-Camacho JJ, García-García I, Santos-Dueñas IM, García-Martínez T, Mauricio JC. Latest Trends in Industrial Vinegar Production and the Role of Acetic Acid Bacteria: Classification, Metabolism, and Applications—A Comprehensive Review. Foods. 2023; 12(19):3705. https://doi.org/10.3390/foods12193705
Chicago/Turabian StyleRomán-Camacho, Juan J., Isidoro García-García, Inés M. Santos-Dueñas, Teresa García-Martínez, and Juan C. Mauricio. 2023. "Latest Trends in Industrial Vinegar Production and the Role of Acetic Acid Bacteria: Classification, Metabolism, and Applications—A Comprehensive Review" Foods 12, no. 19: 3705. https://doi.org/10.3390/foods12193705