Caffeic Acid Phenethyl Ester Encapsulated in Self-Assemble Rice Peptides Nanoparticles: Storage Stability, In Vitro Release, and Their Interaction Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Rice Protein and Peptides
2.3. Particle Size and Polydispersity Index (PDI) Analysis
2.4. Analysis of Internal Forces of RPNPs
2.5. Encapsulation of CAPE by Rice Peptides
2.6. Storage Stability of CAPE-RPNPs
2.7. Release Behavior of CAPE-RPNPs
2.8. Fluorescence Quenching Analysis
2.9. Statistical Analysis
3. Results and Discussion
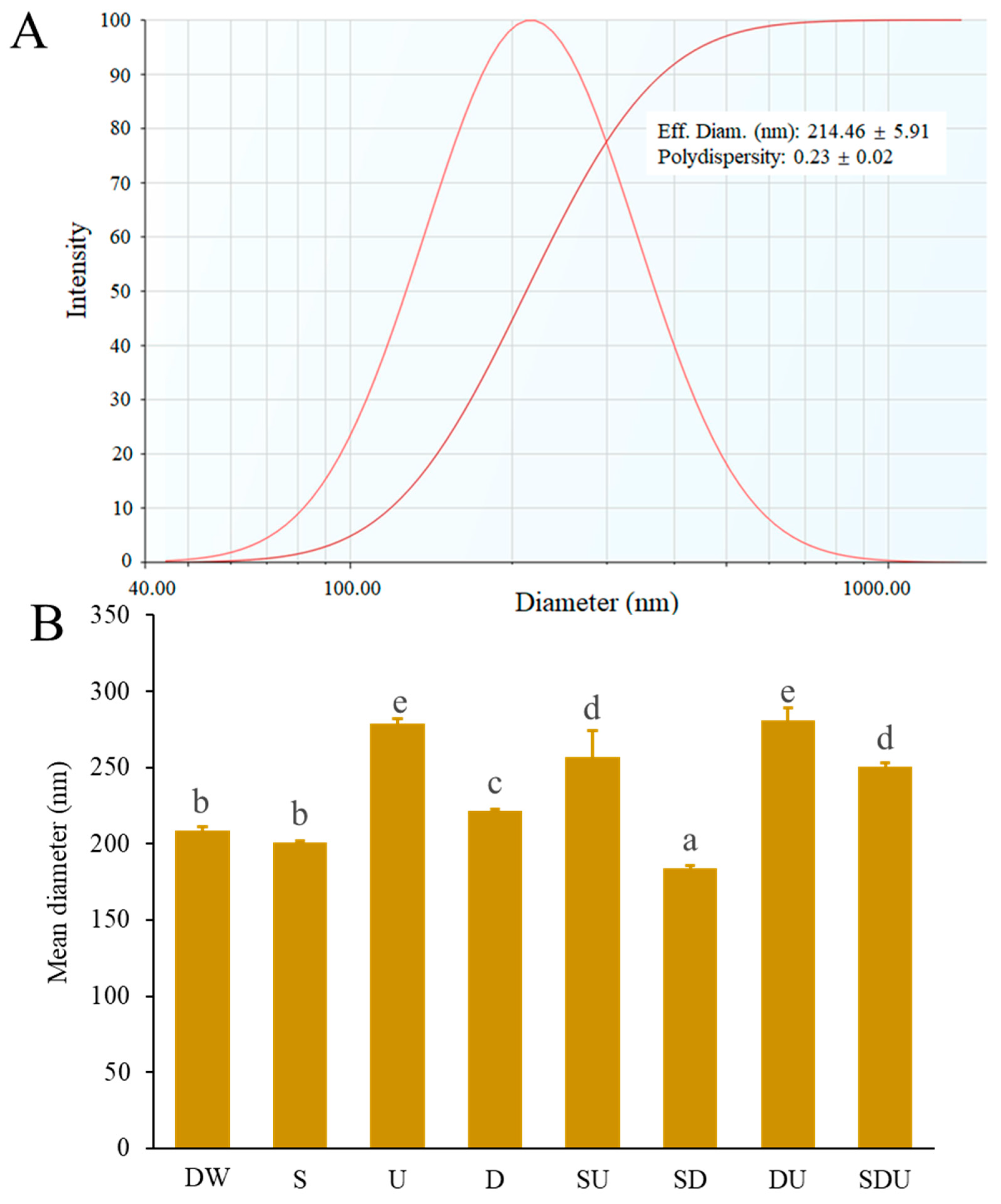
3.1. Effective Diameter, Polydispersity Index (PDI), and Internal Forces of Rice Peptide Nanoparticles (RPNs)
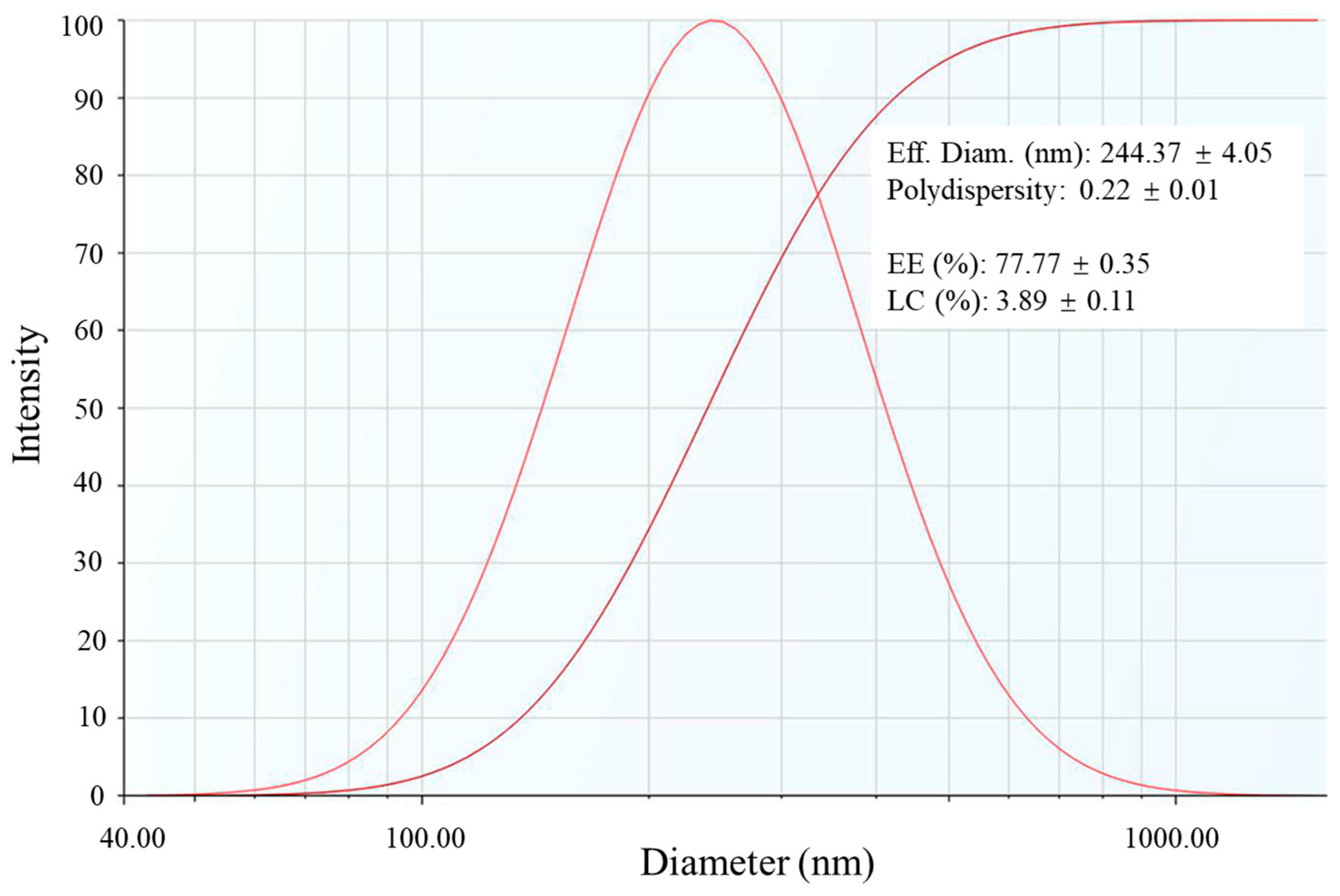
3.2. Encapsulation of CAPE by RPNs
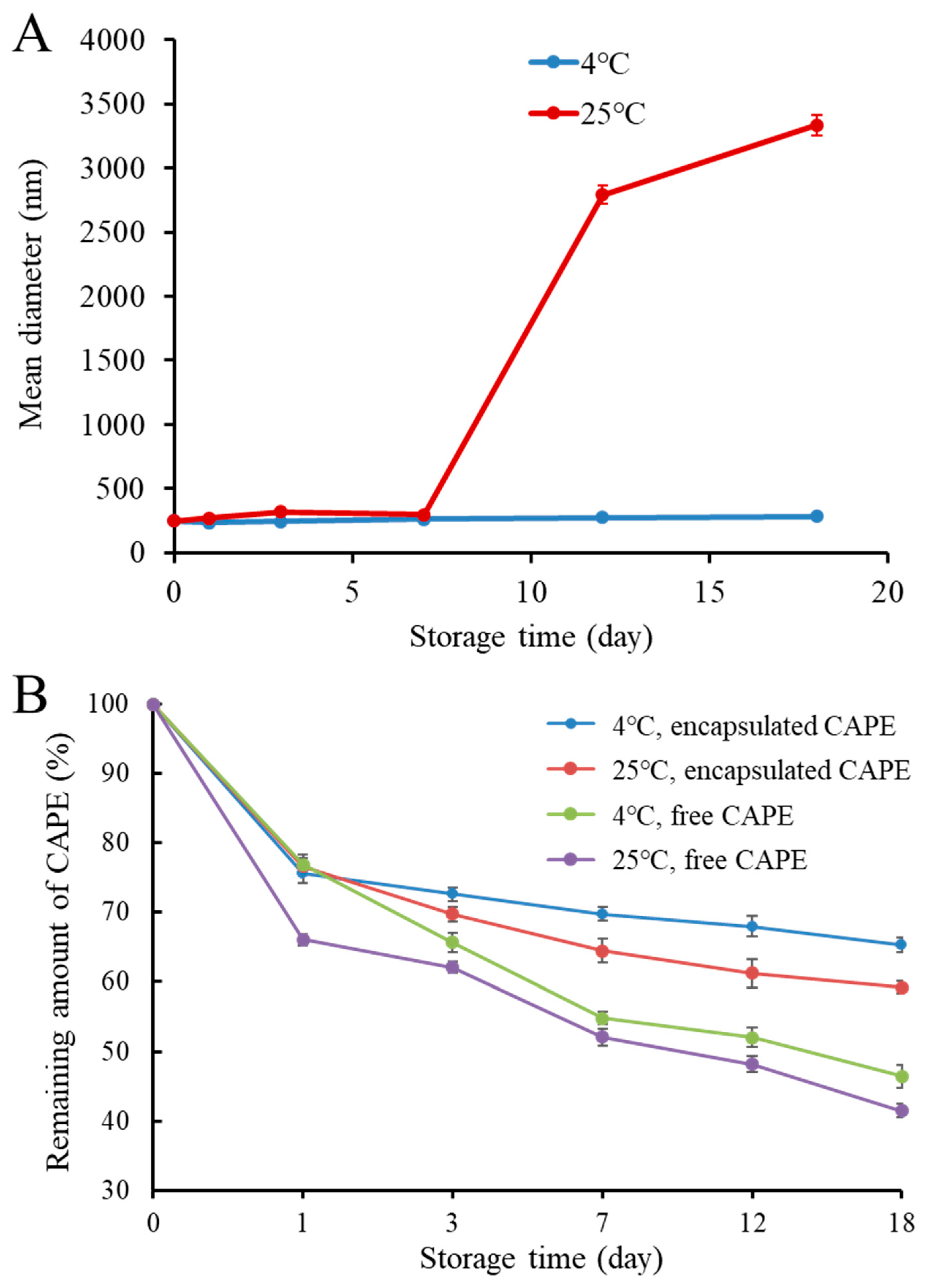
3.3. Storage Stability
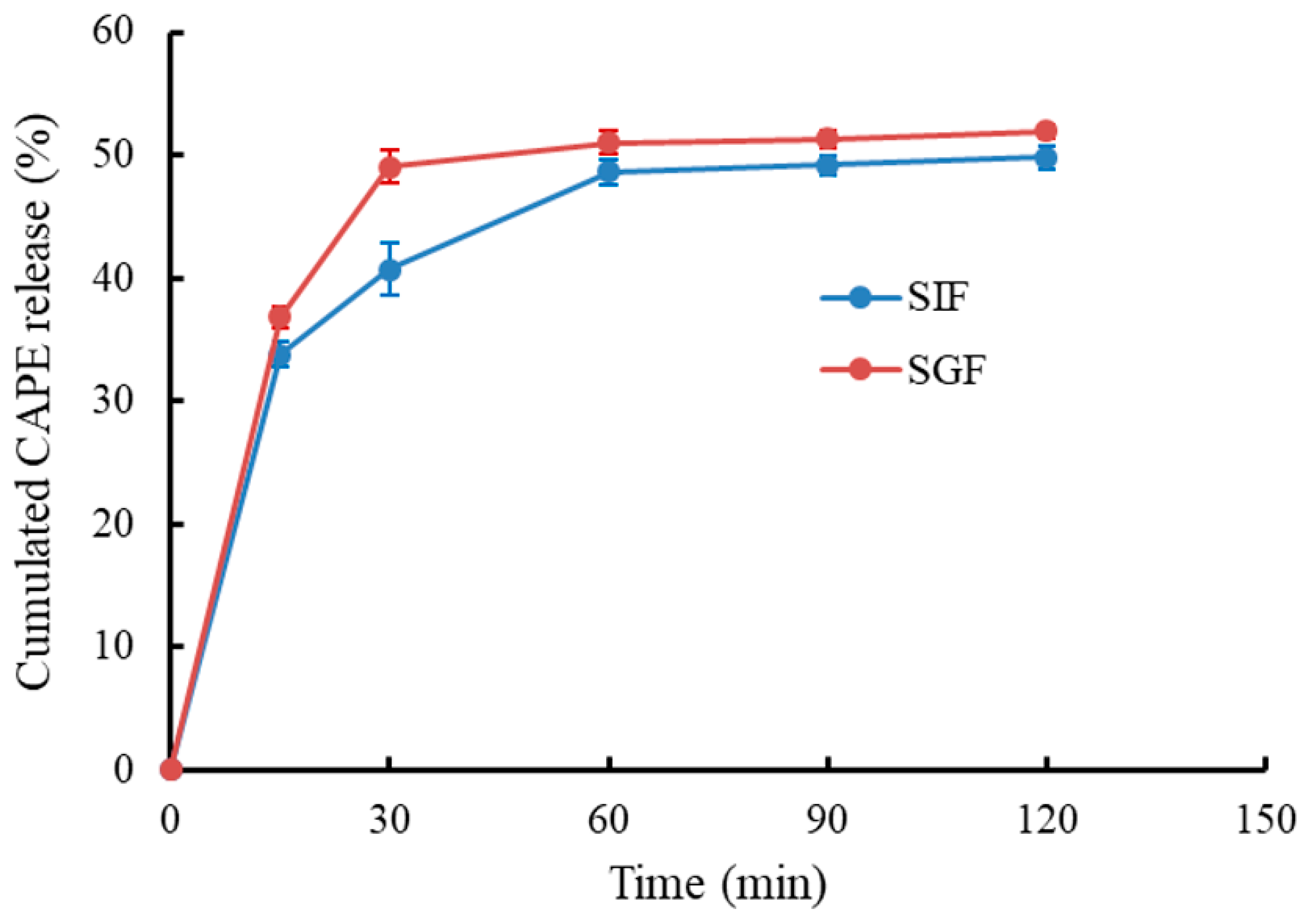
3.4. In Vitro Release of CAPE-RPNs
3.5. Interaction Mechanism between RPNs and CAPE
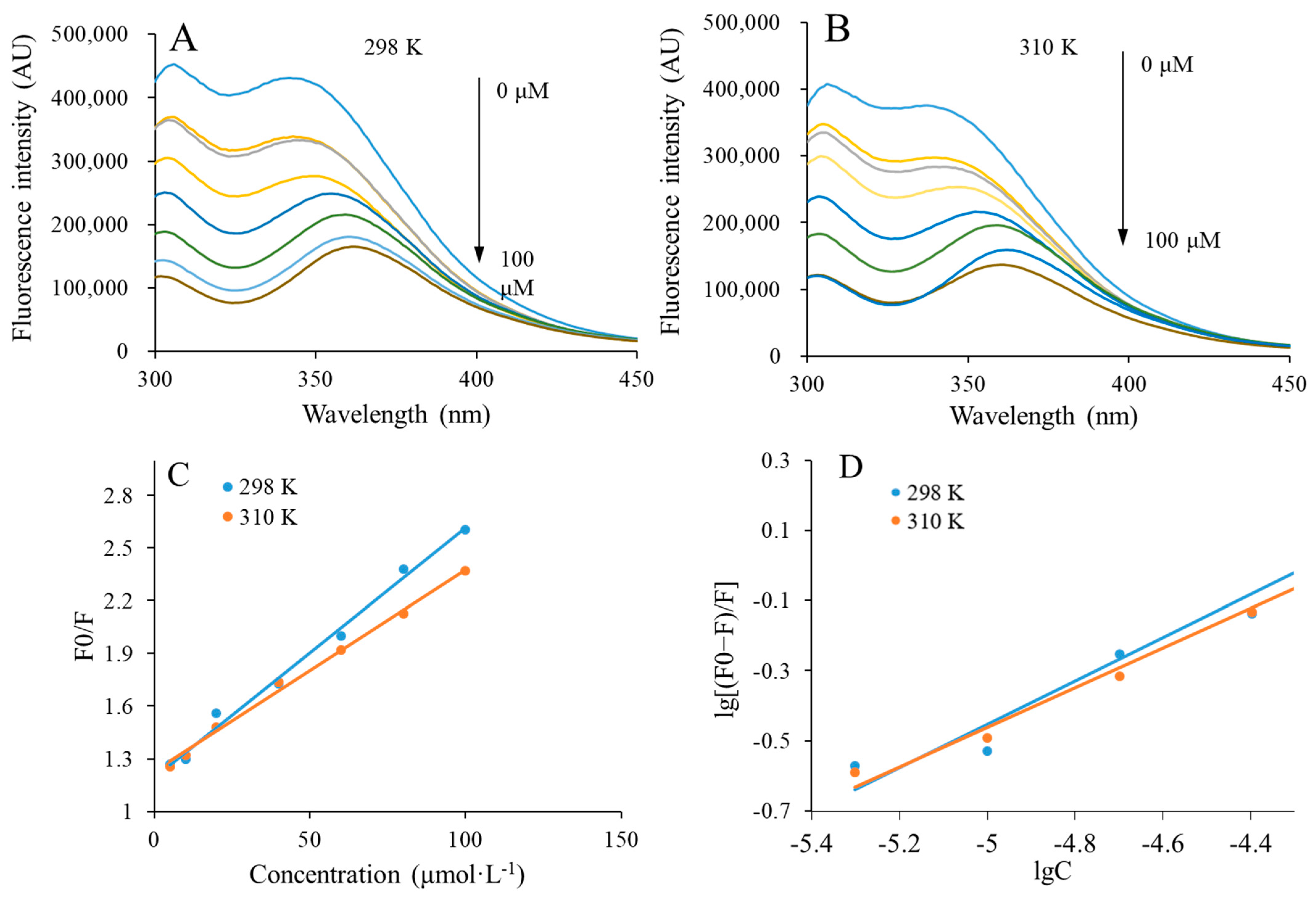
3.5.1. Fluorescence Quenching
3.5.2. Binding Constants and Number of Binding Sites
3.5.3. Thermodynamic Parameters
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, A.Y.; Leible, M.; Lin, J.; Weiss, J.; Zhong, Q.X. Caffeic Acid Phenethyl Ester Loaded in Skim Milk Microcapsules: Physicochemical Properties and Enhanced In Vitro Bioaccessibility and Bioactivity against Colon Cancer Cells. J. Agric. Food Chem. 2020, 68, 14978–14987. [Google Scholar] [CrossRef]
- Nai, X.; Chen, Y.R.; Zhang, Q.; Hao, S.Y.; Xuan, H.Z.; Liu, J. Interaction between caffeic acid phenethyl ester and protease: Monitoring by spectroscopic and molecular docking approaches. Luminescence 2022, 37, 1025–1036. [Google Scholar] [CrossRef]
- Chen, H.Q.; Guan, Y.G.; Baek, S.J.; Zhong, Q.X. Caffeic Acid Phenethyl Ester Loaded in Microemulsions: Enhanced In Vitro Activity against Colon and Breast Cancer Cells and Possible Cellular Mechanisms. Food Biophys. 2019, 14, 80–89. [Google Scholar] [CrossRef]
- Wei, X.L.; Dai, J.; Zhong, Y.; Zhang, D.; Liu, L.; Wang, L.J.; Huang, Y.K.; Chen, P.F.; Zhou, Z.; Chen, X.G.; et al. Caffeic acid phenethyl ester loaded in nano-targeted delivery system with casein: Physicochemical characterization, in vitro release, and binding mechanisms. LWT-Food Sci. Technol. 2021, 150, 10. [Google Scholar] [CrossRef]
- Garrido, E.; Cerqueira, A.S.; Chavarria, D.; Silva, T.; Borges, F.; Garrido, J. Microencapsulation of caffeic acid phenethyl ester and caffeic acid phenethyl amide by inclusion in hydroxypropyl-β-cyclodextrin. Food Chem. 2018, 254, 260–265. [Google Scholar] [CrossRef]
- Ling, M.; Xu, Y.F.; Huang, X.; He, C.W.; Zhou, Z. Phosphorylated walnut protein/chitosan nanocomplexes as promising carriers for encapsulation of caffeic acid phenethyl ester. J. Sci. Food Agric. 2023, 103, 5770–5781. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Liu, K.K.; Wang, X.L.; Shao, Y.W.; Li, X.; Hao, L.M.; Zhang, X.M.; Yi, J.J.; Lu, J.K. Co-assembling nanoparticles of Asiatic acid and Caffeic acid phenethyl ester: Characterization, stability and bioactivity in vitro. Food Chem. 2023, 402, 8. [Google Scholar] [CrossRef]
- Mo, H.R.; Chen, X.W.; Cui, B.; Chen, Y.L.; Chen, M.L.; Xu, Z.; Wen, L.; Cheng, Y.H.; Jiao, Y. Formation and Characterization of Self-Assembled Rice Protein Hydrolysate Nanoparticles as Soy Isoflavone Delivery Systems. Foods 2023, 12, 1523. [Google Scholar] [CrossRef]
- Bao, C.; Li, Z.K.; Liang, S.; Hu, Y.L.; Wang, X.Y.; Fang, B.; Wang, P.J.; Chen, S.N.; Li, Y. Microneedle Patch Delivery of Capsaicin-Containing α-Lactalbumin Nanomicelles to Adipocytes Achieves Potent Anti-Obesity Effects. Adv. Funct. Mater. 2021, 31, 10. [Google Scholar] [CrossRef]
- Du, Z.Y.; Li, Q.; Li, J.G.; Su, E.Y.; Liu, X.; Wan, Z.L.; Yang, X.Q. Self-Assembled Egg Yolk Peptide Micellar Nanoparticles as a Versatile Emulsifier for Food-Grade Oil-in-Water Pickering Nanoemulsions. J. Agric. Food Chem. 2019, 67, 11728–11740. [Google Scholar] [CrossRef]
- Amagliani, L.; O’Regan, J.; Kelly, A.L.; O’Mahony, J.A. The composition, extraction, functionality and applications of rice proteins: A review. Trends Food Sci. Technol. 2017, 64, 1–12. [Google Scholar] [CrossRef]
- Song, H.D.; Wang, Q.Y.; Shao, Z.W.; Wang, X.Y.; Cao, H.W.; Huang, K.; Sun, Q.Q.; Sun, Z.L.; Guan, X. In vitro gastrointestinal digestion of buckwheat (Fagopyrum esculentum Moench) protein: Release and structural characteristics of novel bioactive peptides stimulating gut cholecystokinin secretion. Food Funct. 2023, 14, 7469–7477. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.J.; Song, J.L.; Qi, M.M.; Suo, W.J.; Deng, Y.X.; Liu, Y.; Li, L.X.; Zhang, D.L.; Wang, C.J.; Li, H.J. Modification mechanism of protein in rice adjuncts upon extrusion and its effects on nitrogen conversion during mashing. Food Chem. 2023, 407, 135150. [Google Scholar] [CrossRef]
- Zou, Y.; Zheng, Q.W.; Chen, X.; Ye, Z.W.; Wei, T.; Guo, L.Q.; Lin, J.F. Physicochemical and emulsifying properties of protein isolated from Phlebopus portentosus. LWT-Food Sci. Technol. 2021, 142, 8. [Google Scholar] [CrossRef]
- Song, H.D.; He, A.J.; Guan, X.; Chen, Z.Y.; Bao, Y.Z.; Huang, K. Fabrication of chitosan-coated epigallocatechin-3-gallate (EGCG)-hordein nanoparticles and their transcellular permeability in Caco-2/HT29 cocultures. Int. J. Biol. Macromol. 2022, 196, 144–150. [Google Scholar] [CrossRef]
- Zaky, A.A.; El-Aty, A.M.A.; Ma, A.; Jia, Y. An overview on antioxidant peptides from rice bran proteins: Extraction, identification, and applications. Crit. Rev. Food Sci. Nutr. 2022, 62, 1350–1362. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zuo, Z.Y.; Yu, P.B.; Li, T.; Guang, M.; Chen, Z.X.; Wang, L. Rice peptide nanoparticle as a bifunctional food-grade Pickering stabilizer prepared by ultrasonication: Structural characteristics, antioxidant activity, and emulsifying properties. Food Chem. 2021, 343, 128545. [Google Scholar] [CrossRef]
- Hong, H.; Akbari, A.; Wu, J.P. Small amphipathic peptides are responsible for the assembly of cruciferin nanoparticles. Sci. Rep. 2017, 7, 13. [Google Scholar] [CrossRef]
- Chen, H.M.; Cai, X.X.; Cheng, J.; Wang, S.Y. Self-assembling peptides: Molecule-nanostructure-function and application on food industry. Trends Food Sci. Technol. 2022, 120, 212–222. [Google Scholar] [CrossRef]
- Song, H.D.; Wang, Q.Y.; He, A.J.; Li, S.; Guan, X.; Hu, Y.W.; Feng, S.Y. Antioxidant activity, storage stability and in vitro release of epigallocatechin-3-gallate (EGCG) encapsulated in hordein nanoparticles. Food Chem. 2022, 388, 8. [Google Scholar] [CrossRef]
- Wang, R.H.; Zhang, L.Y.; Chi, Y.J.; Chi, Y. Forces involved in freeze-induced egg yolk gelation: Effects of various bond dissociation reagents on gel properties and protein structure changes. Food Chem. 2022, 371, 9. [Google Scholar] [CrossRef]
- Amagliani, L.; O’Regan, J.; Keny, A.L.; O’Mahony, J.A. Composition and protein profile analysis of rice protein ingredients. J. Food Compos. Anal. 2017, 59, 18–26. [Google Scholar] [CrossRef]
- Lee, H.Y.; Jeong, Y.I.; Kim, E.J.; Lee, K.D.; Choi, S.H.; Kim, Y.J.; Kim, D.H.; Choi, K.C. Preparation of Caffeic Acid Phenethyl Ester-Incorporated Nanoparticles and Their Biological Activity. J. Pharm. Sci. 2015, 104, 144–154. [Google Scholar] [CrossRef]
- Xie, H.J.; Liu, C.Z.; Gao, J.; Shi, J.Y.; Ni, F.F.; Luo, X.; He, Y.; Ren, G.R.; Luo, Z.S. Fabrication of Zein-Lecithin-EGCG complex nanoparticles: Characterization, controlled release in simulated gastrointestinal digestion. Food Chem. 2021, 365, 8. [Google Scholar] [CrossRef]
- Göçer, H.; Gülçin, I. Caffeic acid phenethyl ester (CAPE): Correlation of structure and antioxidant properties. Int. J. Food Sci. Nutr. 2011, 62, 821–825. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Lu, Y.C.; Yang, Y.; Li, S.Y.; Wang, C.; Wang, C.N.; Zhang, T.H. Comparison of non-covalent binding interactions between three whey proteins and chlorogenic acid: Spectroscopic analysis and molecular docking. Food Biosci. 2021, 41, 11. [Google Scholar] [CrossRef]
- Koppal, V.V.; Melavanki, R.; Kusanur, R.; Patil, N.R. Analysis of Fluorescence Quenching of Coumarin Derivative under Steady State and Transient State Methods. J. Fluoresc. 2021, 31, 393–400. [Google Scholar] [CrossRef]
- Wu, X.C.; Zhao, X.J.; Deng, Z.A.; Liang, X.R.; Fang, S. Investigation of interactions between zein and natamycin by fluorescence spectroscopy and molecular dynamics simulation. J. Mol. Liq. 2021, 327, 8. [Google Scholar] [CrossRef]
- Ribeiro, M.; de Sousa, T.; Poeta, P.; Bagulho, A.S.; Igrejas, G. Review of Structural Features and Binding Capacity of Polyphenols to Gluten Proteins and Peptides In Vitro: Relevance to Celiac Disease. Antioxidants 2020, 9, 463. [Google Scholar] [CrossRef]
- Wang, B.L.; Pan, D.Q.; Zhou, K.L.; Lou, Y.Y.; Shi, J.H. Multi-spectroscopic approaches and molecular simulation research of the intermolecular interaction between the angiotensin-converting enzyme inhibitor (ACE inhibitor) benazepril and bovine serum albumin (BSA). Spectroc. Acta Part A-Molec. Biomolec. Spectr. 2019, 212, 15–24. [Google Scholar] [CrossRef]
- Nai, X.; Chen, Y.R.; Hao, S.Y.; Liu, M.; Zhang, Q.; Liu, J.; Li, M.Y.; Kong, J. Temperature, pH and additives effects on the binding of Caffeic acid phenethyl ester to the native state of bovine serum albumin. J. Chem. Thermodyn. 2022, 168, 10. [Google Scholar] [CrossRef]
- Li, H.L.; Wu, F.; Tan, J.; Wang, K.; Zhang, C.P.; Zheng, H.Q.; Hu, F.L. Caffeic acid phenethyl ester exhibiting distinctive binding interaction with human serum albumin implies the pharmacokinetic basis of propolis bioactive components. J. Pharm. Biomed. Anal. 2016, 122, 21–28. [Google Scholar] [CrossRef]
- Qin, J.J.; Yang, M.; Wang, Y.C.; Wa, W.Q.; Zheng, J. Interaction between caffeic acid/caffeic acid phenethyl ester and micellar casein. Food Chem. 2021, 349, 10. [Google Scholar] [CrossRef]
| Temperature (K) | Ksv (L∙mol−1) | Kq (L∙mol−1∙s−1) | Quenching Type | Ka (L∙mol−1) | n | ΔG (kJ∙mol−1) | ΔH (kJ∙mol−1) | ΔS (J∙mol−1∙K−1) |
|---|---|---|---|---|---|---|---|---|
| 298 | 1.42 × 104 | 1.42 × 1012 | static quenching | 0.44 × 103 | 0.62 | −20.79 | −41.65 | −70 |
| 310 | 1.15 × 104 | 1.15 × 1012 | static quenching | 0.23 × 103 | 0.56 | −19.95 | −41.65 | −70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Feng, S.; Song, H. Caffeic Acid Phenethyl Ester Encapsulated in Self-Assemble Rice Peptides Nanoparticles: Storage Stability, In Vitro Release, and Their Interaction Mechanisms. Foods 2024, 13, 755. https://doi.org/10.3390/foods13050755
Wang X, Feng S, Song H. Caffeic Acid Phenethyl Ester Encapsulated in Self-Assemble Rice Peptides Nanoparticles: Storage Stability, In Vitro Release, and Their Interaction Mechanisms. Foods. 2024; 13(5):755. https://doi.org/10.3390/foods13050755
Chicago/Turabian StyleWang, Xinyue, Siyi Feng, and Hongdong Song. 2024. "Caffeic Acid Phenethyl Ester Encapsulated in Self-Assemble Rice Peptides Nanoparticles: Storage Stability, In Vitro Release, and Their Interaction Mechanisms" Foods 13, no. 5: 755. https://doi.org/10.3390/foods13050755




