Comparison of Non-Covalent and Covalent Interactions between Lactoferrin and Chlorogenic Acid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Reagent Configuration
- (1)
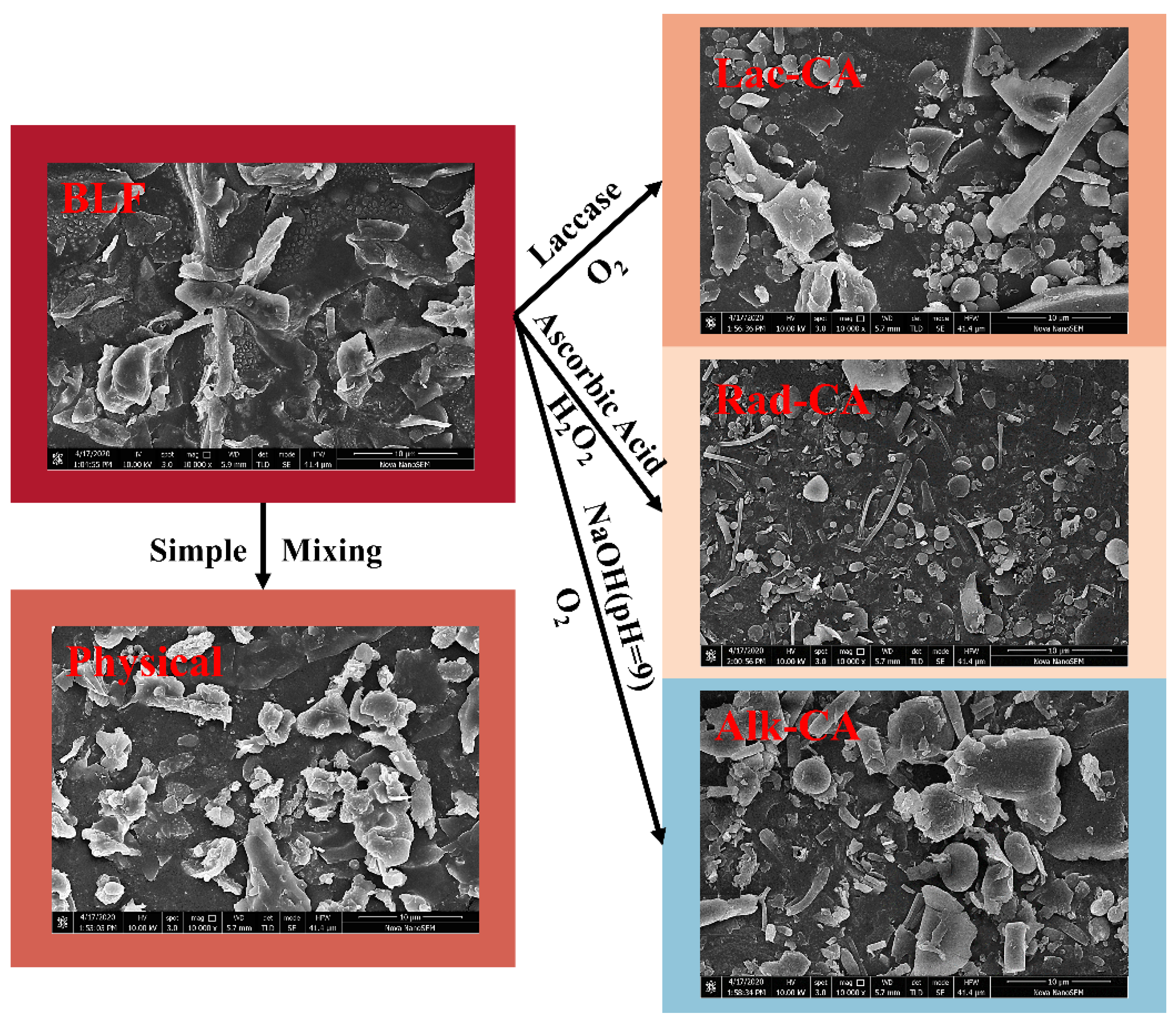
- Oxidation catalyzed by laccase: After adjusting the pH of CA solutions to 4.5 ± 0.5, they were combined with BLF, in that order. The mixture was continually mixed for eight hours while exposed to oxygen at room temperature. After adding laccase (60 U g−1), the mixture was cultured for 16 h. “Lac-CA” was the sample’s recorded name.
- (2)
- Free radical grafting: for three hours, use a magnetic stirrer to weigh, dissolve, and hydrate 60 mg BLF in 5.94 mL deionized water. The mixture was allowed to stand for two hours at room temperature and atmospheric pressure after adding 15 mg ascorbic acid and 0.06 mL of 5 mol L−1 hydrogen peroxide. After adding 0.021 mmol of CA, the reaction took place for 24 h at room temperature. “Rad-CA” was the sample’s record number.
- (3)
- Alkali treatment: after using 0.1 M NaOH to bring the pH of the fully hydrated 1% (w/v) BLF and 0.2% (w/v) CA solutions to 9.0, respectively, the BLF and CA were combined in equal amounts while being continuously agitated (at 200 rpm). For a full day, the sample was continuously agitated and exposed to air at room temperature, which encouraged contact between the solution and oxygen. “Alk-CA” was the sample’s record number.
2.2.2. Determination of Total Phenol Content
2.2.3. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.2.4. UV-Visible Spectrum
2.2.5. Fluorescence Spectrum
2.2.6. Circular Dichroism Experiment
2.2.7. Fourier Transform Infrared Spectroscopy
2.2.8. Particle Size Distribution and Zeta Potential
2.2.9. Scanning Electron Microscopy (SEM)
2.2.10. Molecular Docking and Simulation
2.2.11. DPPH Radical Scavenging Activity
2.2.12. Surface Hydrophobicity
2.2.13. Determination of Solubility
2.2.14. Measurement of Thermal Stability
2.2.15. Determination of Foaming Properties and Emulsifying Properties
2.2.16. Measurement of Biological Accessibility
2.2.17. Statistical Analysis
3. Results and Discussion
3.1. Formation of Covalent and Non-Covalent Complexes
3.1.1. Total Phenol Content
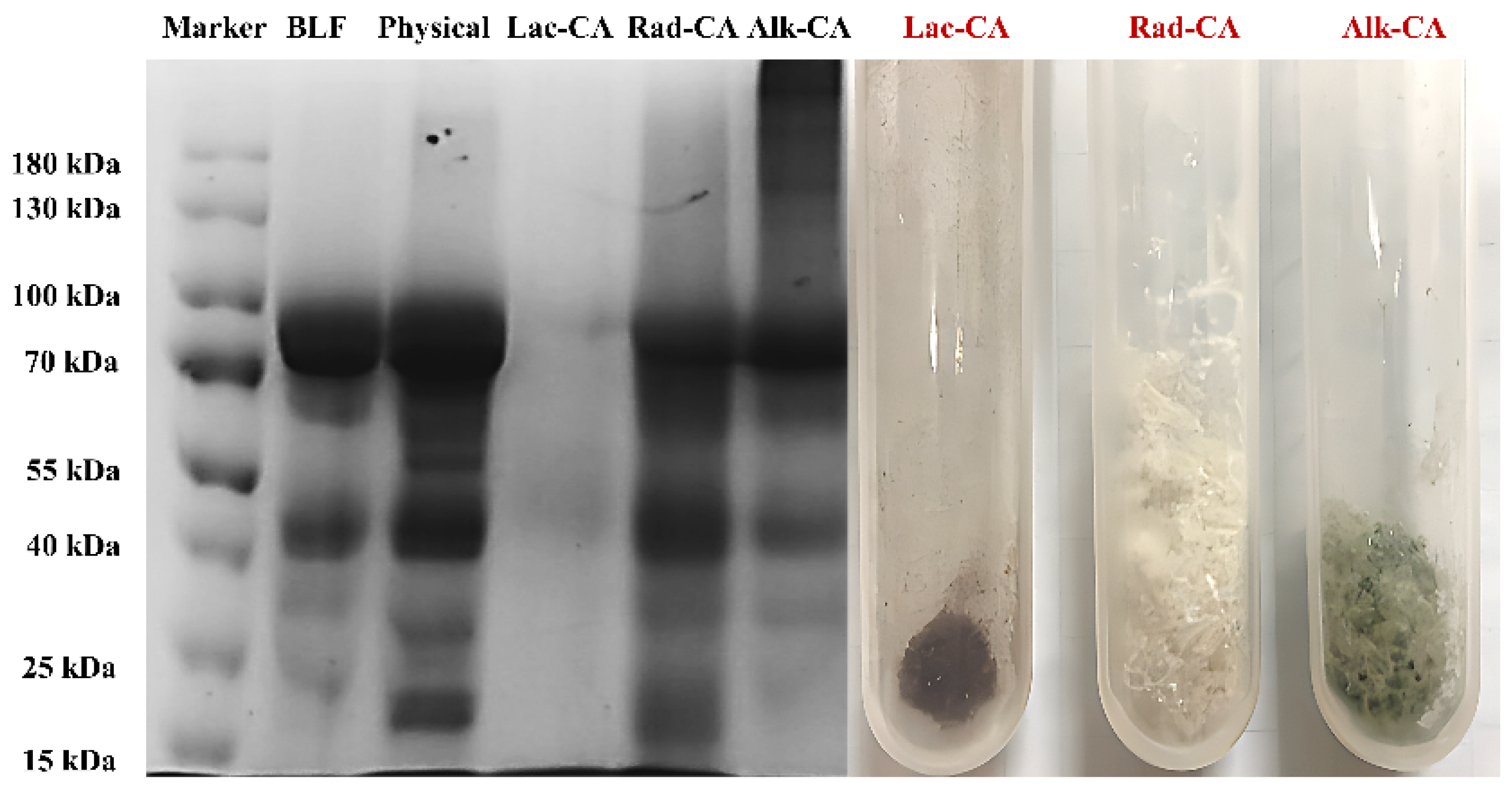
3.1.2. SDS-PAGE
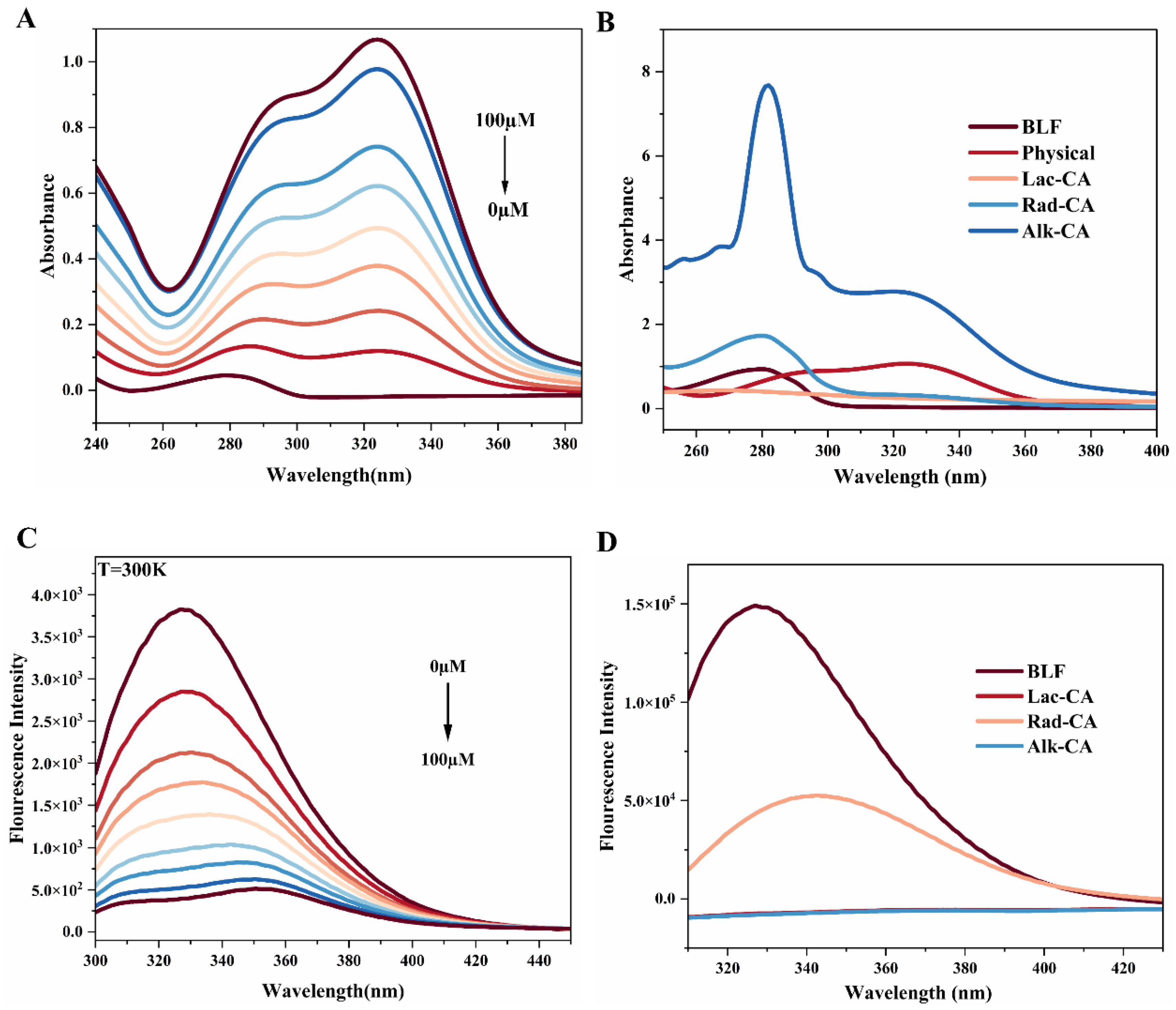
3.2. UV–Visible Spectrum
3.3. Fluorescence Spectrum
3.3.1. Fluorescence Quenching Type of CA-BLF Physical Complex
3.3.2. Binding Constant and Binding Site Analysis of CA-BLF Physical Complex
3.3.3. Thermodynamic Analysis and Interaction Force of CA-BLF Physical Complex
3.3.4. Intrinsic Fluorescence Emission Spectrum of CA-BLF Covalent Complexes
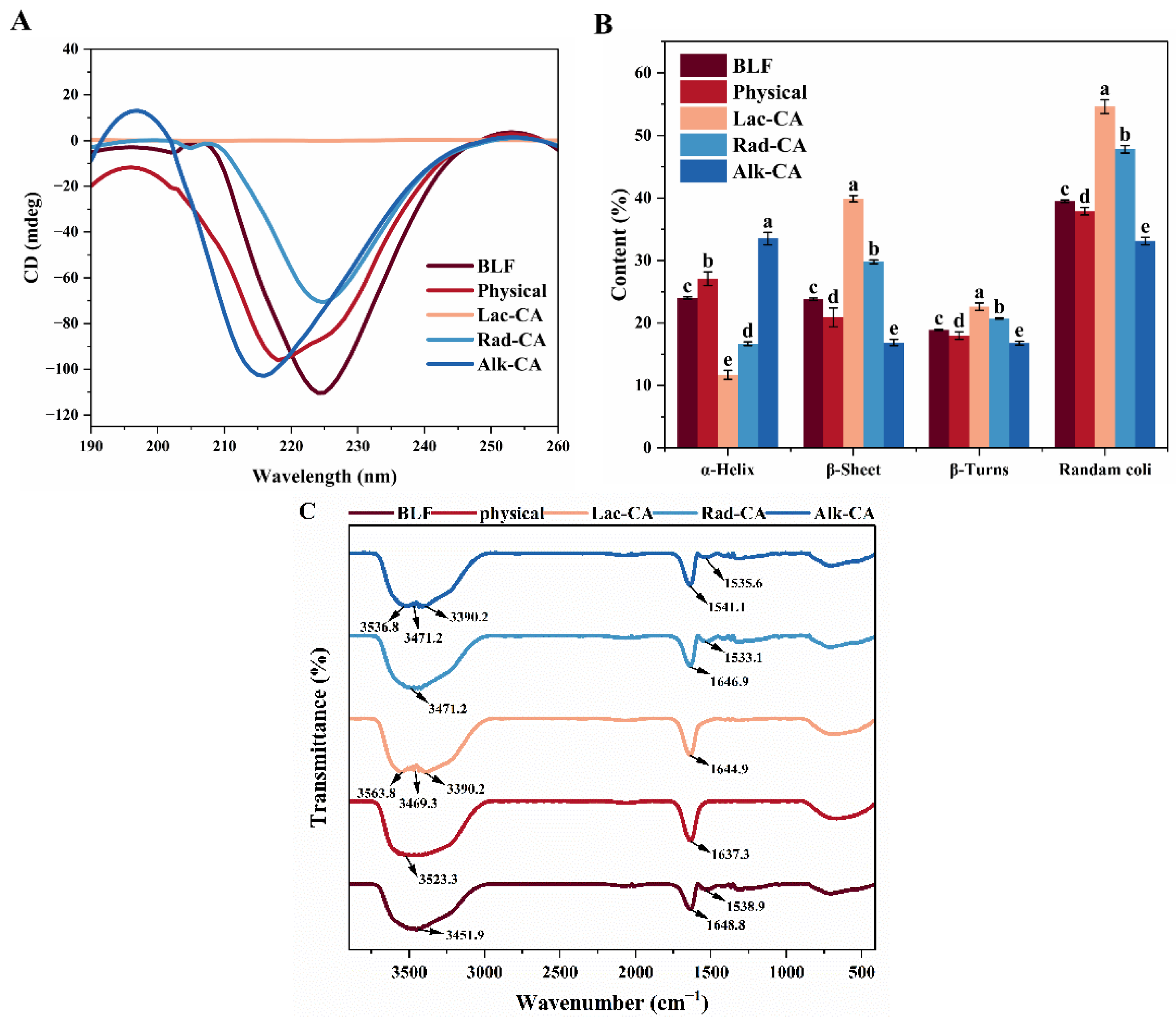
3.4. Circular Dichroism Experiment
3.5. Fourier Transform Infrared Spectroscopy
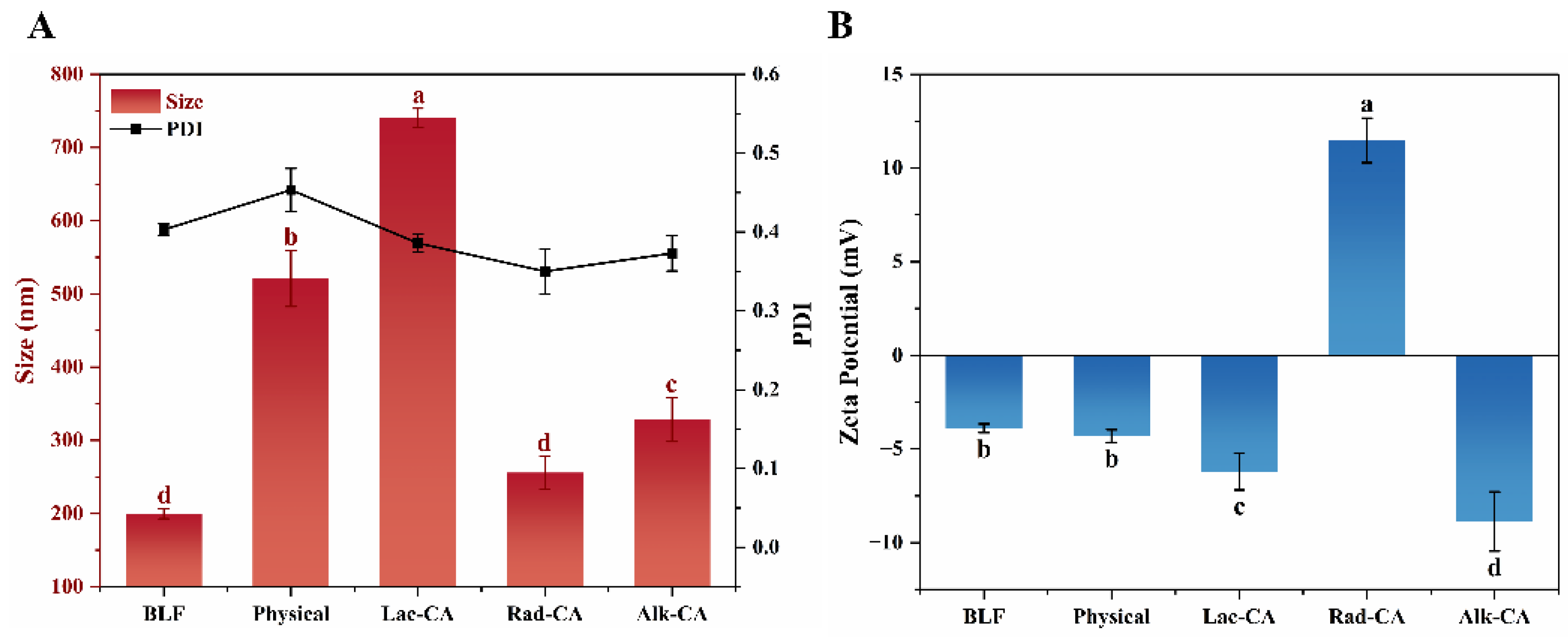
3.6. Particle Size Distribution and Zeta Potential
3.7. SEM
3.8. Binding Mechanisms of Covalent and Non-Covalent Compounds
3.8.1. Binding Mechanisms of Covalent Compounds
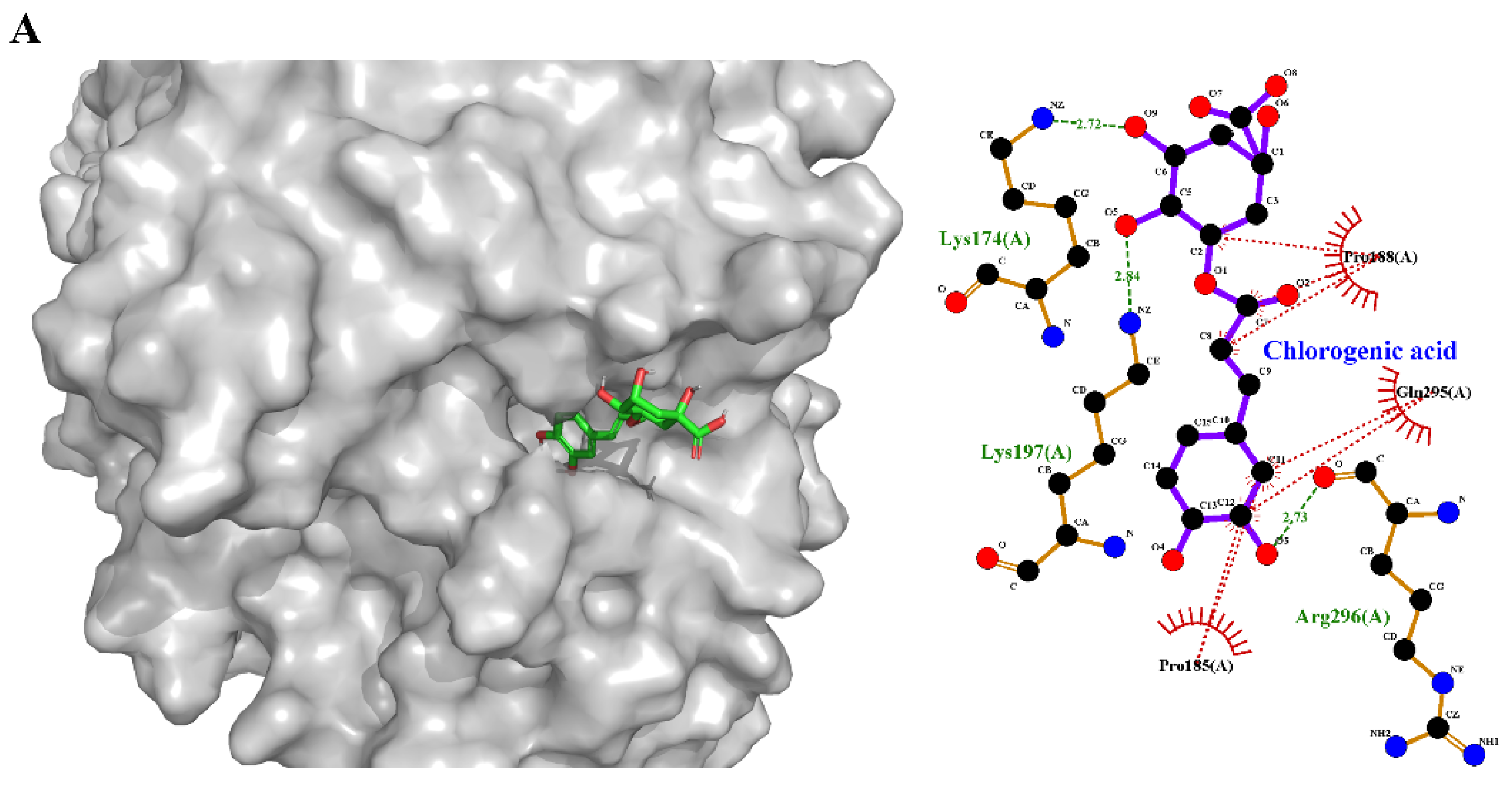
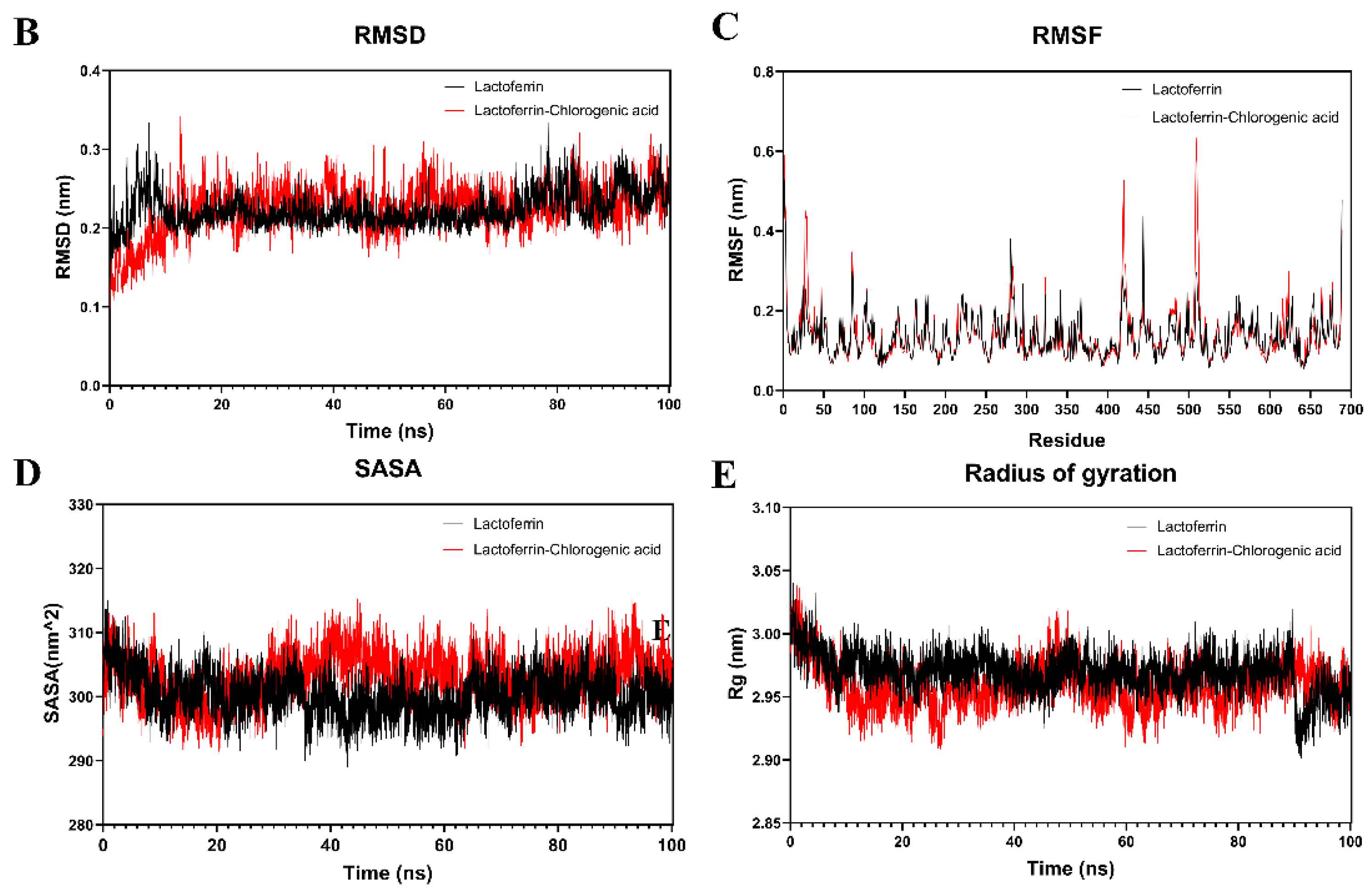
3.8.2. Binding Mechanisms of Non-Covalent Compounds (Molecular Docking and Simulation)
3.9. DPPH Radical Scavenging Activity
3.10. Surface Hydrophobicity (S0)
3.11. Solubility
3.12. Measurement of Thermal Stability
3.13. Foaming Properties and Emulsifying Properties
3.14. Biological Accessibility
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Abd El-Maksoud, A.A.; Abd El-Ghany, I.H.; El-Beltagi, H.S.; Anankanbil, S.; Banerjee, C.; Petersen, S.V.; Pérez, B.; Guo, Z. Adding functionality to milk-based protein: Preparation, and physico-chemical characterization of β-lactoglobulin-phenolic conjugates. Food Chem. 2018, 241, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Ali, M.S.; Al-Lohedan, H.A. Deciphering the interaction of procaine with bovine serum albumin and elucidation of the binding site: A multi spectroscopic and molecular docking study. J. Mol. Liq. 2017, 236, 232–240. [Google Scholar] [CrossRef]
- Bokkhim, H.; Bansal, N.; GrØndahl, L.; Bhandari, B. Physico-chemical properties of different forms of bovine lactoferrin. Food Chem. 2013, 141, 3007–3013. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Balance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Cao, X.; Liu, C.; Zhang, M.; Bi, R.; Fu, M.; Korik, E.; Chen, J.; Gao, J.; Semak, I.; Liu, J. Bovine lactoferrin and Lentinus edodes mycelia polysaccharide complex: The formation and the activity to protect islet beta cells. Int. J. Biol. Macromol. 2021, 191, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chai, L.; Li, H.; Zhang, Y.; Xie, H.M.; Shang, J.; Tian, W.; Yang, P.; Jiang, A.C. Effect of bovine lactoferrin from iron-fortified formulas on diarrhea and respiratory tract infections of weaned infants in a randomized controlled trial. Nutrition 2016, 32, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Czubinski, J.; Dwiecki, K. A review of methods used for investigation of protein–phenolic compound interactions. Int. J. Food Sci. Technol. 2017, 52, 573–585. [Google Scholar] [CrossRef]
- Dai, S.; Xu, T.; Yuan, Y.; Fang, Q.; Lian, Z.; Tian, T.; Tong, X.; Jiang, L.; Wang, H. Combination and precipitation mechanism of soy protein and tea polyphenols. Food Hydrocoll. 2024, 146, 109197. [Google Scholar] [CrossRef]
- Damodaran, S. Amino acids, peptides, and proteins. Fennema’s Food Chem. 2008, 4, 425–439. [Google Scholar]
- Fan, Y.; Liu, Y.; Gao, L.; Zhang, Y.; Yi, J. Oxidative stability and in vitro digestion of menhaden oil emulsions with whey protein: Effects of EGCG conjugation and interfacial cross-linking. Food Chem. 2018, 265, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Cao, D.; Ji, S.; Zhang, X.; Muhoza, B. Tannic acid-assisted cross-linked nanoparticles as a delivery system of eugenol: The characterization, thermal degradation, and antioxidant properties. Food Hydrocoll. 2020, 104, 105717. [Google Scholar] [CrossRef]
- Jia, Y.; Lu, Y.; Wang, X.; Yang, Y.; Zou, M.; Liu Jin, W.; Wang, X.; Pang, G.; Huang, L.; Wang, Z. Mass spectrometry-based quantitative and qualitative analyses reveal N-glycan changes of bovine lactoferrin at different stages of lactation. LWT 2021, 147, 111626. [Google Scholar] [CrossRef]
- Jia, Z.; Zheng, M.; Tao, F.; Chen, W.; Huang, G.; Jiang, J. Effect of covalent modification by (−)-epigallocatechin-3-gallate on physicochemical and functional properties of whey protein isolate. LWT 2016, 66, 305–310. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Zhao, J.; Liu, Y. The effect of non-covalent interaction of chlorogenic acid with whey protein and casein on the physicochemical and radical-scavenging activity of in vitro protein digests. Food Chem. 2018, 268, 334–341. [Google Scholar] [CrossRef]
- Jing, H.; Huang, X.; Jiang, C.; Wang, L.; Du, X.; Ma, C.; Wang, H. Effects of tannic acid on the structure and proteolytic digestion of bovine lactoferrin. Food Hydrocoll. 2021, 117, 106666. [Google Scholar] [CrossRef]
- Khalil, A.; Kashif, M. Interaction studies of levofloxacin with human lysozyme in a ternary complex using multispectroscopic and computational analysis: A circular dichroism method for the quantitation of levofloxacin. J. Mol. Liq. 2023, 370, 121023. [Google Scholar] [CrossRef]
- Khan, J.M.; Malik, A.; Husain, F.M.; Alkaltham, M.S. Molecular interaction of Sunset Yellow with whey protein: Multi-spectroscopic techniques and computational study. J. Mol. Liq. 2022, 345, 117838. [Google Scholar] [CrossRef]
- Kristo, E.; Hazizaj, A.; Corredig, M. Structural changes imposed on whey proteins by UV irradiation in a continuous UV light reactor. J. Agric. Food Chem. 2012, 60, 6204–6209. [Google Scholar] [CrossRef]
- Krolitzki, E.; Schwaminger, S.P.; Pagel, M.; Ostertag, F.; Hinrichs, J.; Berensmeier, S. Current practices with commercial scale bovine lactoferrin production and alternative approaches. Int. Dairy J. 2022, 126, 105263. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Zhang, T.; McClements, D.J.; Liu, X.; Wu, X.; Liu, F. Enzymatic and nonenzymatic conjugates of lactoferrin and (−)-epigallocatechin gallate: Formation, structure, functionality, and allergenicity. J. Agric. Food Chem. 2021, 69, 6291–6302. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Bhattarai, M.; Mikkonen, K.S.; Heinonen, M. Effects of enzymatic hydrolysis of fava bean protein isolate by alcalase on the physical and oxidative stability of oil-in-water emulsions. J. Agric. Food Chem. 2019, 67, 6625–6632. [Google Scholar] [CrossRef]
- Liu, F.; Wang, D.; Sun, C.; McClements, D.J.; Gao, Y. Utilization of interfacial engineering to improve physicochemical stability of β-carotene emulsions: Multilayer coatings formed using protein and protein–polyphenol conjugates. Food Chem. 2016, 205, 129–139. [Google Scholar] [CrossRef]
- Liu, F.; Ma, C.; McClements, D.J.; Gao, Y. Development of polyphenol-protein-polysaccharide ternary complexes as emulsifiers for nutraceutical emulsions: Impact on formation, stability, and bioaccessibility of β-carotene emulsions. Food Hydrocoll. 2016, 61, 578–588. [Google Scholar] [CrossRef]
- Liu, F.; Ma, C.; Gao, Y.; McClements, D.J. Food-grade covalent complexes and their application as nutraceutical delivery systems: A review. Compr. Rev. Food Sci. Food Saf. 2017, 16, 76–95. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, R.; Lönnerdal, B. Assessment of bioactivities of the human milk lactoferrin-osteopontin complex in vitro. J. Nutr. Biochem. 2019, 69, 10–18. [Google Scholar] [CrossRef]
- Nunes, N.M.; Coelho, Y.L.; Castro, J.S.; Vidigal, M.C.T.R.; Mendes, T.A.O.; da Silva, L.H.M.; Pires, A.C.S. Naringenin-lactoferrin binding: Impact on naringenin bitterness and thermodynamic characterization of the complex. Food Chem. 2020, 331, 127337. [Google Scholar] [CrossRef]
- Ochoa, T.J.; Zegarra, J.; Bellomo, S.; Carcamo, C.P.; Cam, L.; Castaneda, A.; Villavicencio, A.; Gonzales, J.; Rueda, M.S.; Turin, C.G.; et al. Randomized Controlled Trial of Bovine Lactoferrin for Prevention of Sepsis and Neurodevelopment Impairment in Infants Weighing Less Than 2000 Grams. J. Pediatr. 2020, 219, 118–125. [Google Scholar] [CrossRef]
- Ojha, H.; Mishra, K.; Hassan, M.I.; Chaudhury, N.K. Spectroscopic and isothermal titration calorimetry studies of binding interaction of ferulic acid with bovine serum albumin. Thermochim. Acta 2012, 548, 56–64. [Google Scholar] [CrossRef]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein–phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Pan, X.; Fang, Y.; Wang, L.; Shi, Y.; Xie, M.; Xia, J.; Pei, F.; Li, P.; Xiong, W.; Shen, X. Covalent interaction between rice protein hydrolysates and chlorogenic acid: Improving the stability of oil-in-water emulsions. J. Agric. Food Chem. 2019, 67, 4023–4030. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.H.; Benjakul, S.; Sae-leaw, T.; Balange, A.K.; Maqsood, S. Protein–polyphenol conjugates: Antioxidant property, functionalities and their applications. Trends Food Sci. Technol. 2019, 91, 507–517. [Google Scholar] [CrossRef]
- Rocha, F.S.; Gomes, A.J.; Lunardi, C.N.; Kaliaguine, S.; Patience, G.S. Experimental methods in chemical engineering: Ultraviolet visible spectroscopy-UV-Vis. Can. J. Chem. Eng. 2018, 96, 2512–2517. [Google Scholar] [CrossRef]
- Rocklin, G.J.; Boyce, S.E.; Fischer, M.; Fish, I.; Mobley, D.L.; Shoichet, B.K.; Dill, K.A. Blind prediction of charged ligand binding affinities in a model binding site. J. Mol. Biol. 2013, 425, 4569–4583. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Gomez-Hens, A.; Valcarcel, M. Analytical applications of synchronous fluorescence spectroscopy. Talanta 1986, 33, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Santa Rosa, L.N.; de Paula Rezende, J.; Coelho, Y.L.; Mendes TA, O.; da Silva LH, M.; dos Santos Pires, A.C. β-lactoglobulin conformation influences its interaction with caffeine. Food Biosci. 2021, 44, 101418. [Google Scholar] [CrossRef]
- Santana-Gálvez, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Chlorogenic acid: Recent advances on its dual role as a food additive and a nutraceutical against metabolic syndrome. Molecules 2017, 22, 358. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Peng, J.; Sagis, L.M.; Landman, J. Air-water interface properties and foam stabilization by mildly extracted lentil protein. Food Hydrocoll. 2024, 147, 109342. [Google Scholar] [CrossRef]
- Sui, X.; Sun, H.; Qi, B.; Zhang, M.; Li, Y.; Jiang, L. Functional and conformational changes to soy proteins accompanying anthocyanins: Focus on covalent and non-covalent interactions. Food Chem. 2018, 245, 871–878. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, F.; Liu, T.; Jing, H.; Huang, Y.; Obadi, M.; Xu, B. Ultrasound-enhanced egg white proteins conjugated with polyphenols: The structure of the polyphenols on their functional properties. LWT 2022, 164, 113600. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Zhang, Y.; Lü, X.; Zhao, L.; Song, Y.; Zhang, L.; Jiang, H.; Zhang, J.; Ge, W. Processing sheep milk by cold plasma technology: Impacts on the microbial inactivation, physicochemical characteristics, and protein structure. LWT 2022, 153, 112573. [Google Scholar] [CrossRef]
- Wu, X.; Lu, Y.; Xu, H.; Lin, D.; He, Z.; Wu, H.; Liu, L.; Gao, K.; Wang, Z.; Lin, X.; et al. Reducing the allergenic capacity of β-lactoglobulin by covalent conjugation with dietary polyphenols. Food Chem. 2018, 256, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Kataoka, M.; Higashino, H.; Sakuma, S.; Sakamoto, T.; Uchimaru, H.; Tsukikawa, H.; Shiramoto, M.; Uchiyama, H.; Tachiki, H.; et al. Measurement of drug concentration in the stomach after intragastric administration of drug solution to healthy volunteers: Analysis of intragastric fluid dynamics and drug absorption. Pharm. Res. 2013, 30, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, Q.; Yang, H.; Shi, H.; Dong, A.; Wang, L.; Yu, S. Progress in infrared spectroscopy as an efficient tool for predicting protein secondary structure. Int. J. Biol. Macromol. 2022, 206, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Lam, T.I.; Yokoyama, W.; Cheng, L.W.; Zhong, F. Beta-carotene encapsulated in food protein nanoparticles reduces peroxyl radical oxidation in Caco-2 cells. Food Hydrocoll. 2015, 43, 31–40. [Google Scholar] [CrossRef]
- Yildirim-Elikoglu, S.; Erdem, Y.K. Interactions between milk proteins and polyphenols: Binding mechanisms, related changes, and the future trends in the dairy industry. Food Rev. Int. 2018, 34, 665–697. [Google Scholar] [CrossRef]
- You, J.; Luo, Y.; Wu, J. Conjugation of ovotransferrin with catechin shows improved antioxidant activity. J. Agric. Food Chem. 2014, 62, 2581–2587. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, Q.; Liu, J.; Lu, X.; Zhu, Y.; Wang, J.; Wang, S. Study of the interaction between 5-sulfosalicylic acid and bovine serum albumin by fluorescence spectroscopy. J. Lumin. 2013, 134, 747–753. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, W.; Gao, J.; Gong, H.; Mao, X. Limited hydrolysis as a strategy to improve the non-covalent interaction of epigallocatechin-3-gallate (EGCG) with whey protein isolate near the isoelectric point. Food Res. Int. 2022, 161, 111847. [Google Scholar] [CrossRef]
- Zhu, B.; Ryan, D.K. Characterizing the interaction between uranyl ion and fulvic acid using regional integration analysis (RIA) and fluorescence quenching. J. Environ. Radioact. 2016, 153, 97–103. [Google Scholar] [CrossRef] [PubMed]

| Samples | Grafting Efficiency (%) |
|---|---|
| Physical complexes | 12.63 ± 1.58 c |
| Laccase catalytic oxidation | 51.39 ± 0.28 b |
| Free radical grafting | 16.82 ± 4.62 c |
| Alkali treatment | 82.40 ± 2.53 a |
| T (K) | Ksv (×104 L mol−1) | Kq (×1012 mol−1 s−1) | KA (×103 L mol−1) | n | ΔH (kJ mol−1) | ΔS (J mol−1 K−1) | ΔG (kJ mol−1) |
|---|---|---|---|---|---|---|---|
| 290 | 7.11 | 7.11 | 5.29 | 1.19 | −1.35 | 85.79 | −26.23 |
| 300 | 7.21 | 7.21 | 5.19 | 1.20 | −27.09 | ||
| 310 | 7.44 | 7.44 | 5.10 | 1.22 | −27.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Al-Wraikat, M.; Hao, C.; Liu, Y. Comparison of Non-Covalent and Covalent Interactions between Lactoferrin and Chlorogenic Acid. Foods 2024, 13, 1245. https://doi.org/10.3390/foods13081245
Li Z, Al-Wraikat M, Hao C, Liu Y. Comparison of Non-Covalent and Covalent Interactions between Lactoferrin and Chlorogenic Acid. Foods. 2024; 13(8):1245. https://doi.org/10.3390/foods13081245
Chicago/Turabian StyleLi, Zekun, Majida Al-Wraikat, Changchun Hao, and Yongfeng Liu. 2024. "Comparison of Non-Covalent and Covalent Interactions between Lactoferrin and Chlorogenic Acid" Foods 13, no. 8: 1245. https://doi.org/10.3390/foods13081245





