Past, Present and Future of Sensors in Food Production
Abstract
:1. Introduction
2. Food Borne Pathogens
3. Biosensors
4. Biosensor Component
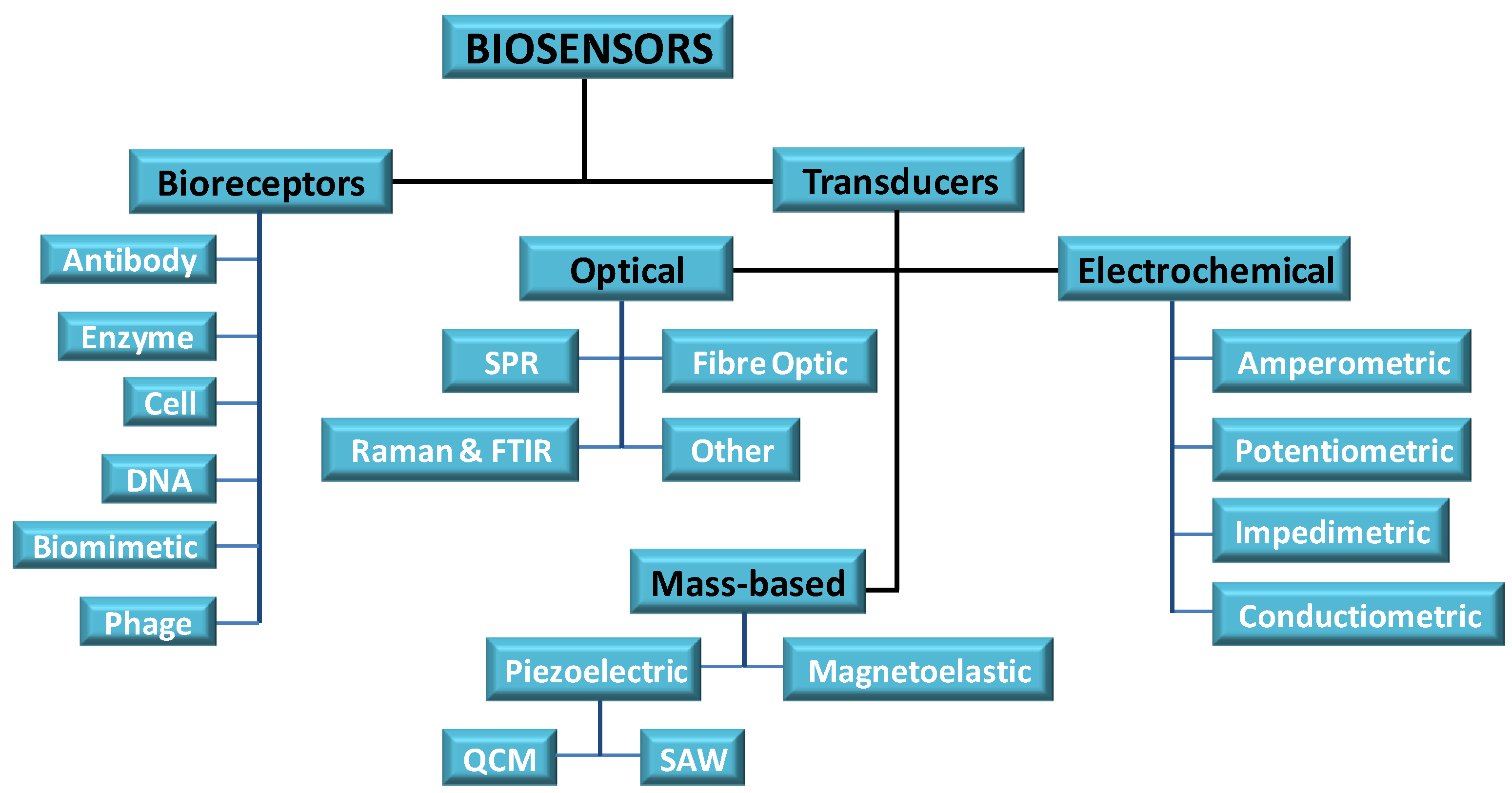
5. Sensor Materials

6. Sensor Designs
7. Microbial Sensing
Microbial Whole Cell Biosensors
8. Nucleic Acid Sensors
9. Sensors Using Bacteriophage
10. Companies Developing and Producing Biosensors
11. Conclusions
Acknowledgements
Conflicts of Interest
References
- Food Quality and Safety Systems—A Training Manual on Food Hygiene and the Hazard Analysis and Critical Control Point (HACCP) System. Food and Agriculture Organisation of the United Nations: Rome, 1998. Available online: http://www.fao.org/docrep/W8088E/W8088E00.htm (accessed on 19 January 2014).
- Arvanitoyannis, I.S.; Varzakas, T.H. Application of ISO 22000 and failure mode and effect analysis (FEMA) for industrial processing of salmon: A case study. Crit. Rev. Food Sci. Nutr. 2008, 48, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Regulation (EC) No 852/2004 of the European Parliament and of the Council of 29 April 2004 on the Hygiene of Foodstuffs. Off. J. Eur. Union 2004, L139, 1.
- Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 Laying Down Specific Hygiene Rules for Food of Animal Origin. Off. J. Eur. Union 2004, L139, 55.
- Chmielewski, R.A.N.; Frank, J.F. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2006, 2, 22–32. [Google Scholar] [CrossRef]
- Cappitelli, F.; Polo, A.; Villa, F. Biofilm formation in food processing environments is still poorly understood and controlled. Food Eng. Rev. 2014, 6, 1–2, 29. [Google Scholar]
- The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2011. EFSA J. 2011, 11, 3129. [CrossRef]
- Commission Regulation (EU) No 218/2014 of 7 March 2014 Amending Annexes to Regulations (EC) No 853/2004 and (EC) No 854/2004 of the European Parliament and of the Council and Commission Regulation (EC) No 2074/2005. Off. J. Eur. Union 2014, L69, 95.
- Web of Science™. Available online: www.webofknowledge.com (accessed on 9 January 2014).
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Chemical Heritage Foundation. Available online: http://www.chemheritage.org/discover/collections/collection-items/scientific-instruments/ysi-blood-glucose-analyzer-model-23a.aspx (accessed on 23 March 2014).
- Thusu, R. Strong Growth Predicted for Biosensor Market. Available online: http://www.sensorsmag.com/specialty-markets/medical/strong-growth-predicted-biosensors-market-7640 (accessed on 23 March 2014).
- Thusu, R. Sensors Facilitating Health Monitoring. Available online: http://www.sensorsmag.com/specialty-markets/medical/sensors-facilitate-health-monitoring-8365 (accessed on 23 March 2014).
- Turiel, E.; Martin-Esteban, A. Molecularly imprinted polymers for sample preparation: A review. Anal. Chim. Acta 2010, 668, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Mosbach, K. Molecularly imprinted polymers and their use in biomimetic sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Turner, A.P.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Guerreiro, A.; Whitcombe, M.J.; Piletska, E.V.; Turner, A.P.F.; Piletsky, S.A. Solid-phase synthesis of molecularly imprinted polymer nanoparticles with a reusable template—“Plastic antibodies”. Adv. Funct. Mater. 2013, 23, 2821–2827. [Google Scholar] [CrossRef]
- Vasapollo, G.; del Sole, R.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, A. Mobile phones democratize and cultivate next-generation imaging, diagnostics and measurement tools. Lab Chip 2014, 14, 3187–3194. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.P. Biosensors: Sense and sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef] [PubMed]
- Doria, G.; Conde, J.; Veigas, B.; Giestas, L.; Almeida, C.; Assuncao, M.; Rosa, J.; Baptista, P.V. Noble metal nanoparticles for biosensing applications. Sensors 2012, 12, 1657–1687. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.G.; Tu, S.I. High-throughput biosensors for multiplexed food-borne pathogen detection. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2011, 4, 151–172. [Google Scholar]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Das, A.P.; Kumar, P.S.; Swain, S. Recent advances in biosensor based endotoxin detection. Biosens. Bioelectron. 2014, 51, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.C.; Bonel, L.; Ezquerra, A.; Hernandez, S.; Bertolin, J.R.; Cubel, C.; Castillo, J.R. Electrochemical affinity biosensors for detection of mycotoxins: A review. Biosens. Bioelectron. 2013, 49, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Kirsch, J.; Simonian, A. Campylobacter spp. detection in the 21st century: A review of the recent achievements in biosensor development. J. Microbiol. Methods 2013, 95, 48–56. [Google Scholar]
- Wang, F.; Yang, Q.; Kase, J.A.; Meng, J.; Clotilde, L.M.; Lin, A.; Ge, B. Current trends in detecting non-O157 Shiga toxin-producing Escherichia coli in food. Foodborne Pathog. Dis. 2013, 10, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Holford, T.R.; Davis, F.; Higson, S.P. Recent trends in antibody based sensors. Biosens. Bioelectron. 2012, 34, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, Z.; Li, X. Advances in pesticide biosensors: Current status, challenges, and future perspectives. Anal. Bioanal. Chem. 2013, 405, 63–90. [Google Scholar] [CrossRef] [PubMed]
- Mortari, A.; Lorenzelli, L. Recent sensing technologies for pathogen detection in milk: A review. Biosens. Bioelectron. 2014, 60, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.S.; Ragavan, K.V. Biosensors in food processing. J. Food Sci. Technol. 2013, 50, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Ezzati Nazhad Dolatabadi, J.; de la Guardia, M. Nanomaterial-based electrochemical immunosensors as advanced diagnostic tools. Anal. Methods 2014, 6, 3891–3900. [Google Scholar]
- Kim, G.; Park, S.B.; Moon, J.-H.; Lee, S. Detection of pathogenic Salmonella with nanobiosensors. Anal. Methods 2013, 5, 5717–5723. [Google Scholar] [CrossRef]
- Arshak, K.; Velusamy, V.; Korostynska, O.; Oliwa-Stasiak, K.; Adley, C. Conducting polymers and their applications to biosensors: Emphasizing on foodborne pathogen detection. Sensors J. IEEE 2009, 9, 1942–1951. [Google Scholar] [CrossRef]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Elfeky, S.A. Fluorescent sensor for bacterial recognition. Spectrochim. Acta Mol. Biomol. Spectros. 2013, 108, 338–341. [Google Scholar] [CrossRef]
- Faridbod, F.; Norouzi, P.; Dinarvand, R.; Ganjali, M.R. Developments in the field of conducting and non-conducting polymer based potentiometric membrane sensors for ions over the past decade. Sensors 2008, 8, 2331–2412. [Google Scholar] [CrossRef]
- Inzelt, G. Rise and rise of conducting polymers. J. Solid State Electrochem. 2011, 15, 1711–1718. [Google Scholar]
- Arshak, K.; Adley, C.; Moore, E.; Cunniffe, C.; Campion, M.; Harris, J. Characterisation of polymer nanocomposite sensors for quantification of bacterial cultures. Sens. Actuators B Chem. 2007, 126, 226–231. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. Conducting polymer based DNA biosensor for the detection of the Bacillus cereus group species. In SPIE7315, Sensing for Agriculture and Food Quality and Safety; Kim, M.S., Tu, S.-I., Chao, K., Eds.; International Society for Optics and Photonics: Orlando, FL, USA, 2009. [Google Scholar]
- Oliwa-Stasiak, K.; Kolaj-Robin, O.; Adley, C.C. Development of Real-Time PCR assays for detection and quantification of Bacillus cereus group species: Differentiation of B. weihenstephanensis and rhizoid B. pseudomycoides isolates from milk. Appl. Environ. Microbiol. 2011, 77, 80–88. [Google Scholar]
- Kreno, L.E.; Leong, K.; Farha, O.K.; Allendorf, M.; van Duyne, R.P.; Hupp, J.T. Metal-organic framework materials as chemical sensors. Chem. Rev. 2012, 112, 1105–1125. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.T.; Li, X.; Xu, F.G.; Tan, H.L.; Li, Z.; Sun, L.L.; Song, Y.H. Metal organic framework-derived anthill-like Cu@carbon nanocomposites for nonenzymatic glucose sensor. Anal. Methods 2014, 6, 1550–1557. [Google Scholar] [CrossRef]
- Xu, H.; Rao, X.; Gao, J.; Yu, J.; Wang, Z.; Dou, Z.; Cui, Y.; Yang, Y.; Chen, B.; Qian, G. A luminescent nanoscale metal-organic framework with controllable morphologies for spore detection. Chem. Commun. 2012, 48, 7377–7379. [Google Scholar] [CrossRef]
- Rozand, C. Paper-based analytical devices for point-of-care infectious disease testing. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Gu, Y.; Beck, C.; Iqbal, Z.; Federici, J.F. Reversible chromatic sensor fabricated by inkjet printing TCDA-ZnO on a paper substrate. Sens. Actuators B Chem. 2014, 193, 10–18. [Google Scholar] [CrossRef]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. A paper based analytical device for the colorimetric detection of food borne pathogens. In Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Seattle, DC, USA, 2–6 October 2011.
- Vaseashta, A.; Dimova-Malinovska, D. Nanostructured and nanoscale devices, sensors and detectors. Sci. Tech. Adv. Mater. 2005, 6, 312–318. [Google Scholar] [CrossRef]
- Kang, X.; Pang, G.; Chen, Q.; Liang, X. Fabrication of Bacillus cereus electrochemical immunosensor based on double-layer gold nanoparticles and chitosan. Sens. Actuators. B Chem. 2013, 177, 1010–1016. [Google Scholar] [CrossRef]
- Dou, W.; Tang, W.; Zhao, G. A disposable electrochemical immunosensor arrays using 4-channel screen-printed carbon electrode for simultaneous detection of Escherichia coli O157:H7 and Enterobacter sakazakii. Electrochim. Acta 2013, 97, 79–85. [Google Scholar] [CrossRef]
- Chen, J.-K.; Zhou, G.-Y.; Chang, C.-J.; Cheng, C.-C. Label-free detection of DNA hybridization using nanopillar arrays based optical biosensor. Sens. Actuators. B Chem. 2014, 194, 10–18. [Google Scholar] [CrossRef]
- Wang, Y.; Ping, J.; Ye, Z.; Wu, J.; Ying, Y. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label-free detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2013, 49, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dou, W.C.; Zhan, X.J.; Zhao, G.Y. A novel immunosensor for Enterobacter sakazakii based on multiwalled carbon nanotube/ionic liquid/thionine modified electrode. Electrochim. Acta 2012, 61, 73–77. [Google Scholar] [CrossRef]
- Dong, J.; Zhao, H.; Xu, M.; Ma, Q.; Ai, S. A label-free electrochemical impedance immunosensor based on AuNPs/PAMAM-MWCNT-Chi nanocomposite modified glassy carbon electrode for detection of Salmonella Typhimurium in milk. Food Chem. 2013, 141, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Salam, F.; Uludag, Y.; Tothill, I.E. Real-time and sensitive detection of Salmonella Typhimurium using an automated quartz crystal microbalance (QCM) instrument with nanoparticles amplification. Talanta 2013, 115, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Pérez-López, B.; Merkoçi, A. Nanomaterials based biosensors for food analysis applications. Trends Food Sci. Technol. 2011, 22, 625–639. [Google Scholar]
- Gilmartin, N.; O’Kennedy, R. Nanobiotechnologies for the detection and reduction of pathogens. Enzyme Microb. Technol. 2012, 50, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Wu, V.C.H.; Chuang, Y.-C.; Lin, C.-S. Using oligonucleotide-functionalized Au nanoparticles to rapidly detect foodborne pathogens on a piezoelectric biosensor. J. Microbiol. Methods 2008, 73, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.J.; Huang, J.L.; Meng, W.J.; Shen, M.; Jiao, X.A. A reusable capacitive immunosensor for detection of Salmonella spp. Based on grafted ethylene diamine and self-assembled gold nanoparticle monolayers. Anal. Chim. Acta 2009, 647, 159–166. [Google Scholar]
- Biacore. Available online: https://www.biacore.com/lifesciences/index.html (accessed on 14 January 2014).
- Leonard, P.; Hearty, S.; Quinn, J.; O’Kennedy, R. A generic approach for the detection of whole Listeria monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens. Bioelectron. 2004, 19, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Biosensing Instruments Ltd. Available online: www.biosensingusa.com (accessed on 16 April 2014).
- Taylor, A.D.; Ladd, J.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Shankar, P.M.; Mutharasan, R. A review of fiber-optic biosensors. Sens. Actuators. B Chem. 2007, 125, 688–703. [Google Scholar] [CrossRef]
- Narsaiah, K.; Jha, S.N.; Bhardwaj, R.; Sharma, R.; Kumar, R. Optical biosensors for food quality and safety assurance—A review. J. Food Sci. Technol. 2012, 49, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.H.; Bhunia, A.K. Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 2013, 33, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Adley, C.; Ryan, M.P. Conductometric biosensor for high throughput screening of pathogens in food. In High Throughput Screening for Food Safety Assessment: Biosensor Technologies, Hyperspectral Imaging and Practical Applications; Bhunia, A.K., Kim, M.S., Taitt, C.R., Eds.; Elsevier: Cambridge, UK, 2014; pp. 315–326. [Google Scholar]
- Yang, X.; Zitova, A.; Kirsch, J.; Fergus, J.W.; Overfelt, R.A.; Simonian, A.L. Portable and remote electrochemical sensing system for detection of tricresyl phosphate in gas phase. Sens. Actuators. B Chem. 2012, 161, 564–569. [Google Scholar] [CrossRef]
- Zhai, D.; Liu, B.; Shi, Y.; Pan, L.; Wang, Y.; Li, W.; Zhang, R.; Yu, G. Highly sensitive glucose sensor based on Pt nanoparticle/polyaniline hydrogel heterostructures. ACS Nano 2013, 7, 3540–3546. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-L.; Li, Y. A QCM immunosensor for Salmonella detection with simultaneous measurements of resonant frequency and motional resistance. Biosens. Bioelectron. 2005, 21, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Mecea, V.M. Is quartz crystal microbalance really a mass sensor? Sens. Actuators A Phys. 2006, 128, 270–277. [Google Scholar] [CrossRef]
- BCC Research. Surface Acoustic Wave (SAW) Devices: Technologies and Global Markets. Available online: http://www.bccresearch.com/report/download/report/ias039a (accessed on 11 May 2014).
- Rocha-Gaso, M.I.; March-Iborra, C.; Montoya-Baides, A.; Arnau-Vives, A. Surface generated acoustic wave biosensors for the detection of pathogens: A review. Sensors 2009, 9, 5740–5769. [Google Scholar]
- Reyes, P.I.; Li, J.; Duan, Z.; Yang, X.; Cai, Y.; Huang, Q.; Lu, Y. ZnO surface acoustic wave sensors built on zein-coated flexible food packages. Sens. Lett. 2013, 11, 539–544. [Google Scholar] [CrossRef]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuators. B Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Barthelmebs, L.; Calas-Blanchard, C.; Istamboulie, G.; Marty, J.-L.; Noguer, T. Biosenosrs as analytical tools in food fermentation industry. In Bio-Farms for Nutraceuticals; Functional Food and Safety Control by Biosensors; Giardia, M.T., Rea, G., Berra, B., Eds.; Landes Bioscience: Austin, TX, USA, 2010; pp. 293–307. [Google Scholar]
- Ikeda, T.; Kato, K.; Maeda, M.; Tatsumi, H.; Kano, K.; Matsushita, K. Electrocatalytic properties of Acetobacter aceti cells immobilized on electrodes for the quinone-mediated oxidation of ethanol. J. Electroanal. Chem. 1997, 430, 197–204. [Google Scholar] [CrossRef]
- Lei, Y.; Mulchandani, P.; Chen, W.; Mulchandani, A. Direct determination of p-nitrophenyl substituent organophosphorus nerve agents using a recombinant Pseudomonas putida JS444-modified clark oxygen electrode. J. Agric. Food Chem. 2004, 53, 524–527. [Google Scholar] [CrossRef]
- Nakamura, H.; Suzuki, K.; Ishikuro, H.; Kinoshita, S.; Koizumi, R.; Okuma, S.; Gotoh, M.; Karube, I. A new BOD estimation method employing a double-mediator system by ferricyanide and menadione using the eukaryote Saccharomyces cerevisiae. Talanta 2007, 72, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Kara, S.; Keskinler, B.; Erhan, E. A novel microbial BOD biosensor developed by the immobilization of P. syringae in micro-cellular polymers. J. Chem. Technol. Biotechnol. 2009, 84, 511–518. [Google Scholar]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.S.; Hsieh, K.; Soh, H.T.; Plaxco, K.W. Electrochemical real-time nucleic acid amplification: Towards point-of-care quantification of pathogens. Trends Biotechnol. 2013, 31, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Pedrero, M.; Campuzano, S.; Pingarron, J.M. Electroanalytical sensors and devices for multiplexed detection of foodborne pathogen microorganisms. Sensors 2009, 9, 5503–5520. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, Y.; Fung, Y. Flow analysis coupled with PQC/DNA biosensor for assay of E. coli based on detecting DNA products from PCR amplification. Biosens. Bioelectron. 2006, 22, 506–512. [Google Scholar]
- Rodriguez, M.I.; Alocilja, E.C. Embedded DNA-polypyrrole biosensor for rapid detection of Escherichia coli. IEEE Sens. J. 2005, 5, 733–736. [Google Scholar] [CrossRef]
- Wu, V.C.; Chen, S.H.; Lin, C.S. Real-time detection of Escherichia coli O157:H7 sequences using a circulating-flow system of quartz crystal microbalance. Biosens. Bioelectron. 2007, 22, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Lermo, A.; Campoy, S.; Barbe, J.; Hernandez, S.; Alegret, S.; Pividori, M.I. In situ DNA amplification with magnetic primers for the electrochemical detection of food pathogens. Biosens. Bioelectron. 2007, 22, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhou, X.; Xing, D. Nano-magnetic primer based electrochemiluminescence-polymerase chain reaction (NMPE-PCR) assay. Biosens. Bioelectron. 2012, 31, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Pöhlmann, C.; Wang, Y.; Humenik, M.; Heidenreich, B.; Gareis, M.; Sprinzl, M. Rapid, specific and sensitive electrochemical detection of foodborne bacteria. Biosens. Bioelectron. 2009, 24, 2766–2771. [Google Scholar]
- Mairhofer, J.; Roppert, K.; Ertl, P. Microfluidic systems for pathogen sensing: A review. Sensors 2009, 9, 4804–4823. [Google Scholar] [CrossRef] [PubMed]
- Berdat, D.; Martin Rodriguez, A.C.; Herrera, F.; Gijs, M.A. Label-free detection of DNA with interdigitated micro-electrodes in a fluidic cell. Lab Chip 2008, 8, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Yeung, S.W.; Lee, T.M.; Cai, H.; Hsing, I.M. A DNA biochip for on-the-spot multiplexed pathogen identification. Nucleic Acids Res. 2006, 34, e118. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.H.; Ramalingam, N.; Xian-Dui, D.; Ngin, T.S.; Xianting, Z.; Lai Kuan, A.T.; Peng Huat, E.Y.; Hai-Qing, G. Real-time PCR microfluidic devices with concurrent electrochemical detection. Biosens. Bioelectron. 2009, 24, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.K.; Chand, R.; Han, D.; Jang, Y.C.; Ra, G.S.; Kim, J.S.; Nahm, B.H.; Kim, Y.S. An integrated PCR microfluidic chip incorporating aseptic electrochemical cell lysis and capillary electrophoresis amperometric DNA detection for rapid and quantitative genetic analysis. Lab Chip 2012, 12, 4455–4464. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Nahar, S.; Safavieh, M.; Zourob, M. Real-time electrochemical detection of pathogen DNA using electrostatic interaction of a redox probe. Analyst 2013, 138, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Amaya-Gonzalez, S.; de-los-Santos-Alvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castanon, M.J. Aptamer-based analysis: A promising alternative for food safety control. Sensors 2013, 13, 16292–16311. [Google Scholar]
- Zelada-Guillen, G.A.; Bhosale, S.V.; Riu, J.; Rius, F.X. Real-time potentiometric detection of bacteria in complex samples. Anal. Chem. 2010, 82, 9254–9260. [Google Scholar]
- Ning, Y.; Li, W.; Duan, Y.; Yang, M.; Deng, L. High specific DNAzyme-aptamer sensor for Salmonella paratyphi a using single-walled nanotubes-based dual fluorescence-spectrophotometric methods. J. Biomol. Screen. 2014, 19, 1099–1106. [Google Scholar]
- Jyoti, A.; Vajpayee, P.; Singh, G.; Patel, C.B.; Gupta, K.C.; Shanker, R. Identification of environmental reservoirs of nontyphoidal salmonellosis: Aptamer-assisted bioconcentration and subsequent detection of Salmonella Typhimurium by quantitative polymerase chain reaction. Environ. Sci. Technol. 2011, 45, 8996–9002. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Loessner, M.J. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014, 4, e28137. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Bacteriophages: Biosensing tools for multi-drug resistant pathogens. Analyst 2014, 139, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Nanosphere. Available online: www.nanosphere.us (accessed on 10 May 2014).
- 3M. Available online: http://www.3m.com/ (accessed on 12 May 2014).
- Neogen. Available online: www.neogen.com (accessed on 12 May 2014).
- Serosep. Available online: www.serosep.com (accessed on 12 May 2014).
- Veredus Laboratories. Available online: www.vereduslabs.com (accessed on 1 May 2014).
- BIO phage PHARMA Inc. Available online: http://www.biophagepharma.net/index.php/en/ (accessed on 10 May 2014).
- Easy Life Science. Available online: http://www.elice.fr/ (accessed on 1 May 2014).
- STRATOPHASE™. Available online: http://www.stratophase.com/ (accessed on 11 April 2014).
- Available Real-Time PCR Cyclers. Available online: http://cyclers.gene-quantification.info/ (accessed on 12 May 2015).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Adley, C.C. Past, Present and Future of Sensors in Food Production. Foods 2014, 3, 491-510. https://doi.org/10.3390/foods3030491
Adley CC. Past, Present and Future of Sensors in Food Production. Foods. 2014; 3(3):491-510. https://doi.org/10.3390/foods3030491
Chicago/Turabian StyleAdley, Catherine C. 2014. "Past, Present and Future of Sensors in Food Production" Foods 3, no. 3: 491-510. https://doi.org/10.3390/foods3030491



