Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits
Abstract
:1. Introduction
2. Genomic Characteristics of Latilactobacillus curvatus
3. Physiological and Biochemical Properties of Latilactobacillus curvatus
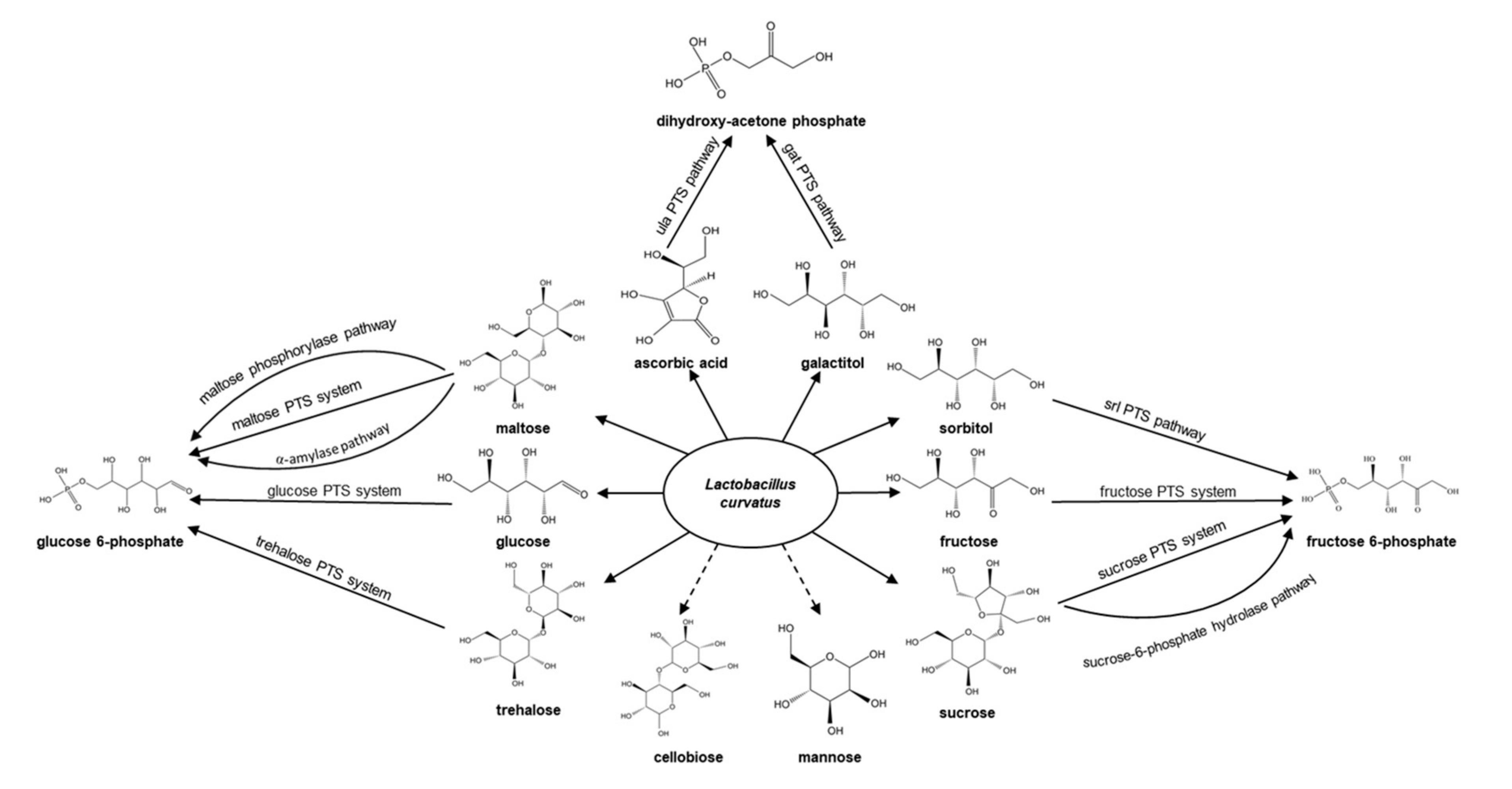
3.1. Carbohydrate Utilization
3.2. Antibiotic Resistance
3.3. Auto-Aggregation and Co-Aggregation Capacity
3.4. Resistance to Gastrointestinal Tract Conditions
3.5. Generation and Degradation of Biogenic Amines
3.6. Production of Bacteriocin
4. Applications of Latilactobacillus curvatus in Fermented Meat Products and Food Packaging
4.1. Starter for Meat Products
4.2. Food Packaging
5. Probiotic Function of Latilactobacillus curvatus
5.1. Obesity
5.2. Dyslipidemia
5.3. Others
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bourdichon, F.; Berger, B.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; et al. Safety demonstration of microbial food cultures (MFC) in fermented food products. Bull. Int. Dairy Fed. 2012, 455, 62. [Google Scholar]
- EFSA Panel on Biological Hazards. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J. 2013, 11, 3449. [Google Scholar] [CrossRef] [Green Version]
- Hammes, W.P.; Hertel, C. Lactobacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–76. [Google Scholar]
- Lucquin, I.; Zagorec, M.; Champomier-Verges, M.; Chaillou, S. Fingerprint of lactic acid bacteria population in beef carpaccio is influenced by storage process and seasonal changes. Food Microbiol. 2012, 29, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Desmonts, M.H.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kask, S.; Adamberg, K.; Orłowski, A.; Vogensen, F.K.; Møller, P.L.; Ardö, Y.; Paalme, T. Physiological properties of Lactobacillus paracasei, L. danicus and L. curvatus strains isolated from Estonian semi-hard cheese. Food Res. Int. 2003, 36, 1037–1046. [Google Scholar] [CrossRef]
- Vogelxy, R.F.; Lohmann, M.; Nguyen, M. MolecuIar characterization of Lactobacillus curvatus and Lact. sake isolated from sauerkraut and their application in sausage fermentations. J. Appl. Microbiol. 1993, 74, 295–300. [Google Scholar] [CrossRef]
- Nakano, K.; Shiroma, A.; Tamotsu, H.; Ohki, S.; Shimoji, M.; Ashimine, N.; Shinzato, M.; Minami, M.; Nakanishi, T.; Teruya, K.; et al. First Complete Genome Sequence of the Skin-Improving Lactobacillus curvatus Strain FBA2, Isolated from Fermented Vegetables, Determined by PacBio Single-Molecule Real-Time Technology. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef] [Green Version]
- Michel, E.; Monfort, C.; Deffrasnes, M.; Guezenec, S.; Lhomme, E.; Barret, M.; Sicard, D.; Dousset, X.; Onno, B. Characterization of relative abundance of lactic acid bacteria species in French organic sourdough by cultural, qPCR and MiSeq high-throughput sequencing methods. Int. J. Food. Microbiol. 2016, 239, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Bulgasem, B.Y.; Lani, M.N.; Hassan, Z.; Wan Yusoff, W.M.; Fnaish, S.G. Antifungal Activity of Lactic Acid Bacteria Strains Isolated from Natural Honey against Pathogenic Candida Species. Mycobiology 2016, 44, 302–309. [Google Scholar] [CrossRef] [Green Version]
- Koleva, Z.; Dedov, I.; Kizheva, J.; Lipovanska, R.; Moncheva, P.; Hristova, P. Lactic acid microflora of the gut of snail Cornu aspersum. Biotechnol. Biotechnol. Equip. 2014, 28, 627–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zommiti, M.; Connil, N.; Hamida, J.B.; Ferchichi, M. Probiotic Characteristics of Lactobacillus curvatus DN317, a Strain Isolated from Chicken Ceca. Probiotics Antimicrob. Proteins 2017, 9, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Dal Bello, F.; Walter, J.; Hammes, W.P.; Hertel, C. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 2003, 45, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Kandler, O.; Abo-Elnaga, I.G. On the taxonomy of genus Lactobacillus Beijerinck I. Subgenus Streptobacterium Orla Jensen. Zent. fur Bakteriol. Parasitenkd. Infekt. Hyg. Zweite Nat. Abt. Allg. Landwirtsch. Tech. Mikrobiol. 1965, 119, 1–36. [Google Scholar]
- Hammes, W. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Lett. 1990, 87, 165–173. [Google Scholar] [CrossRef]
- Zheng, J.S.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; Toole, P.W.O.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimon, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef]
- Berthier, F.; Ehrlich, S.D. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int. J. Syst. Bacteriol. 1999, 49, 997–1007. [Google Scholar] [CrossRef]
- Petrick, H.A.R.; Ambrosio, R.E.; Holzapfel, W.H. Isolation of a DNA Probe for Lactobacillus curvatus. Appl. Environ. Microbiol. 1988, 54, 405–408. [Google Scholar] [CrossRef] [Green Version]
- De Souza Barbosa, M.; Todorov, S.D.; Ivanova, I.; Chobert, J.M.; Haertle, T.; de Melo Franco, B.D.G. Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiol. 2015, 46, 254–262. [Google Scholar] [CrossRef] [Green Version]
- Tichaczek, P.S.; Nissen-Meyer, J.; Nes, I.F.; Vogel, R.F.; Hammes, W.P. Characterization of the Bacteriocins Curvacin A from Lactobacillus curvatus LTH1174 and Sakacin P from L. sake LTH673. Syst. Appl. Microbiol. 1992, 15, 460–468. [Google Scholar] [CrossRef]
- Torriani, S.; Reenen, C.A.V.; Klein, G.; Reuter, G.; Dellaglio, F.; Dicks, L.M.T. Lactobacillus curvatus subsp. curvatus subsp. nov. and Lactobacillus curvatus subsp. melibiosus subsp. nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., New Subspecies of Lactobacillus curvatus Abo-Elnaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara, and Fukami 1934 (Klein et al. 1996, Emended Descriptions), Respectively. Int. J. Syst. Bacteriol. 1996, 46, 1158–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koort, J.; Vandamme, P.; Schillinger, U.; Holzapfel, W.; Bjorkroth, J. Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int. J. Syst. Evol. Microbiol. 2004, 54, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Hebert, E.M.; Saavedra, L.; Taranto, M.P.; Mozzi, F.; Magni, C.; Nader, M.E.; Font de Valdez, G.; Sesma, F.; Vignolo, G.; Raya, R.R. Genome sequence of the bacteriocin-producing Lactobacillus curvatus strain CRL705. J. Bacteriol. 2012, 194, 538–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teran, L.C.; Coeuret, G.; Raya, R.; Zagorec, M.; Champomier-Verges, M.C.; Chaillou, S. Phylogenomic Analysis of Lactobacillus curvatus Reveals Two Lineages Distinguished by Genes for Fermenting Plant-Derived Carbohydrates. Genome Biol. Evol. 2018, 10, 1516–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenbach, L.; Janssen, D.; Ehrmann, M.A.; Vogel, R.F. Comparative genomics of Lactobacillus curvatus enables prediction of traits relating to adaptation and strategies of assertiveness in sausage fermentation. Int. J. Food. Microbiol. 2018, 286, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Ercolini, D.; La Storia, A.; Casaburi, A.; Villani, F. Development of polythene films for food packaging activated with an antilisterial bacteriocin from Lactobacillus curvatus 32Y. J. Appl. Microbiol. 2004, 97, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Kong, B.; Chen, Q.; Han, Q.; Diao, X. N-nitrosoamine inhibition and quality preservation of Harbin dry sausages by inoculated with Lactobacillus pentosus, Lactobacillus curvatus and Lactobacillus sake. Food Control 2017. [Google Scholar] [CrossRef]
- Fadda, S.; Lopez, C.; Vignolo, G. Role of lactic acid bacteria during meat conditioning and fermentation: Peptides generated as sensorial and hygienic biomarkers. Meat Sci. 2010, 86, 66–79. [Google Scholar] [CrossRef]
- Jo, S.G.; Noh, E.J.; Lee, J.Y.; Kim, G.; Choi, J.H.; Lee, M.E.; Song, J.H.; Chang, J.Y.; Park, J.H. Lactobacillus curvatus WiKim38 isolated from kimchi induces IL-10 production in dendritic cells and alleviates DSS-induced colitis in mice. J. Microbiol. 2016, 54, 503–509. [Google Scholar] [CrossRef]
- Yoo, S.R.; Kim, Y.J.; Park, D.Y.; Jung, D.J.; Jeon, S.M.; Ahn, Y.T.; Huh, C.S.; McGrego, R.; Choi, M.S. Probiotics, L. Plantarum and L. Curvatus In Combination Alter Hepatic Lipid Metabolism and Suppress Diet-Induced Obesity. Obesity 2013, 21, 2571–2578. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.Y.; Kim, M.; Ahn, Y.T.; Sim, J.H.; Choi, I.D.; Lee, S.H.; Lee, J.H. The triglyceride-lowering effect of supplementation with dual probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032: Reduction of fasting plasma lysophosphatidylcholines in nondiabetic and hypertriglyceridemic subjects. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Park, D.Y.; Ahn, Y.T.; Park, S.H.; Huh, C.S.; Yoo, S.R.; Yu, R.; Sung, M.K.; McGregor, R.A.; Choi, M.S. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE 2013, 8, e59470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuki, R.; Sakata, S.; Nakao, R.; Oishi, K.; Nakamura, Y. Lactobacillus curvatus CP2998 Prevents Dexamethasone-Induced Muscle Atrophy in C2C12 Myotubes. J. Nutr. Sci. Vitaminol. 2019, 65, 455–458. [Google Scholar] [CrossRef] [Green Version]
- Chaillou, S.; Champomier-Verges, M.C.; Cornet, M.; Crutz-Le Coq, A.M.; Dudez, A.M.; Martin, V.; Beaufils, S.; Darbon-Rongere, E.; Bossy, R.; Loux, V.; et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Jung, M.Y.; Song, J.H.; Lee, M.; Chang, J.Y. Complete Genome Sequence of Lactobacillus curvatus Strain WiKim38 Isolated from Kimchi. Genome Announc. 2017, 5, 503–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teran, L.C.; Coeuret, G.; Raya, R.; Champomier-Verges, M.C.; Chaillou, S. Draft Genome Sequence of Lactobacillus curvatus FLEC03, a Meat-Borne Isolate from Beef Carpaccio Packaged in a Modified Atmosphere. Genome Announc. 2017, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyoui, D.; Mikami, N.; Yamamoto, H.; Kawarai, T.; Ogihara, H. Complete Genome Sequence of Lactobacillus curvatus NFH-Km12, Isolated from the Japanese Traditional Fish Fermented Food Kabura-zushi. Microbiol. Resour. Announc. 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jans, C.; Lagler, S.; Lacroix, C.; Meile, L.; Stevens, M.J.A. Complete Genome Sequences of Lactobacillus curvatus KG6, L. curvatus MRS6, and Lactobacillus sakei FAM18311, Isolated from Fermented Meat Products. Genome Announc. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- Inglin, R.C.; Meile, L.; Stevens, M.J.A. Draft Genome Sequences of 43 Lactobacillus Strains from the Species L. curvatus, L. fermentum, L. paracasei, L. plantarum, L. rhamnosus, and L. sakei, Isolated from Food Product. Genome Announc. 2017, 5, e00632-17. [Google Scholar] [CrossRef] [Green Version]
- Cousin, F.C.; Lynch, S.M.; Harris, H.M.B.; McCann, A.; Lynch, D.B.; Neville, B.A.; Irisawa, T.; Okada, S.; Endo, A.; O’Toole, P.W. Detection and Genomic Characterization of Motility in Lactobacillus curvatus: Confirmation of Motility in a Species outside the Lactobacillus salivarius Clade. Appl. Environ. Microbiol. 2014, 81, 1297–1308. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Nyquist, O.L.; McLeod, A.; Brede, D.A.; Snipen, L.; Aakra, A.; Nes, I.F. Comparative genomics of Lactobacillus sakei with emphasis on strains from meat. Mol. Genet. Genomics 2011, 285, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Verge, M.-C.C.; Manuel, Z.; Françoise, M.-D.; Perez-Martinez, G.; Zagorec, M.; Ehrlich, S.D. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 1999, 2, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Wiame, E.; Lamosa, P.; Santos, H.; Van Schaftingen, E. Identification of glucoselysine-6-phosphate deglycase, an enzyme involved in the metabolism of the fructation product glucoselysine. Biochem. J. 2005, 392, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Stentz, R.; Zagorec, M. Ribose utilization in Lactobacillus sakei: Analysis of the regulation of the rbs operon and putative involvement of a new transporter. J. Microbiol. Biotechnol. 1999, 1, 165–173. [Google Scholar]
- Yew, W.S.; Gerlt, J.A. Utilization of L-ascorbate by Escherichia coli K-12: Assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 2002, 184, 302–306. [Google Scholar] [CrossRef] [Green Version]
- Salminen, S.; Wright, A.v.; Morelli, L.; Marteau, P.; Brassart, D.; Vos, W.M.d.; Fonden, R.; Saxelin, M.; Collins, K.; Mogensen, G.; et al. Demonstration of safety of probiotics—A review. Int. J. Food Microbiol. 1998, 44, 93–106. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef]
- Hong, S.W.; Kim, J.H.; Bae, H.J.; Ham, J.S.; Yoo, J.G.; Chung, K.S.; Oh, M.H. Selection and characterization of broad-spectrum antibacterial substance-producing Lactobacillus curvatus PA40 as a potential probiotic for feed additives. Anim. Sci. J. 2018, 89, 1459–1467. [Google Scholar] [CrossRef]
- Ahmadova, A.; Todorov, S.D.; Hadji-Sfaxi, I.; Choiset, Y.; Rabesona, H.; Messaoudi, S.; Kuliyev, A.; Franco, B.D.; Chobert, J.M.; Haertle, T. Antimicrobial and antifungal activities of Lactobacillus curvatus strain isolated from homemade Azerbaijani cheese. Anaerobe 2013, 20, 42–49. [Google Scholar] [CrossRef]
- Ammor, M.S.; Florez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Schwarz, F.V.; Teuber, M.; Levy, S.B. Mdt(A), a new efflux protein conferring multiple antibiotic resistance in Lactococcus lactis and Escherichia coli. Antimicrob. Agents Chemother. 2001, 45, 1109–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.S.; Pillidge, C.J.; Gopal, P.K.; Gill, H.S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005, 98, 211–217. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, T.T.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.H.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic. Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.F.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.Y.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 2007, 45, 454–460. [Google Scholar] [CrossRef]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.P.; Gómez de Cadiñanos, L.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Corzo, G.; Gilliland, S.E. Bile Salt Hydrolase Activity of Three Strains of Lactobacillus acidophilus. J. Dairy Sci. 1999, 82, 472. [Google Scholar] [CrossRef]
- Garcia-Ruiz, A.; Gonzalez de Llano, D.; Esteban-Fernandez, A.; Requena, T.; Bartolome, B.; Moreno-Arribas, M.V. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.E.; Proctor, V.A.; Goetsch, S.J. Egg-white lysozyme as a food preservative: An overview. World Poultry Sci. J. 1991, 47, 141. [Google Scholar] [CrossRef]
- Fernandez, M.F.; Boris, S.; Barbes, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 2003, 94, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, S.E.; Staley, T.E.; Bush, L.J. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 1984, 67, 3045–3051. [Google Scholar] [CrossRef]
- Jin, J.H.; Zhang, B.; Guo, H.; Cui, J.; Jiang, L.; Song, S.; Sun, M.; Ren, F.Z. Mechanism Analysis of Acid Tolerance Response of Bifidobacterium longum subsp. longum BBMN 68 by Gene Expression Profile Using RNA-Sequencing. PLoS ONE 2012, 7, e50777. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Ren, F.; Liu, S.; Zhao, L.; Guo, H.; Hou, C. Enhanced acid tolerance in Bifidobacterium longum by adaptive evolution: Comparison of the genes between the acid-resistant variant and wild-type strain. J. Microbiol. Biotechnol. 2016, 26, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Mathara, J.M.; Schillinger, U.; Kutima, P.M.; Mbugua, S.K.; Guigas, C.; Franz, C.; Holzapfel, W.H. Functional properties of Lactobacillus plantarum strains isolated from Maasai traditional fermented milk products in Kenya. Curr. Microbiol. 2008, 56, 315–321. [Google Scholar] [CrossRef]
- Erkkilä, S.; Petäjä, E. Screening of commercial meat starter cultures at low pH and in the presence of bile salts for potential probiotic use. Meat Sci. 2000, 55, 297–300. [Google Scholar] [CrossRef]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Bai, X.; Byun, B.Y.; Mah, J.H. Formation and destruction of biogenic amines in Chunjang (a black soybean paste) and Jajang (a black soybean sauce). Food Chem. 2013, 141, 1026–1031. [Google Scholar] [CrossRef]
- Santos, M.H.S. Biogenic amines: Their importance in foods. Int. J. Food Microbiol. 1996, 29, 213–231. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Polo, L.; Parodo, I. Biogenic amines in wines from three Spanish regions. J. Agric. Food Chem. 2005, 53, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, C.A.; Conte-Júnior, C.A.; Canto, A.C.; Monteiro, M.L.G.; Costa-Lima, B.; Cruz, A.G.D.; Mársico, E.T.; Franco, R.M. Biogenic amines as bacterial quality indicators in different poultry meat species. Lwt Food Sci. Technol. 2015, 60, 15–21. [Google Scholar] [CrossRef]
- Guarcello, R.; De Angelis, M.; Settanni, L.; Formiglio, S.; Gaglio, R.; Minervini, F.; Moschetti, G.; Gobbetti, M. Selection of Amine-Oxidizing Dairy Lactic Acid Bacteria and Identification of the Enzyme and Gene Involved in the Decrease of Biogenic Amines. Appl. Environ. Microbiol. 2016, 82, 6870–6880. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wen, X.; Wen, Z.; Chen, S.; Wang, L.; Wei, X. Evaluation of the Biogenic Amines Formation and Degradation Abilities of Lactobacillus curvatus From Chinese Bacon. Front. Microbiol. 2018, 9, 1015. [Google Scholar] [CrossRef] [Green Version]
- Pachlová, V.; Buňková, L.; Flasarová, R.; Salek, R.N.; Dlabajová, A.; Butor, I.; Buňka, F. Biogenic amine production by nonstarter strains of Lactobacillus curvatus and Lactobacillus paracasei in the model system of Dutch-type cheese. LWT 2018, 97, 730–735. [Google Scholar] [CrossRef]
- Landete, J.M.; Pardo, I.; Ferrer, S. Tyramine and phenylethylamine production among lactic acid bacteria isolated from wine. Int. J. Food Microbiol. 2007, 115, 364–368. [Google Scholar] [CrossRef]
- Latorre-Moratalla, M.L.; Bover-Cid, S.; Bosch-Fuste, J.; Vidal-Carou, M.C. Influence of technological conditions of sausage fermentation on the aminogenic activity of L. curvatus CTC273. Food Microbiol. 2012, 29, 43–48. [Google Scholar] [CrossRef]
- Bardócz, S. Polyamines in food and their consequences for food quality and human health. Trends Food Sci. Technol. 1996, 6, 341–346. [Google Scholar] [CrossRef]
- Freiding, S.; Gutsche, K.A.; Ehrmann, M.A.; Vogel, R.F. Genetic screening of Lactobacillus sakei and Lactobacillus curvatus strains for their peptidolytic system and amino acid metabolism, and comparison of their volatilomes in a model system. Syst. Appl. Microbiol. 2011, 34, 311–320. [Google Scholar] [CrossRef]
- Cid, S.B.; Miguelez-Arrizado, M.J.; Becker, B.; Holzapfel, W.H.; Vidal-Carou, M.C. Amino acid decarboxylation by Lactobacillus curvatus CTC273 affected by the pH and glucose availability. Food Microbiol. 2008, 25, 269–277. [Google Scholar] [CrossRef]
- Dapkevicius, M.L.N.E.; Nout, M.J.R.; Rombouts, F.M.; Houben, J.H.; Wymenga, W. Biogenic amine formation and degradation by potential fish silage starter microorganisms. Int. J. Food Microbiol. 2000, 57, 107–114. [Google Scholar] [CrossRef]
- García-Ruiz, A.; González-Rompinelli, E.M.; Bartolomé, B.; Moreno-Arribas, M.V. Potential of wine-associated lactic acid bacteria to degrade biogenic amines. Int. J. Food. Microbiol. 2011, 148, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, ecology, and application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balciunas, E.M.; Castillo Martinez, F.A.; Todorov, S.D.; Franco, B.D.G.D.M.; Converti, A.; Oliveira, R.P.D.S. Novel biotechnological applications of bacteriocins: A review. Food Control 2013, 32, 134–142. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing Innate Immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Verluyten, J.; Messens, W.; Vuyst, L.D. Sodium chloride reduces production of curvacin A, a bacteriocin produced by Lactobacillus curvatus strain LTH 1174, originating from fermented sausage. Appl. Environ. Microbiol. 2004, 70, 2271–2278. [Google Scholar] [CrossRef] [Green Version]
- Mechoud, M.A.; Álvarez, O.E.; Cayré, M.E.; Castro, M.P.; Minahk, C.; Saavedra, L. Sakacin G is the main responsible bacteriocin for the anti-listerial activity of meat-borne Lactobacillus curvatus ACU-1. Ann. Microbiol. 2017, 67, 615–621. [Google Scholar] [CrossRef]
- Bouttefroy, A.; Linder, M.; Milliere, J.B. Predictive models of the combined effects of curvaticin 13, NaCl and pH on the behaviour of Listeria monocytogenes ATCC 15313 in broth. J. Appl. Microbiol. 2000, 88, 919–929. [Google Scholar] [CrossRef]
- Massani, M.B.; Molina, V.; Sanchez, M.; Renaud, V.; Eisenberg, P.; Vignolo, G. Active polymers containing Lactobacillus curvatus CRL705 bacteriocins: Effectiveness assessment in Wieners. Int. J. Food Microbiol. 2014, 178, 7–12. [Google Scholar] [CrossRef]
- Xiraphi, N.; Georgalaki, M.; Driessche, G.V.; Devreese, B.; Beeumen, J.V.; Tsakalidou, E.; Metaxopoulos, J.; Drosinos, E.H. Purification and characterization of curvaticin L442, a bacteriocin produced by Lactobacillus curvatus L442. Antonie Van Leeuwenhoek 2006, 89, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa bacteriocins: Diversity and new developments. Int. J. Mol. Sci. 2012, 13, 16668–16707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellano, P.; Belfiore, C.; Fadda, S.; Vignolo, G. A review of bacteriocinogenic lactic acid bacteria used as bioprotective cultures in fresh meat produced in Argentina. Meat Sci. 2008, 79, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Rivas, F.P.; Castro, M.P.; Vallejo, M.; Marguet, E.; Campos, C.A. Sakacin Q produced by Lactobacillus curvatus ACU-1: Functionality characterization and antilisterial activity on cooked meat surface. Meat Sci. 2014, 97, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.P.; Palavecino, N.Z.; Herman, C.; Garro, O.A.; Campos, C.A. Lactic acid bacteria isolated from artisanal dry sausages: Characterization of antibacterial compounds and study of the factors affecting bacteriocin production. Meat Sci. 2011, 87, 321–329. [Google Scholar] [CrossRef]
- Garve, K.I.; Murian, P.M. Purification and Partial Amino Acid Sequence of Curvaticin FS47, a Heat-Stable Bacteriocin Produced by Lactobacillus curvatus FS4. Appl. Environ. Microbiol. 1994, 60, 2191–2195. [Google Scholar] [CrossRef] [Green Version]
- Zommiti, M.; Almohammed, H.; Ferchichi, M. Purification and Characterization of a Novel Anti-Campylobacter Bacteriocin Produced by Lactobacillus curvatus DN317. Probiotics Antimicrob. Proteins 2016, 8, 191–201. [Google Scholar] [CrossRef]
- Casaburi, A.; Martino, V.D.; Ferranti, P.; Picariello, L.; Villani, F. Technological properties and bacteriocins production by Lactobacillus curvatus 54M16 and its use as starter culture for fermented sausage manufacture. Food Control 2016, 59, 31–45. [Google Scholar] [CrossRef]
- Gómez-Sala, B.; Muñoz-Atienza, E.; Diep, D.B.; Feito, J.; del Campo, R.; Nes, I.F.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Biotechnological potential and in vitro safety assessment of Lactobacillus curvatus BCS35, a multibacteriocinogenic strain isolated from dry-salted cod (Gadus morhua). LWT 2019, 112. [Google Scholar] [CrossRef]
- Dortu, C.; Huch, M.; Holzapfel, W.H.; Franz, C.M.; Thonart, P. Anti-listerial activity of bacteriocin-producing Lactobacillus curvatus CWBI-B28 and Lactobacillus sakei CWBI-B1365 on raw beef and poultry meat. Lett. Appl. Microbiol. 2008, 47, 581–586. [Google Scholar] [CrossRef]
- Tiehaczek, P.S.; Vogel, R.F.; Hammes, W.P. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTHl174. Arch. Microbiol. 1993, 160, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Fremaux, C.; Cenatiempo, Y.; Berjeaud, J.M. Sakacin g, a new type of antilisterial bacteriocin. Appl. Environ. Microbiol. 2002, 68, 6416–6420. [Google Scholar] [CrossRef] [Green Version]
- Papagianni, M.; Anastasiadou, S. Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb. Cell Fact. 2009, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammes, W.P.; Hertel, C. Selection and improvement of lactic acid bacteria used in meat and sausage fermentation. Dairy Sci. Technol. 1996, 76, 159–168. [Google Scholar] [CrossRef]
- Vogel, R.F.; Pohle, B.S.; Tichaczek, P.S.; Hammes, W.P. The Competitive Advantage of Lactobacillus curvatus LTH 1174 in Sausage Fermentations is Caused by Formation of Curvacin A. Syst. Appl. Microbiol. 1993, 16, 457–462. [Google Scholar] [CrossRef]
- Giello, M.; La Storia, A.; De Filippis, F.; Ercolini, D.; Villani, F. Impact of Lactobacillus curvatus 54M16 on microbiota composition and growth of Listeria monocytogenes in fermented sausages. Food Microbiol. 2018, 72, 1–15. [Google Scholar] [CrossRef]
- Vogel, B.F.; Hansen, L.T.; Mordhorst, H.; Gram, L. The survival of Listeria monocytogenes during long term desiccation is facilitated by sodium chloride and organic material. Int. J. Food Microbiol. 2010, 140, 192–200. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Dong, P.; Liang, R.; Mao, Y.; Qiu, S.; Luo, X. Bio-protective potential of lactic acid bacteria: Effect of Lactobacillus sakei and Lactobacillus curvatus on changes of the microbial community in vacuum-packaged chilled beef. Asian Australas. J. Anim. Sci. 2018, 31, 585–594. [Google Scholar] [CrossRef] [Green Version]
- Stella, S.; Bernardi, C.; Cattaneo, P. Evaluation of the in vitro antimicrobial activity of mixtures of Lactobacillus sakei and L. curvatus isolated from Argentine meat and their application on vacuum-packed beef. Ital. J. Food Sci. 2016, 28, 612–624. [Google Scholar]
- Chen, Q.; Kong, B.; Sun, Q.; Dong, F.; Liu, Q. Antioxidant potential of a unique LAB culture isolated from Harbin dry sausage: In vitro and in a sausage model. Meat Sci. 2015, 110, 180–188. [Google Scholar] [CrossRef]
- Maere, H.D.; Fraeye, I.; Mey, E.D.; Dewulf, L.; Michiels, C.; Paelinck, H.; Chollet, S. Formation of naturally occurring pigments during the production of nitrite-free dry fermented sausages. Meat Sci. 2016, 114, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, S.S.; Duedahl-Olesen, L.; Christensen, T.; Olesen, P.T.; Granby, K. Dietary exposure to volatile and non-volatile N-nitrosamines from processed meat products in Denmark. Food Chem. Toxicol. 2015, 80, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Xia, W.; Ge, C. Effect of mixed starter cultures fermentation on the characteristics of silver carp sausages. World J. Microb. Biot. 2006, 23, 1021–1031. [Google Scholar] [CrossRef]
- Nie, X.; Lin, S.; Zhang, Q. Proteolytic characterisation in grass carp sausage inoculated with Lactobacillus plantarum and Pediococcus pentosaceus. Food Chem. 2014, 145, 840–844. [Google Scholar] [CrossRef]
- Kim, S.H.; Kang, K.H.; Kim, S.H.; Lee, S.; Lee, S.H.; Ha, E.S.; Sung, N.J.; Kim, J.G.; Chung, M.J. Lactic acid bacteria directly degrade N-nitrosodimethylamine and increase the nitrite-scavenging ability in kimchi. Food Control 2017, 71, 101–109. [Google Scholar] [CrossRef]
- Fadda, S.; Sanz, Y.; Vignolo, G.; Aristoy, M.C.; Toldra, F.; Oliver, G. Hydrolysis of pork muscle sarcoplasmic proteins by Lactobacillus curvatus and Lactobacillus sake. Appl. Environ. Microbiol. 1999, 65, 578–584. [Google Scholar] [CrossRef] [Green Version]
- Joerger, R.D. Antimicrobial films for food applications: A quantitative analysis of their effectiveness. Packag. Technol. Sci. 2007, 20, 231–273. [Google Scholar] [CrossRef]
- Massani, M.B.; Fernandez, M.R.; Ariosti, A.; Eisenberg, P.; Vignolo, G. Development and characterization of an active polyethylene film containing Lactobacillus curvatus CRL705 bacteriocins. Food Addit. Contam. Part A 2008, 25, 1424–1430. [Google Scholar] [CrossRef]
- Massani, M.B.; Vignolo, G.M.; Eisenberg, P.; Morando, P.J. Adsorption of the bacteriocins produced by Lactobacillus curvatus CRL705 on a multilayer-LLDPE film for food-packaging applications. LWT Food Sci. Technol. 2013, 53, 128–138. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Backhed, F.; Ding, H.; Wang, T. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, W.; Li, H.; Zhao, L. Gut microbiota: A potential new territory for drug targeting. Nat. Rev. Drug Discov. 2008, 7, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komaroff, A.L. The Microbiome and Risk for Obesity and Diabetes. JAMA 2017, 317, 355–356. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Kim, S.S. Probiotics and Prebiotics: Present Status and Future Perspectives on Metabolic Disorders. Nutrients 2016, 8, 173. [Google Scholar] [CrossRef] [Green Version]
- Backhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef] [Green Version]
- Jeung, W.H.; Nam, W.; Kim, H.J.; Kim, J.Y.; Nam, B.; Jang, S.S.; Lee, J.L.; Sim, J.H.; Park, S.D. Oral Administration of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 with Cinnamomi Ramulus Extract Reduces Diet-Induced Obesity and Modulates Gut Microbiota. Prev. Nutr. Food Sci. 2019, 24, 136–143. [Google Scholar] [CrossRef]
- Meehan, C.J.; Beiko, R.G. A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-associated Bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, D.K.; Renuka; Puniya, M.; Shandilya, U.K.; Dhewa, T.; Kumar, N.; Kumar, S.; Puniya, A.K.; Shukla, P. Gut Microbiota Modulation and Its Relationship with Obesity Using Prebiotic Fibers and Probiotics: A Review. Front. Microbiol. 2017, 8, 563. [Google Scholar] [CrossRef]
- Jeung, W.H.; Shim, J.J.; Woo, S.W.; Sim, J.H.; Lee, J.L. Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 Cell Extracts Inhibit Adipogenesis in 3T3-L1 and HepG2 Cells. J. Med. Food 2018, 21, 876–886. [Google Scholar] [CrossRef]
- Jung, S.; Lee, Y.J.; Kim, M.; Kim, M.; Kwak, J.H.; Lee, J.-W.; Ahn, Y.-T.; Sim, J.-H.; Lee, J.H. Supplementation with two probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032, reduced body adiposity and Lp-PLA2 activity in overweight subjects. J. Funct. Foods 2015, 19, 744–752. [Google Scholar] [CrossRef]
- Hunter, P.M.; Hegele, R.A. Functional foods and dietary supplements for the management of dyslipidaemia. Nat. Rev. Endocrinol. 2017, 13, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.C.; Lajo, T.; Carrion, J.M.; Cune, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, H.Y.; Kim, M.; Chae, J.S.; Ahn, Y.T.; Sim, J.H.; Choi, I.D.; Lee, S.H.; Lee, J.H. Supplementation with two probiotic strains, Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032, reduces fasting triglycerides and enhances apolipoprotein A-V levels in non-diabetic subjects with hypertriglyceridemia. Atherosclerosis 2015, 241, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.K.; Heeren, J.; Olivecrona, G.; Merkel, M. Apolipoprotein A-V; a potent triglyceride reducer. Atherosclerosis 2011, 219, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.D.; Kim, S.H.; Jeong, J.W.; Lee, D.E.; Huh, C.S.; Hong, S.S.; Sim, J.H.; Ahn, Y.T. Triglyceride-Lowering Effects of Two Probiotics, Lactobacillus plantarum KY1032 and Lactobacillus curvatus HY7601, in a Rat Model of High-Fat Diet-Induced Hypertriglyceridemia. J. Microbiol. Biotechnol. 2016, 26, 483–487. [Google Scholar] [CrossRef]
- Park, M.Y.; Kim, J.; Kim, S.; Whang, K.Y. Lactobacillus curvatus KFP419 and Leuconostoc mesenteroides subsp. mesenteroides KDK411 Isolated from Kimchi Ameliorate Hypercholesterolemia in Rats. J. Med. Food 2018, 21, 647–653. [Google Scholar] [CrossRef]

| Strain | Source | Genome Size (Mb) | GC Content (%) | Number of CDS | Accession Number | Sequencing Status (Sequencing Technology) | Reference |
|---|---|---|---|---|---|---|---|
| L. curvatus FBA2 | Fermented vegetables | 1.849 | 42.10 | 1711 | CP016028 | Complete (PacBio RS II platform) | [8] |
| L. curvatus Wikim38 | Kimchi | 1.940 | 41.93 | 1885 | CP017124 | Complete (PacBio RS II platform) | [37] |
| L. curvatus Wikim52 | Kimchi | 1.987 | 42.00 | 1875 | CP016602 | Complete (PacBio RS II platform) | NP |
| L. curvatus KG6 | Meat | 2.002 | 42.03 | 1970 | CP022475 | Complete (PacBio RS II platform) | [40] |
| L. curvatus MRS6 | Meat | 2.114 | 41.70 | 1975 | CP022474 | Complete (PacBio RS II platform) | [40] |
| L. curvatus NFH-Km12 | Traditional Japanese fermented fish | 1.989 | 41.81 | 1946 | AP018699 | Illumina MiSeq pair-end | [39] |
| L. curvatus TMW 1.421 | Sausage | 1.994 | 41.97 | 1961 | CP016221 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.439 | Sausage | 1.948 | 42.04 | 1939 | CP015489 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.624 | Sausage | 2.132 | 41.63 | 2148 | CP015490 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.595 | Starter culture | 2.032 | 41.95 | 1991 | CP016470 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.1381 | Starter culture | 1.949 | 42.05 | 1993 | CP015493 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.1390 | Starter culture | 1.977 | 42.07 | 1949 | CP015494 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.401 | Sauerkraut | 1.886 | 42.00 | 1830 | CP016216 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.407 | Sauerkraut | 1.886 | 42.01 | 1831 | CP016218 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.27 | Unknown | 2.056 | 41.86 | 2027 | CP016467 | Complete (PacBio RS II platform) | [27] |
| L. curvatus TMW 1.167 | Unknown | 1.951 | 42.03 | 1940 | CP016472 | Complete (PacBio RS II platform) | [27] |
| L. curvatus FLEC03 | Beef | 1.902 | 41.70 | 1926 | GCA_900178545.1 | Draft (Illumina MiSeq pair-end) | [38] |
| L. curvatus RI-124 | Meat | 1.810 | 42.00 | 1838 | MKDR00000000 | Draft (Illumina MiSeq pair-end) | [41] |
| L. curvatus RI-193 | Meat | 1.805 | 42.00 | 1862 | MKGD00000000 | Draft (Illumina MiSeq pair-end) | [41] |
| L. curvatus RI-198 | Meat | 1.804 | 42.00 | 1848 | MKGC00000000 | Draft (Illumina MiSeq pair-end) | [41] |
| L. curvatus RI-406 | Meat | 2.001 | 41.70 | 2020 | MKDG00000000 | Draft (Illumina MiSeq pair-end) | [41] |
| L. curvatus CRL705 | Argentinean fermented sausages | 1.838 | 41.90 | 1830 | AGBU01000000 | Draft (454 GS Titanium pyrosequencing) | [25] |
| L. curvatus NRIC0822 | Kabura-zushi | 1.945 | 41.80 | 1831 | GCA_000805355.1 | Draft (Illumina HiSeq pair-end) | [42] |
| L. curvatus DSM20019 | Milk | 1.917 | 41.99 | 1828 | GCA_004101845.1 | Draft (Ion Torrent PGM) | NP |
| Bacteriocin-Producing Strain | Bacteriocin | Source | Active Against | Reference |
|---|---|---|---|---|
| L. curvatus LTH1174 | Curvacin A | Fermented sausages | Enterococcus faecalis, Listeriamonocytogenes | [88] |
| L. curvatus SB13 | Curvaticin 13 | Semidry sausages | L. monocytogenes, Staphylococcus aureus | [90] |
| L. curvatus FS47 | Curvaticin FS47 | Beef | L. monocytogenes | [97] |
| L. curvatus L422 | Curvaticin L422 | Fermented sausages | L. monocytogenes | [92] |
| L. curvatus CRL705 | Lactocin 705 | Argentine fermented sausage | L. monocytogenes | [91] |
| L. curvatus DN317 | Curvaticin DN317 | Chicken Ceca | Campylobacter jejuni, L. monocytogenes, Bacillus subtilis | [98] |
| L. curvatus 54M16 | Sakacin X, P, T | Fermented sausages | Staphylococci, Enterobacteriaceae | [99] |
| L. curvatus A61 | Curvacin A | Azerbaijani cheese | L. monocytogenes, B. cereus | [53] |
| L. curvatus BCS35 | SakacinP-H12Y, Sakacin X | Dry-salted cod | L. monocytogenes | [100] |
| L. curvatus ACU-1 | Sakacin G, P, Q | Argentine fermented sausage | L. monocytogenes | [89] |
| L. curvatus MBSa2 | Sakacin P, X | Salami | L. monocytogenes | [21] |
| L. curvatus CWBI-B28 | Sakacin P | Raw poultry meat | L. monocytogenes | [101] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yu, L.; Qiao, N.; Xiao, Y.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits. Foods 2020, 9, 1366. https://doi.org/10.3390/foods9101366
Chen Y, Yu L, Qiao N, Xiao Y, Tian F, Zhao J, Zhang H, Chen W, Zhai Q. Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits. Foods. 2020; 9(10):1366. https://doi.org/10.3390/foods9101366
Chicago/Turabian StyleChen, Ying, Leilei Yu, Nanzhen Qiao, Yue Xiao, Fengwei Tian, Jianxin Zhao, Hao Zhang, Wei Chen, and Qixiao Zhai. 2020. "Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits" Foods 9, no. 10: 1366. https://doi.org/10.3390/foods9101366






