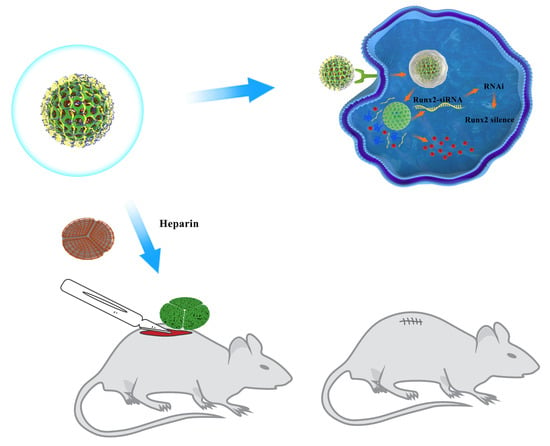
Biofunctionalized Decellularized Tissue-Engineered Heart Valve with Mesoporous Silica Nanoparticles for Controlled Release of VEGF and RunX2-siRNA against Calcification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Processing, Cell Culture, and Calcification Model In Vitro
2.2. RunX2-siRNA against Valvular Interstitial Cells Calcification
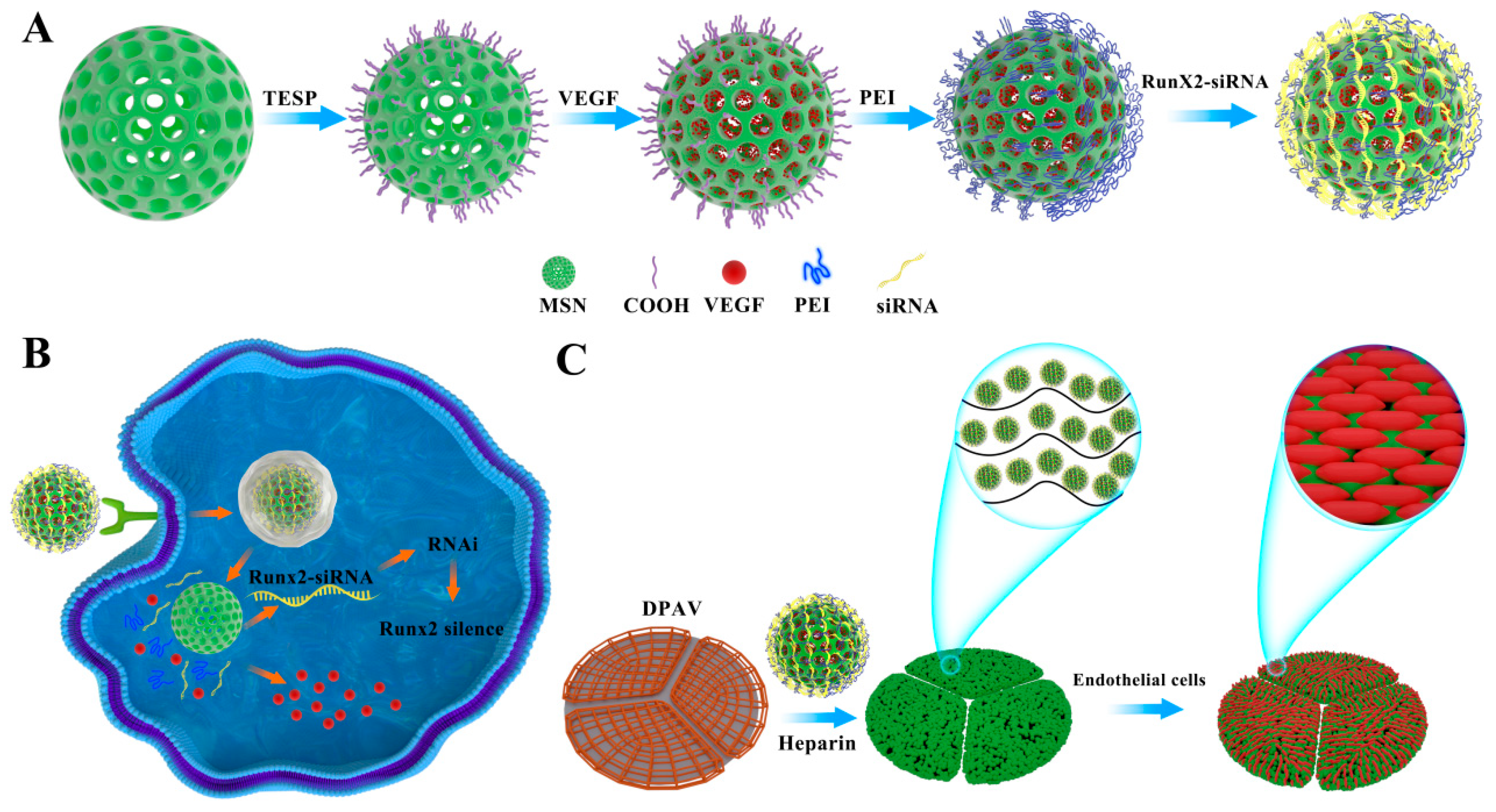
2.3. Preparation of MSN@VEGF-PEI-siRNA Delivery System
2.4. Encapsulation Efficiency and In Vitro Release Study
2.5. Cytotoxicity Assay
2.6. Cellular Uptake Study
2.7. Inflammatory Factor Detection
2.8. Efficiency of the MSN@VEGF-PEI-siRNA Delivery System for Gene Silencing in VICs
2.9. Endothelialization Rate of the MSN@VEGF -PEI-siRNA Delivery System
2.10. Preparation and Characterization of the DPAV Modified with MSN@VEGF -PEI-siRNA
2.11. In Vitro Release Assay of MSN@VEGF-PEI-siRNA Nanocomposite-Modified DPAV
2.12. Hemolysis Tests
2.13. Cell Seeding
2.14. Biomechanical Characterization of MSN@VEGF-PEI-siRNA Nanocomposite-Modified DPAV
2.15. CD31 Immunofluorescence Staining
2.16. Statistical Analysis
3. Results and Discussion
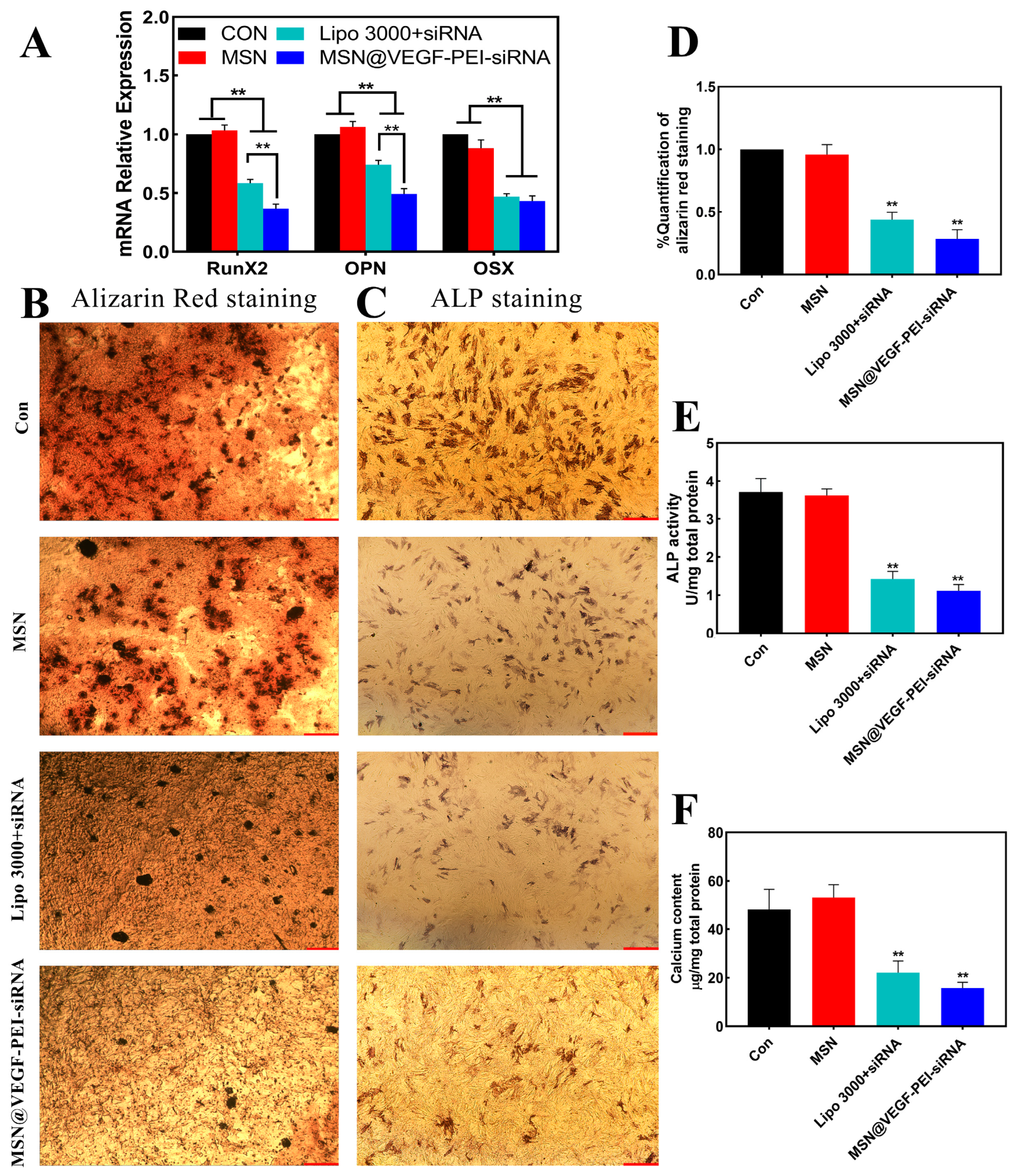
3.1. Anti-Calcification Research of RunX2-siRNA
3.2. Loading VEGF and RunX2-siRNA in the MSN and Characterization
3.3. Characterize the Anti-Calcification Property of MSN@VEGF-PEI-siRNA Nanocomposite with Valvular Interstitial Cells
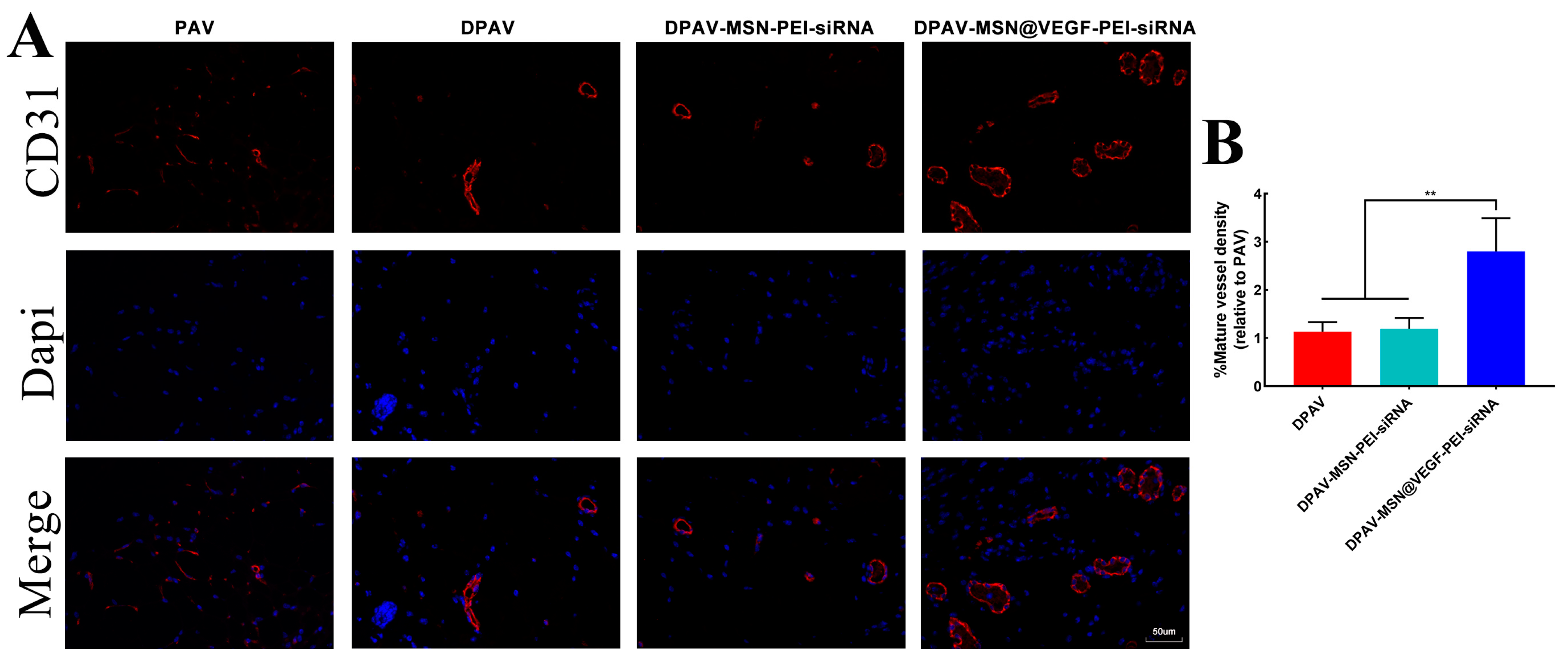
3.4. Characterize the Endothelialization Rate of the MSN@VEGF-PEI-siRNA Nanocomposite
3.5. Immobilize MSN@VEGF-PEI-siRNA Nanocomposite into DPAV and Characterization
3.6. Biomechanical Characterization of MSN@VEGF-PEI-siRNA Nanocomposite-Modified DPAV
3.7. Endothelialization of the MSN@VEGF-PEI-siRNA Nanocomposite-Modified DPAV In Vivo
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindroos, M.; Kupari, M.; Heikkilä, J.; Tilvis, R. Prevalence of aortic valve abnormalities in the elderly: An echocardiographic study of a random population sample. J. Am. Coll. Cardiol. 1993, 21, 1220–1225. [Google Scholar] [CrossRef] [Green Version]
- Otto C, M.; Lind, B.K.; Kitzman, D.W.; Gersh, B.J.; Siscovick, D.S. Association of aortic-valve sclerosis with cardiovascular mortality and morbidity in the elderly. N. Engl. J. Med. 1999, 341, 142–147. [Google Scholar] [CrossRef]
- Donato, M.; Ferri, N.; Lupo, M.G.; Faggin, E.; Rattazzi, M. Current Evidence and Future Perspectives on Pharmacological Treatment of Calcific Aortic Valve Stenosis. Int. J. Mol. Sci. 2020, 21, 8263. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Mequanint, K. Prosthetic aortic heart valves: Modeling and design. Med. Eng. Phys. 2011, 33, 131–147. [Google Scholar] [CrossRef]
- Brennan, J.M.; Edwards, F.H.; Zhao, Y. Long-term safety and effectiveness of mechanical versus biologic aortic valve prostheses in older patients: Results from the Society of Thoracic Surgeons Adult Cardiac Surgery National Database. Circulation 2013, 127, 1647–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, C.; Croce, B.; Cao, C. Tissue and mechanical heart valves. Ann. Cardiothorac. Surg. 2015, 4, 399. [Google Scholar]
- Boroumand, S.; Asadpour, S.; Akbarzadeh, A.; Faridi-Majidi, R.; Ghanbari, H. Heart valve tissue engineering: An overview of heart valve decellularization processes. Regen. Med. 2018, 13, 41–54. [Google Scholar] [CrossRef]
- Nachlas, A.; Li, S.; Davis, M.E. Developing a Clinically Relevant Tissue Engineered Heart Valve-A Review of Current Approaches. Adv. Healthc. Mater. 2017, 6, 1700918. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, Z.; Jo, H.; Nerem, R.M. Differences in valvular and vascular cell responses to strain in osteogenic media. Biomaterials 2011, 32, 2885–2893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somers, P.; De, S.F.; Cornelissen, M.; Thierens, H.; Van, N.G. Decellularization of heart valve matrices: Search for the ideal balance. Artif. Cells Blood Substit. Immobil. Biotechnol. 2012, 40, 151–162. [Google Scholar] [CrossRef]
- Simon, P.; Kasimir, M.T.; Seebacher, G. Early failure of the tissue engineered porcine heart valve SYNERGRAFT in pediatric patients. Eur. J. Cardiothorac. Surg. 2003, 23, 1002–1006, discussion 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.H.; Simmons, C.A. Cell-matrix interactions in the pathobiology of calcific aortic valve disease: Critical roles for matricellular, matricrine, and matrix mechanics cues. Circ. Res. 2011, 108, 1510–1524. [Google Scholar] [CrossRef]
- Sevostyanova, V.V.; Antonova, L.V.; Velikanova, E.A. Endothelialization of Polycaprolactone Vascular Graft under the Action of Locally Applied Vascular Endothelial Growth Factor. Bull. Exp. Biol. Med. 2018, 165, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Hu, S.; Ding, J.; Xu, J.; Shi, J.; Dong, N. Tissue engineering of heart valves: PEGylation of decellularized porcine aortic valve as a scaffold for in vitro recellularization. Biomed. Eng. Online 2013, 12, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Ding, J.; Zhu, Z. Surface biofunctionalization of the decellularized porcine aortic valve with VEGF-loaded nanoparticles for accelerating endothelialization. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 97, 632–643. [Google Scholar] [CrossRef]
- Ye, X.; Wang, H.; Zhou, J. The effect of Heparin-VEGF multilayer on the biocompatibility of decellularized aortic valve with platelet and endothelial progenitor cells. PLoS ONE 2013, 8, e54622. [Google Scholar] [CrossRef]
- Paris, J.L.; Vallet-Regí, M. Mesoporous Silica Nanoparticles for Co-Delivery of Drugs and Nucleic Acids in Oncology: A Review. Pharmaceutics 2020, 12, 526. [Google Scholar] [CrossRef]
- Castillo, R.R.; Vallet-Regí, M. Functional Mesoporous Silica Nanocomposites: Biomedical applications and Biosafety. Int. J. Mol. Sci. 2019, 20, 929. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, G.; Sagarzazu, A.; Cordova, A. Comparative study of two silica mesoporous materials (SBA-16 and SBA-15) modified with a hydroxyapatite layer for clindamycin controlled delivery. Microporous Mesoporous Mater. 2018, 256, 251–265. [Google Scholar] [CrossRef]
- Sandra, M.; Guillermo, A.; Laura, G.; María, V.; Blanca, G.; Luque-Garcia, J.L. Cancer cell targeting and therapeutic delivery of silver nanoparticles by mesoporous silica nanocarriers: Insights into the action mechanisms using quantitative proteomics. Nanoscale 2019, 11, 4531–4545. [Google Scholar]
- Ricci, V.; Zonari, D.; Cannito, S. Hyaluronated mesoporous silica nanoparticles for active targeting: Influence of conjugation method and hyaluronic acid molecular weight on the nanovector properties. J. Colloid. Interface Sci. 2018, 516, 484–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, F.; Ahmed, K.; Airoldi, C. Amine bridges grafted mesoporous silica, as a prolonged/controlled drug release system for the enhanced therapeutic effect of short life drugs. Mater. Sci. Eng. C 2017, 72, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Carmona, M.; Lozano, D.; Colilla, M.; Vallet-Regí, M. Lectin-conjugated pH-responsive mesoporous silica nanoparticles for targeted bone cancer treatment. Acta Biomater. 2018, 65, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yao, W.; Sun, W. A self-targeting and controllable drug delivery system by fabricating with multi-stimuli responsive chitosan-based thin film layer on mesoporous silica nanoparticles. Int. J. Biol. Macromol. 2018, 122. [Google Scholar]
- Wu, M.; Meng, Q.; Chen, Y. Large-pore ultrasmall mesoporous organosilica nanoparticles: Micelle/precursor co-templating assembly and nuclear-targeted gene delivery. Adv. Mater. 2015, 27, 215–222. [Google Scholar] [CrossRef]
- Varache, M.; Bezverkhyy, I.; Weber, G. Loading of Cisplatin into Mesoporous Silica Nanoparticles: Effect of Surface Functionalization. Langmuir 2019, 35, 8984–8995. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhou, J.; Ding, J.; Xu, J.; Zhong, H.; Lei, S. A novel approach to prepare a tissue engineering decellularized valve scaffold with poly(ethylene glycol)-poly(epsilon-caprolactone). RSC Adv. 2016, 6, 14427–14438. [Google Scholar] [CrossRef]
- Masjedi, S.; Amarnath, A.; Baily, K.M.; Ferdous, Z. Comparison of calcification potential of valvular interstitial cells isolated from individual aortic valve cusps. Cardiovasc. Pathol. 2016, 25, 185–194. [Google Scholar] [CrossRef]
- Jang, W.G.; Kim, E.J.; Kim, D.K. BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J. Biol. Chem. 2012, 287, 905–915. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.; Deshpande, P.P.; Navarro, G.; Dodwadkar, N.S.; Torchilin, V.P. Lipid modified triblock PAMAM-based nanocarriers for siRNA drug co-delivery. Biomaterials 2013, 34, 1289–1301. [Google Scholar] [CrossRef] [Green Version]
- Tsunoda, S.; Mazda, O.; Oda, Y. Sonoporation using microbubble BR14 promotes pDNA/siRNA transduction to murine heart. Biochem. Biophys. Res. Commun. 2005, 336, 118–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishida, T.; Asada, H.; Gojo, S. Sequence-specific gene silencing in murine muscle induced by electroporation-mediated transfer of short interfering RNA. J. Gene Med. 2004, 6, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, D.V.; Blanchard, K.; Shaw, L. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 2005, 41, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Kim, S.W. Efficient siRNA Delivery with Non-viral Polymeric Vehicles. Pharm. Res. 2009, 26, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.C.; Mozumdar, S.; Huang, L. Lipid-based systemic delivery of siRNA. Adv. Drug Deliv. Rev. 2009, 61, 721–731. [Google Scholar] [CrossRef] [Green Version]
- Simeoni, F.; Morris, M.C.; Heitz, F.; Divita, G. Peptide-based strategy for siRNA delivery into mammalian cells. Methods Mol. Biol. 2005, 309, 251–260. [Google Scholar]
- Kim, B.; Park, J.H.; Sailor, M.J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019, 31, e1903637. [Google Scholar] [CrossRef] [Green Version]
- Meng, H.; Mai, W.X.; Zhang, H. Codelivery of an optimal drug/siRNA combination using mesoporous silica nanoparticles to overcome drug resistance in breast cancer in vitro and in vivo. ACS Nano 2013, 7, 994–1005. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, X.; Liu, T. Highly effective antiangiogenesis via magnetic mesoporous silica-based siRNA vehicle targeting the VEGF gene for orthotopic ovarian cancer therapy. Int. J. Nanomed. 2015, 10, 2579–2594. [Google Scholar]
- Li, X.; Chen, Y.; Wang, M.; Ma, Y.; Xia, W.; Gu, H. A mesoporous silica nanoparticle—PEI—Fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials 2013, 34, 1391–1401. [Google Scholar] [CrossRef]
- Morry, J.; Ngamcherdtrakul, W.; Gu, S. Targeted Treatment of Metastatic Breast Cancer by PLK1 siRNA Delivered by an Antioxidant Nanoparticle Platform. Mol. Cancer Ther. 2017, 16, 763–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngamcherdtrakul, W.; Morry, J.; Gu, S. Cationic Polymer Modified Mesoporous Silica Nanoparticles for Targeted SiRNA Delivery to HER2+ Breast Cancer. Adv. Funct. Mater. 2015, 25, 2646–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aigner, A. Delivery systems for the direct application of siRNAs to induce RNA interference (RNAi) in vivo. J. Biomed. Biotechnol. 2006, 2006, 71659. [Google Scholar] [CrossRef] [Green Version]
- Ge, Q.; Filip, L.; Bai, A.; Nguyen, T.; Eisen, H.N.; Chen, J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA 2004, 101, 8676–8681. [Google Scholar] [CrossRef]
- Urban-Klein, B.; Werth, S.; Abuharbeid, S.; Czubayko, F.; Aigner, A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005, 12, 461–466. [Google Scholar] [CrossRef] [Green Version]
- Knetsch, M.L.; Koole, L.H. VEGF-E enhances endothelialization and inhibits thrombus formation on polymeric surfaces. J. Biomed. Mater. Res. A 2010, 93, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Poh, C.K.; Shi, Z.; Lim, T.Y.; Neoh, K.G.; Wang, W. The effect of VEGF functionalization of titanium on endothelial cells in vitro. Biomaterials 2010, 31, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J. Angiogenesis-dependent diseases. Semin. Oncol. 2001, 28, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.R.; Fonseca, F.N.; Paese, K. Poly(ϵ-caprolactone) microcapsules and nanocapsules in drug delivery. Expert Opin. Drug Deliv. 2013, 10, 623–638. [Google Scholar] [CrossRef]
- Mendelson, K.; Schoen, F.J. Heart valve tissue engineering: Concepts, approaches, progress, and challenges. Ann. Biomed. Eng. 2006, 34, 1799–1819. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, D.; Stock, U.A.; Hoerstrup, S.P. Tissue engineering of heart valves using decellularized xenogeneic or polymeric starter matrices. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1505–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutsch, M.; Meinhart, J.; Zilla, P. Long-term experience in autologous in vitro endothelialization of infrainguinal ePTFE grafts. J. Vasc. Surg. 2009, 49, 352–362, discussion 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichhorn, S.; Baier, D.; Horst, D. Pressure shift freezing as potential alternative for generation of decellularized scaffolds. Int. J. Biomater. 2013, 2013, 693793. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Zhu, X.; Liu, J.; Zhou, J. Biofunctionalized Decellularized Tissue-Engineered Heart Valve with Mesoporous Silica Nanoparticles for Controlled Release of VEGF and RunX2-siRNA against Calcification. Bioengineering 2023, 10, 859. https://doi.org/10.3390/bioengineering10070859
Yu W, Zhu X, Liu J, Zhou J. Biofunctionalized Decellularized Tissue-Engineered Heart Valve with Mesoporous Silica Nanoparticles for Controlled Release of VEGF and RunX2-siRNA against Calcification. Bioengineering. 2023; 10(7):859. https://doi.org/10.3390/bioengineering10070859
Chicago/Turabian StyleYu, Wenpeng, Xiaowei Zhu, Jichun Liu, and Jianliang Zhou. 2023. "Biofunctionalized Decellularized Tissue-Engineered Heart Valve with Mesoporous Silica Nanoparticles for Controlled Release of VEGF and RunX2-siRNA against Calcification" Bioengineering 10, no. 7: 859. https://doi.org/10.3390/bioengineering10070859