Uncertainty-Aware Convolutional Neural Network for Identifying Bilateral Opacities on Chest X-rays: A Tool to Aid Diagnosis of Acute Respiratory Distress Syndrome
Abstract
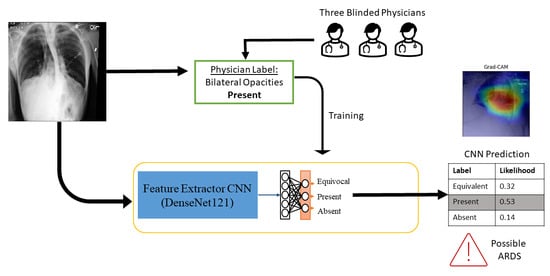
:1. Introduction
2. Materials and Methods
2.1. Dataset Generation
2.2. Training the Convolutional Neural Network
2.2.1. Image Preprocessing
2.2.2. Network Architecture and Training Parameters
2.2.3. Cross-Entropy Loss with Uncertainty Aware Probability Scores
2.3. Performance Metrics
2.4. Model Calibration
2.5. Model Interpretability
2.6. External Dataset Validation
2.7. Comparison with Labels Derived from Radiology Notes
3. Results
3.1. Data Characterization
3.2. CNN Performance Comparison
3.3. Model Calibration
3.4. External Validation on MIMIC-CXR
3.5. Visualization of Saliency Maps
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARDS | Acute Respiratory Distress Syndrome |
| CNN | Convolutional Neural Network |
| ML | Machine Learning |
| EMR | Electronic Medical Record |
| CXR | Chest X-rays |
References
- Ware, L.B.; Matthay, M.A. The acute respiratory distress syndrome. N. Engl. J. Med. 2000, 342, 1334–1349. [Google Scholar] [CrossRef] [PubMed]
- The ARDS Definition Task Force. Acute Respiratory Distress Syndrome. JAMA 2012, 307, 2526–2533. [Google Scholar] [CrossRef]
- Bellani, G.; Laffey, J.G.; Pham, T.; Fan, E.; Brochard, L.; Esteban, A.; Gattinoni, L.; van Haren, F.; Larsson, A.; McAuley, D.F.; et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. JAMA 2016, 315, 788–800. [Google Scholar] [CrossRef]
- Kerchberger, V.E.; Brown, R.M.; Semler, M.W.; Zhao, Z.; Koyama, T.; Janz, D.R.; Bastarache, J.A.; Ware, L.B. Impact of Clinician Recognition of Acute Respiratory Distress Syndrome on Evidenced-Based Interventions in the Medical ICU. Crit. Care Explor. 2021, 3, e0457. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Raheem, B.; Kamaleswaran, R. OnAI-Comp: An Online AI Experts Competing Framework for Early Sepsis Detection. IEEE/ACM Trans. Comput. Biol. Bioinf. 2021, 19, 3595–3603. [Google Scholar] [CrossRef] [PubMed]
- van Wyk, F.; Khojandi, A.; Mohammed, A.; Begoli, E.; Davis, R.L.; Kamaleswaran, R. A minimal set of physiomarkers in continuous high frequency data streams predict adult sepsis onset earlier. Int. J. Med. Inf. 2019, 122, 55–62. [Google Scholar] [CrossRef]
- Futoma, J.; Simons, M.; Doshi-Velez, F.; Kamaleswaran, R. Generalization in Clinical Prediction Models: The Blessing and Curse of Measurement Indicator Variables. Crit. Care Explor. 2021, 3, e0453. [Google Scholar] [CrossRef]
- Liu, Z.; Khojandi, A.; Mohammed, A.; Li, X.; Chinthala, L.K.; Davis, R.L.; Kamaleswaran, R. HeMA: A hierarchically enriched machine learning approach for managing false alarms in real time: A sepsis prediction case study. Comput. Biol. Med. 2021, 131, 104255. [Google Scholar] [CrossRef]
- Farzaneh, N.; Ansari, S.; Lee, E.; Ward, K.R.; Sjoding, M.W. Collaborative strategies for deploying artificial intelligence to complement physician diagnoses of acute respiratory distress syndrome. npj Digit. Med. 2023, 6, 62. [Google Scholar] [CrossRef]
- Reamaroon, N.; Sjoding, M.W.; Gryak, J.; Athey, B.D.; Najarian, K.; Derksen, H. Automated detection of acute respiratory distress syndrome from chest X-rays using Directionality Measure and deep learning features. Comput. Biol. Med. 2021, 134, 104463. [Google Scholar] [CrossRef]
- Singhal, L.; Garg, Y.; Yang, P.; Tabaie, A.; Wong, A.I.; Mohammed, A.; Chinthala, L.; Kadaria, D.; Sodhi, A.; Holder, A.L.; et al. eARDS: A multi-center validation of an interpretable machine learning algorithm of early onset Acute Respiratory Distress Syndrome (ARDS) among critically ill adults with COVID-19. PLoS ONE 2021, 16, e0257056. [Google Scholar] [CrossRef] [PubMed]
- Sjoding, M.W.; Taylor, D.; Motyka, J.; Lee, E.; Co, I.; Claar, D.; McSparron, J.I.; Ansari, S.; Kerlin, M.P.; Reilly, J.P.; et al. Deep learning to detect acute respiratory distress syndrome on chest radiographs: A retrospective study with external validation. Lancet Digit. Health 2021, 3, e340–e348. [Google Scholar] [CrossRef] [PubMed]
- Sjoding, M.W.; Hofer, T.P.; Co, I.; Courey, A.; Cooke, C.R.; Iwashyna, T.J. Interobserver Reliability of the Berlin ARDS Definition and Strategies to Improve the Reliability of ARDS Diagnosis. Chest 2018, 153, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Bellani, G.; Pham, T.; Laffey, J.G. Missed or delayed diagnosis of ARDS: A common and serious problem. Intensive Care Med. 2020, 46, 1180–1183. [Google Scholar] [CrossRef] [Green Version]
- Brady, A.P. Error and discrepancy in radiology: Inevitable or avoidable? Insights Imaging 2017, 8, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Busardò, F.P.; Frati, P.; Santurro, A.; Zaami, S.; Fineschi, V. Errors and malpractice lawsuits in radiology: What the radiologist needs to know. Radiol. Med. 2015, 120, 779–784. [Google Scholar] [CrossRef]
- Song, X.; Weister, T.J.; Dong, Y.; Kashani, K.B.; Kashyap, R. Derivation and Validation of an Automated Search Strategy to Retrospectively Identify Acute Respiratory Distress Patients Per Berlin Definition. Front. Med. 2021, 8, 614380. [Google Scholar] [CrossRef]
- Honavar, S. Electronic medical records—The good, the bad and the ugly. Indian J. Ophthalmol. 2020, 68, 417–418. [Google Scholar] [CrossRef]
- Maley, J.H.; Thompson, B.T. Embracing the Heterogeneity of ARDS. Chest 2019, 155, 453–455. [Google Scholar] [CrossRef] [Green Version]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.-y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holste, G.; Wang, S.; Jiang, Z.; Shen, T.C.; Shih, G.; Summers, R.M.; Peng, Y.; Wang, Z. Long-Tailed Classification of Thorax Diseases on Chest X-ray: A New Benchmark Study. In Data Augmentation, Labelling, and Imperfections, Proceedings of the Second MICCAI Workshop, DALI 2022, Held in Conjunction with MICCAI 2022, Singapore, 22 September 2022; Springer: Cham, Switzerland, 2022; Volume 13567. [Google Scholar] [CrossRef]
- Vardhan, A.; Makhnevich, A.; Omprakash, P.; Hirschorn, D.; Barish, M.; Cohen, S.L.; Zanos, T.P. A radiographic, deep transfer learning framework, adapted to estimate lung opacities from chest X-rays. Bioelectron. Med. 2023, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Makhnevich, A.; Sinvani, L.; Cohen, S.L.; Feldhamer, K.H.; Zhang, M.; Lesser, M.L.; McGinn, T.G. The Clinical Utility of Chest Radiography for Identifying Pneumonia: Accounting for Diagnostic Uncertainty in Radiology Reports. Am. J. Roentgenol. 2019, 213, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Makhnevich, A.; Sinvani, L.; Feldhamer, K.H.; Zhang, M.; Richardson, S.; McGinn, T.G.; Cohen, S.L. Comparison of Chest Radiograph Impressions for Diagnosing Pneumonia: Accounting for Categories of Language Certainty. J. Am. Coll. Radiol. 2022, 19, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Karimi, D.; Dou, H.; Warfield, S.K.; Gholipour, A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis. Med. Image Anal. 2020, 65, 101759. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Darapaneni, N.; Ranjan, A.; Bright, D.; Trivedi, D.; Kumar, K.; Kumar, V.; Paduri, A.R. Pneumonia Detection in Chest X-rays using Neural Networks. arXiv 2022, arXiv:2204.03618. [Google Scholar]
- Ukwuoma, C.C.; Qin, Z.; Heyat, M.B.B.; Akhtar, F.; Bamisile, O.; Muaad, A.Y.; Addo, D.; Al-antari, M.A. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest X-ray images. J. Adv. Res. 2023, 48, 191–211. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Pang, G.; Liao, Z.; Verjans, J.; Li, W.; Sun, Z.; He, J.; Li, Y.; Shen, C.; et al. Viral Pneumonia Screening on Chest X-ray Images Using Confidence-Aware Anomaly Detection. IEEE Trans. Med. Imaging 2021, 40, 879–890. [Google Scholar] [CrossRef]
- Showkatian, E.; Salehi, M.; Ghaffari, H.; Reiazi, R.; Sadighi, N. Deep learning-based automatic detection of tuberculosis disease in chest X-ray images. Pol. J. Radiol. 2022, 87, 118–124. [Google Scholar] [CrossRef]
- Xu, T.; Yuan, Z. Convolution Neural Network With Coordinate Attention for the Automatic Detection of Pulmonary Tuberculosis Images on Chest X-rays. IEEE Access 2022, 10, 86710–86717. [Google Scholar] [CrossRef]
- Nabulsi, Z.; Sellergren, A.; Jamshy, S.; Lau, C.; Santos, E.; Kiraly, A.P.; Ye, W.; Yang, J.; Pilgrim, R.; Kazemzadeh, S.; et al. Deep learning for distinguishing normal versus abnormal chest radiographs and generalization to two unseen diseases tuberculosis and COVID-19. Sci. Rep. 2021, 11, 15523. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Emadi, N.A.; et al. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef] [PubMed]
- Yamac, M.; Ahishali, M.; Degerli, A.; Kiranyaz, S.; Chowdhury, M.E.; Gabbouj, M. Convolutional Sparse Support Estimator-Based COVID-19 Recognition from X-ray Images. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 1810–1820. [Google Scholar] [CrossRef]
- Liu, F.; Zang, C.; Shi, J.; He, W.; Liang, Y.; Li, L. An Improved COVID-19 Lung X-ray Image Classification Algorithm Based on ConvNeXt Network. Int. J. Image Graph. 2023, 2450036. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Rousan, L.A.; Elobeid, E.; Karrar, M.; Khader, Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020, 20, 245. [Google Scholar] [CrossRef]
- Gour, M.; Jain, S. Automated COVID-19 detection from X-ray and CT images with stacked ensemble convolutional neural network. Biocybern. Biomed. Eng. 2022, 42, 27–41. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Zhong, A.; Li, X.; Wu, D.; Ren, H.; Kim, K.; Kim, Y.; Buch, V.; Neumark, N.; Bizzo, B.; Tak, W.Y.; et al. Deep metric learning-based image retrieval system for chest radiograph and its clinical applications in COVID-19. Med. Image Anal. 2021, 70, 101993. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Sadi, M.S.; Islam, M.M. EMCNet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers. Inform. Med. Unlocked 2021, 22, 100505. [Google Scholar] [CrossRef] [PubMed]
- Nahiduzzaman, M.; Goni, M.O.F.; Hassan, R.; Islam, M.R.; Syfullah, M.K.; Shahriar, S.M.; Anower, M.S.; Ahsan, M.; Haider, J.; Kowalski, M. Parallel CNN-ELM: A multiclass classification of chest X-ray images to identify seventeen lung diseases including COVID-19. Expert Syst. Appl. 2023, 229, 120528. [Google Scholar] [CrossRef] [PubMed]
- Nasser, A.A.; Akhloufi, M.A. A Review of Recent Advances in Deep Learning Models for Chest Disease Detection Using Radiography. Diagnostics 2023, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef]
- Singh, S.; Rawat, S.S.; Gupta, M.; Tripathi, B.K.; Alanzi, F.; Majumdar, A.; Khuwuthyakorn, P.; Thinnukool, O. Deep Attention Network for Pneumonia Detection Using Chest X-ray Images. Comput. Mater. Contin. 2023, 74, 1673–1691. [Google Scholar] [CrossRef]
- Yuan, Z.; Yan, Y.; Sonka, M.; Yang, T. Large-scale Robust Deep AUC Maximization: A New Surrogate Loss and Empirical Studies on Medical Image Classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021. [Google Scholar]
- Zhao, J.; Li, M.; Shi, W.; Miao, Y.; Jiang, Z.; Ji, B. A deep learning method for classification of chest X-ray images. J. Phys. Conf. Ser. 2021, 1848, 012030. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Yao, L.; Poblenz, E.; Dagunts, D.; Covington, B.; Bernard, D.; Lyman, K. Learning to diagnose from scratch by exploiting dependencies among labels. arXiv 2017, arXiv:1710.10501. [Google Scholar]
- Islam, M.Z.; Islam, M.M.; Asraf, A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked 2020, 20, 100412. [Google Scholar] [CrossRef]
- Guan, Q.; Huang, Y. Multi-label chest X-ray image classification via category-wise residual attention learning. Pattern Recognit. Lett. 2020, 130, 259–266. [Google Scholar] [CrossRef]
- Heidari, M.; Mirniaharikandehei, S.; Khuzani, A.Z.; Danala, G.; Qiu, Y.; Zheng, B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020, 144, 104284. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, S.; Wang, X. Enhancement Guidance Network for Classification of Pneumonia in Chest X-rays. In Proceedings of the 2022 4th International Conference on Robotics, Intelligent Control and Artificial Intelligence, Dongguan China, 16–18 December 2022; pp. 940–945. [Google Scholar] [CrossRef]
- Motamed, S.; Rogalla, P.; Khalvati, F. Data augmentation using Generative Adversarial Networks (GANs) for GAN-based detection of Pneumonia and COVID-19 in chest X-ray images. Inform. Med. Unlocked 2021, 27, 100779. [Google Scholar] [CrossRef] [PubMed]
- Gulakala, R.; Markert, B.; Stoffel, M. Rapid diagnosis of Covid-19 infections by a progressively growing GAN and CNN optimisation. Comput. Methods Programs Biomed. 2023, 229, 107262. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P.; Bluethgen, C.; Delbrouck, J.B.; der Sluijs, R.V.; Połacin, M.; Chaves, J.M.Z.; Abraham, T.M.; Purohit, S.; Langlotz, C.P.; Chaudhari, A. RoentGen: Vision-Language Foundation Model for Chest X-ray Generation. arXiv 2022, arXiv:2211.12737. [Google Scholar]
- Pai, K.C.; Chao, W.C.; Huang, Y.L.; Sheu, R.K.; Chen, L.C.; Wang, M.S.; Lin, S.H.; Yu, Y.Y.; Wu, C.L.; Chan, M.C. Artificial intelligence–aided diagnosis model for acute respiratory distress syndrome combining clinical data and chest radiographs. Digit. Health 2022, 8, 1–15. [Google Scholar] [CrossRef]
- Reamaroon, N.; Sjoding, M.W.; Lin, K.; Iwashyna, T.J.; Najarian, K. Accounting for label uncertainty in machine learning for detection of acute respiratory distress syndrome. IEEE J. Biomed. Health Inform. 2019, 23, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Kim, K.; Wu, D.; Ren, H.; Tak, W.Y.; Park, S.Y.; Lee, Y.R.; Kang, M.K.; Park, J.G.; Kim, B.S.; et al. Deep Learning-Based Four-Region Lung Segmentation in Chest Radiography for COVID-19 Diagnosis. Diagnostics 2022, 12, 101. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Müller, R.; Kornblith, S.; Google, G.H.; Toronto, B. When Does Label Smoothing Help? In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M. The diagnostic odds ratio: A single indicator of test performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Computer Vision—ECCV 2014, Proceedings of the 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Eyre, H.; Chapman, A.B.; Peterson, K.S.; Shi, J.; Alba, P.R.; Jones, M.M.; Box, T.L.; DuVall, S.L.; Patterson, O.V. Launching into clinical space with medspaCy: A new clinical text processing toolkit in Python. In Proceedings of the 2021 Annual Symposium, San Diego, CA, USA, 30 October–3 November 2021. [Google Scholar]
- Guérin, C.; Reignier, J.; Richard, J.C.; Beuret, P.; Gacouin, A.; Boulain, T.; Mercier, E.; Badet, M.; Mercat, A.; Baudin, O.; et al. Prone Positioning in Severe Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2013, 368, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- The Acute Respiratory Distress Syndrome Network. Ventilation with Lower Tidal Volumes as Compared with Traditional Tidal Volumes for Acute Lung Injury and the Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2000, 342, 1301–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellani, G.; Laffey, J.G.; Pham, T.; Madotto, F.; Fan, E.; Brochard, L.; Esteban, A.; Gattinoni, L.; Bumbasirevic, V.; Piquilloud, L.; et al. Noninvasive Ventilation of Patients with Acute Respiratory Distress Syndrome. Insights from the LUNG SAFE Study. Am. J. Respir. Crit. Care Med. 2017, 195, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Ganapathy, A.; Adhikari, N.K.; Spiegelman, J.; Scales, D.C. Routine chest X-rays in intensive care units: A systematic review and meta-analysis. Crit. Care 2012, 16, R68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Socha, M.; Prażuch, W.; Suwalska, A.; Foszner, P.; Tobiasz, J.; Jaroszewicz, J.; Gruszczynska, K.; Sliwinska, M.; Nowak, M.; Gizycka, B.; et al. Pathological changes or technical artefacts? The problem of the heterogenous databases in COVID-19 CXR image analysis. Comput. Methods Programs Biomed. 2023, 240, 107684. [Google Scholar] [CrossRef] [PubMed]
- DeGrave, A.J.; Janizek, J.D.; Lee, S.I. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 2021, 3, 610–619. [Google Scholar] [CrossRef]







| Internal Dataset (Emory University) | External Dataset (MIMIC-CXR) | ||
|---|---|---|---|
| Patients | 663 | 952 | |
| Chest X-rays | 7825 | 1639 | |
| Age—median (IQR) | 57 (43–67) | 65 (53–76) | |
| Sex | Male | 315 (48%) | 532 (56%) |
| Female | 348 (52%) | 420 (44%) | |
| Race | Caucasian/White | 304 (46%) | 605 (63%) |
| African American/Black | 251 (38%) | 153 (16%) | |
| Asian | 14 (2%) | 44 (5%) | |
| Other | 94 (14%) | 150 (16%) | |
| CXR Labels | Bilateral Opacities Present | 4227 (54%) | 1009 (61.5%) |
| Bilateral Opacities Absent | 1788 (23%) | 442 (27%) | |
| Equivocal | 1810 (23%) | 188 (11.5%) | |
| Experiment | Loss Function | AUROC | AUPRC | F-Score | Precision | Sensitivity | Specificity | Diagnostic Odds Ratio | Balanced Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| Two-Class Model | Cross-Entropy Loss | 0.819 (0.791, 0.844) | 0.553 (0.521, 0.584) | 0.707 (0.679, 0.735 | 0.678 (0.671, 0.726) | 0.724 (0.694, 0.754) | 0.809 (0.781, 0.836) | 11.141 (8.545, 14.765) | 0.767 (0.736, 0.795) |
| Focal Loss | 0.825 (0.802, 0.852) | 0.568 (0.539, 0.595) | 0.715 (0.684, 0.744) | 0.706 (0.674, 0.735) | 0.730 (0.698, 0.732) | 0.816 (0.787, 0.843) | 11.967 (8.979, 16.144) | 0.773 (0.743, 0.788) | |
| Two-Class ModelDE | Cross-Entropy Loss | 0.884 (0.864, 0.904) | 0.758 (0.736, 0.779) | 0.734 (0.689, 0.743) | 0.777 (0.750, 0.807) | 0.714 (0.689, 0.743) | 0.923 (0.904, 0.941) | 29.917 (22.455, 43.151) | 0.819 (0.796, 0.842) |
| Focal Loss | 0.875 (0.855, 0.896) | 0.756 (0.731, 0.789) | 0.774 (0.746, 0.799) | 0.775 (0.750, 0.803) | 0.772 (0.742, 0.801) | 0.868 (0.843, 0.893) | 22.235 (16.398, 31.375) | 0.820 (0.793, 0.847) | |
| Three-Class Model | Focal Loss | 0.810 (0.785, 0.837) | 0.845 (0.803, 0.8475) | 0.739 (0.712, 0.766) | 0.738 (0.711, 0.766) | 0.740 (0.712, 0.767) | 0.712 (0.666, 0.757) | 7.006 (5.011, 10.191) | 0.726 (0.689, 0.762) |
| Cross-Entropy Loss | 0.790 (0.764, 0.818) | 0.819 (0.0.792, 0.834) | 0.688 (0.659, 0.717) | 0.716 (0.691, 0.744) | 0.707 (0.681, 0.732) | 0.851 (0.819, 0.883) | 13.769 (9.948, 19.926) | 0.776 (0.750, 0.808) | |
| Cross-Entropy Loss with probability targets | 0.828 (0.803, 0.853) | 0.874 (0.861, 0.887) | 0.746 (0.720, 0.775) | 0.755 (0.731, 0.782) | 0.761 (0.735, 0.786) | 0.842 (0.812, 0.875) | 17.211 (12.106, 25.976) | 0.802 (0.773, 0.831) |
| Experiment | Loss Function | MCE |
|---|---|---|
| Two-Class Model | Cross-Entropy Loss | 0.424 |
| Focal Loss | 0.385 | |
| Two-Class Model: Disregarding Equivocal | Cross-Entropy Loss | 0.479 |
| Focal Loss | 0.318 | |
| Three-Class Model | Cross-Entropy Loss | 0.408 |
| Focal Loss | 0.240 | |
| Cross-Entropy Loss with probability targets | 0.150 |
| Experiment | AUROC | AUPRC | F-Score | Precision | Sensitivity | Specificity | Diagnostic Odds Ratio | Balanced Accuracy |
|---|---|---|---|---|---|---|---|---|
| Three-Class Model: Cross-Entropy Loss with Probability Targets | 0.834 (0.811, 0.858) | 0.898 (0.873, 0.917) | 0.658 (0.627, 0.685) | 0.729 (0.704, 0.749) | 0.727 (0.703, 0.747) | 0.955 (0.929, 0.973) | 2.180 (1.801, 1.00) | 0.841 (0.816, 0.860) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, M.; Davis, C.M.; Gowda, N.R.; Foster, D.G.; Mondal, A.; Coopersmith, C.M.; Kamaleswaran, R. Uncertainty-Aware Convolutional Neural Network for Identifying Bilateral Opacities on Chest X-rays: A Tool to Aid Diagnosis of Acute Respiratory Distress Syndrome. Bioengineering 2023, 10, 946. https://doi.org/10.3390/bioengineering10080946
Arora M, Davis CM, Gowda NR, Foster DG, Mondal A, Coopersmith CM, Kamaleswaran R. Uncertainty-Aware Convolutional Neural Network for Identifying Bilateral Opacities on Chest X-rays: A Tool to Aid Diagnosis of Acute Respiratory Distress Syndrome. Bioengineering. 2023; 10(8):946. https://doi.org/10.3390/bioengineering10080946
Chicago/Turabian StyleArora, Mehak, Carolyn M. Davis, Niraj R. Gowda, Dennis G. Foster, Angana Mondal, Craig M. Coopersmith, and Rishikesan Kamaleswaran. 2023. "Uncertainty-Aware Convolutional Neural Network for Identifying Bilateral Opacities on Chest X-rays: A Tool to Aid Diagnosis of Acute Respiratory Distress Syndrome" Bioengineering 10, no. 8: 946. https://doi.org/10.3390/bioengineering10080946