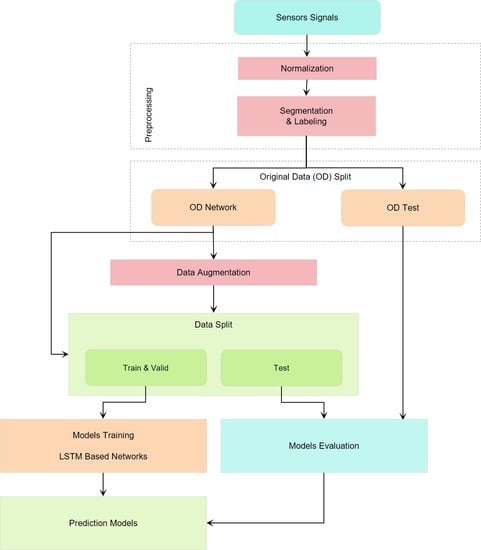
This section describes the methods applied in this study, including those for data collection, data preprocessing, data augmentation, LSTM architecture and optimization, and evaluation of the prediction models.
2.1. Dataset Descriptions
The datasets used in this study were (1) 35-mer adenosine, (2) 31-mer oestradiol, and (3) 35-mer oestradiol. The datasets contained several time-series signals representing the drain current of three different aptasensors.
Table 1 describes three key features of the sensors used for data collection to provide a quick and brief comparison of the datasets. The distinguishing features of these datasets were two main components of their sensors: their target analytes and their bioreceptors. However, their transducers, another main component used for these aptasensors, were carbon nanotube (CNT) field-effect transistors (FETs). Explaining all of details of the functionalization of these sensors is beyond the scope of this paper. However, detailed information on the 35-mer adenosine sensor, including transistor fabrication and aptamer functionalization, can be found in [
24].
As the sensing protocols for the drain current measurements might provide a clear insight into the registered signals, the following explains the method of measuring the signals. The sensing protocols for measuring the 31-mer and 35-mer oestradiol sensors’ responses were similar, but they were different from those of the adenosine sensors.
Table 2 summarizes and compares the sensing protocols of the adenosine and oestradiol datasets.
The sensing responses for adenosine and oestradiol were measured in time intervals of 1 and 1.081 s with a standard deviation of , respectively, with similar gate and drain voltages, i.e., V and mV. The buffer selected for the adenosine sensor was 2 mM Tris-HCI, and that for the oestradiol sensors was 0.05 × PBS (phosphate-buffered saline) with 5% ethanol (EtOH).
Regarding the adenosine sensor, the initial load for each measurement was 110 M of 2 mM Tris-HCI in a polydimethylsiloxane (PDMS) well, which lasted for 1000 s. Then, the adenosine solution was added to the PDMS well every 500 s in successively greater concentrations, considering the adenosine concentration in the PDMS well before each addition. The process of adding the adenosine solution increased the adenosine concentration, which varied from 1 pM to 10 M in the PDMS well.
Regarding the oestradiol sensors, the initial load for each measurement was 100 L of 0.05 × PBS 5% EtOH in the well, which lasted for 300 s. Then, in the next 300 s, 20 L of 0.05 × PBS 5% EtOH was added, while the oestradiol concentration did not increase. Then, the oestradiol solution was added to the well every 300 s in successively greater concentrations, considering the oestradiol concentration in the well before each addition. In addition, each time the oestradiol concentration was increased, a solution of 20 L of 0.05 × PBS 5% EtOH was added to the well. The process of adding the oestradiol solution increased the oestradiol concentration, which varied from 1 nM to 10 M in the well.
2.2. Contextual Outlier Detection
A contextual outlier, also known as a conditional anomaly, is defined as a data instance whose pattern does not conform to that of other well-defined data instances with similar contextual information [
27,
28]. Regarding the experiments related to this study, factors that could cause contextual outliers were background noises in the lab, the use of broken transistors, issues in immobilizing the aptamers on carbon nanotube surfaces, issues in fabricating the sensing interface, and so on.
The patterns of signals affected by these factors deviate from the patterns of well-defined and normal signals. The purpose of removing outliers is to eliminate non-informative signals or segments. As there were a few signals in the datasets, removing the outliers was performed with prior knowledge of the biosensors’ behaviors and through data visualization.
In this paper, the signals were preprocessed with data normalization before being fed into the DL models. It needs to be mentioned that data normalization was applied to the entire signal. Data normalization or feature scaling puts all of the signals in a dataset on the same scale and prevents a feature from dominating and controlling the others. The data normalization applied in this paper was a Z-score scaling that used the mean () and standard deviation () of a signal.
Suppose that
is an entire signal, where the
n is the number of data points within the given signal or the length of the signal. Then, Equation (
1) shows the new signal
created by Z-score scaling.
Segmentation and Labeling
After rescaling the signal, it was split into different segments. Each segment was a part of the signal for which the concentration of analyte remained constant from its beginning to its end. Then, each segment was labeled with its corresponding analyte concentration. This means that the labels for the three datasets were: No Analyte, 1 nM, 10 nM, 100 nM, 1
M, and 10
M. As shown in
Table 3, these six labels and concentrations were considered as the six different classes, in the same order.
2.3. Data Split
The data fed into the DL model needed to be split into three subsets, namely, the training, validation, and test set, with the proportions of 60%, 20%, and 20%, respectively. The training set was used to extract meaningful information and find the optimal parameters, the validation set was used to tune the parameters, and the test set was used to assess the model’s performance [
13].
In this paper, the original data were split into two sets—network and test sets—with proportions of approximately 70% and 30%, respectively. These sets were named the original data (OD) network and OD test set, respectively. The former set was used to make DL models based on original data and for data augmentation. The latter, the OD test set, assessed the DL models and acted as a control group. The reasons for considering this OD test set were to prove the functionality of the data augmentation method in making the prediction models and to avoid biased results. In order to complete the information related to the data split, it must be mentioned that the augmented data and OD networks were randomly shuffled and separated again into the training set (60%), validation set (20%), and network test set (20%).
2.4. Data Augmentation
In machine learning, small amounts of training data might cause overfitting and might not be enough for training models [
29]. The need for data augmentation is more critical for real-world data, since acquiring large enough real-world datasets has not always been possible due to cost or time limitations. Generating synthetic data, which is also known as data augmentation, is a solution for overcoming the problem of insufficient data samples [
30] or compensating for datasets with imbalanced classes [
31]. Data augmentation helps to increase the generalization capability of an ML prediction model and improve the model’s performance by increasing the variability of the training and validation data samples [
32,
33].
In this paper, we utilized a data augmentation method to increase the size of the available datasets. Suppose the
and
are two preprocessed segments from an identical dataset with similar analyte concentrations. Then,
is an augmented segment generated with the following Equation (
2):
where
and is a normally distributed random number generated by the
function in MATLAB R2021b.
2.5. Background of LSTM
This subsection explains long short-term memory (LSTM) and its architecture. Then, the unidirectional and bidirectional LSTM structures, as well as their similarities and differences, are discussed.
The advantage of using an LSTM network over a recurrent neural network (RNN) is that LSTM can capture the temporal dependency of input sequences during the training process [
21,
34]. An LSTM network is an RNN that prevents the long-term dependency problem by utilizing gates and calculating a hidden state with an enhanced function [
17]. The building blocks of LSTM networks are LSTM cells, which means that an LSTM layer consists of recurrently connected cells [
16,
17,
34]. An LSTM cell, or an LSTM hidden unit, consists of four parts: the forget gate, input gate, output gate, and a cell candidate.
Figure 1 presents the structure of a cell. This cell decides to ignore or remember something in its memory by using a gating mechanism. The role of the three gates is to selectively control and transfer needed information into and out of the cell [
34]. This figure can also be considered an LSTM layer consisting of only one memory cell or hidden unit, where
and
, respectively, are the input and output of the LSTM layer.
Consider
as a sequence input into the memory block at time step
t; the forget gate selects which data to erase and which data to remember. As shown in Equation (
3), these decisions are made by the
layer:
The input gate is responsible for controlling the level at which the cell state is updated by using another
layer. As shown in Equation (
4), the
layer of the input gate decides which data need to be updated. In the next step, as shown in Equation (
5), the cell candidate (
) is responsible for adding information to the cell state by using the
layer. Now, the cell state is ready to be updated with the combination of the forget and input gates and new candidate values of
. Equation (
6) describes the mathematical formula for calculating the cell state:
The output gate, which is shown in Equation (
7), utilizes a
layer to decide which part of the cell state contributes to the output. Now, the hidden state or output of the memory cell is ready to be calculated. The output gate and the cell state are contributors to the hidden state. Equation (
8) presents its mathematical formula:
Note that , , , and refer to the input weight matrices for the input gate, forget gate, output gate, and cell value, respectively, and , , , and are the recurrent weights for the gates and the cell value in the same order. Their corresponding bias vectors are , , , and .
Moreover, it can be seen that the cell state and gate activation functions (AFs) are, respectively,
(Equation (
9)) and
(Equation (
10)); these map the nonlinearity and make decisions:
An LSTM layer in a deep neural network consists of a set of LSTM cells. LSTM layers can be categorized into unidirectional LSTM (ULSTM) and bidirectional LSTM (BLSTM) layers.
Figure 2 represents a ULSTM structure. It can be said that a ULSTM structure is an RNN that uses LSTM cells instead. The unfolded figure of the ULSTM shows that the output of each cell is the input for the next cell in the same layer. It should be mentioned that an LSTM block refers to several LSTM cells or hidden units.
Figure 3 depicts a BLSTM structure consisting of forward and backward layers. The unfolded figure shows that the forward layer moves in a positive temporal direction, while the backward movement is in a negative temporal direction. In addition, the outputs from both the forward and backward LSTM cells are joined and concatenated as the layer’s output.
Figure 4 presents the flow of information in an LSTM layer during different time steps. In this figure,
N is the length of the sequential input for the LSTM layer,
L is the number of hidden units in the LSTM layer, and
T is the length of the training set. Note that in this figure,
can be considered as just the forward movement (
) in the ULSTM layer or as a concatenation of both forward (
) and backward (
) movements in the BLSTM layer.
2.6. LSTM Architecture
In this work, two LSTM networks were employed to classify the analyte concentrations. The objective was to classify the input data into six different concentration classes: 0 M, 1 nM, 10 nM, 100 nM, 1 M , and 10 M. The target outputs of each class were labeled in a binary vector format, where the desired class was labeled with “1” and the others were labeled with “0”. Recall that the input data were the corresponding concentration segments of the signals, as well as the original and/or augmented segments.
Figure 5 visualizes the architectures of both networks. The networks comprised five successive layers: a sequential input layer, an LSTM layer, a fully connected layer, a softmax layer, and a classification layer. The only difference between the networks was in their LSTM layers. The LSTM layer in the first network was a unidirectional LSTM layer, while this was a bidirectional LSTM layer in the second network. It should be taken into consideration that the fully connected (FC) layers were affected by the previous LSTM layers and the number of output classes in the classification layer.
Table 4 describes and compares the layers and the properties of the unidirectional and bidirectional LSTM networks depicted in
Figure 5. Recall that the size of the input layer entering the networks was equal to the length of the segments and was considered as one sequence, and the output size of the networks (
m) was identical to the number of classes in the data. It should be mentioned that all of the input weights, the recurrent weight, and the bias matrices were concatenated together to form the input weights (
), recurrent weights (
), and bias (
).
2.8. Evaluation Metrics
After training the prediction models, the classification performance of the neural networks needed to be assessed with relevant metrics. In this paper, the LSTM networks’ performances were assessed using two standard metrics: overall accuracy (ACC) and macro F1-score (MF1) [
21,
36]. In this work, we created a confusion matrix using the predictions from the test data, and then the overall accuracy and the macro F1-score were calculated with the confusion matrix.
The overall accuracy was calculated from the sum of the diagonal numbers of the confusion matrix divided by the total number of elements. In other words, the overall accuracy (Equation (
11)) was the proportion of correctly classified elements among all elements in the test set.
Before mentioning the method for calculating the macro F1-score, its building blocks—recall, precision, and F1-score—need to be defined. The recall, which is the true positive rate (TPR) and is presented in Equation (
12), is the number of correctly classified positive elements among all positive elements. Precision, which is the positive predicted value (PPV) and is presented in Equation (
13), is the number of correctly classified positive elements among the elements classified as positives by the model. Then, the harmonic mean of the recall and precision is called the F1-score (Equation (
14)). The macro F1-score (Equation (
15)) is the mean of the class-wise F1-score of each concentration.
where
m is the number of classes in a given dataset.