Uttarakhand Medicinal Plants Database (UMPDB): A Platform for Exploring Genomic, Chemical, and Traditional Knowledge
Abstract
:1. Introduction
2. Results and Discussion
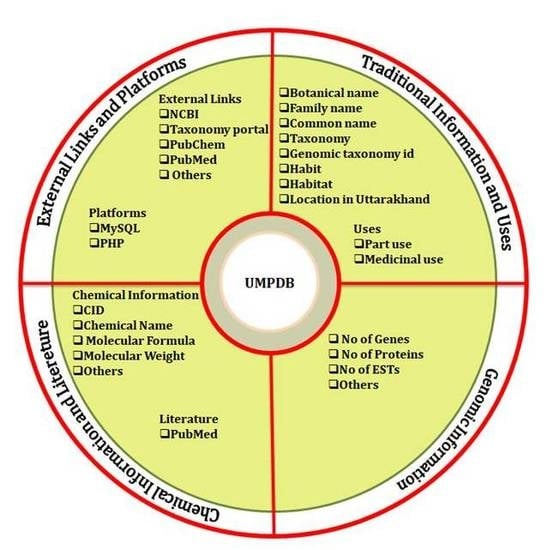
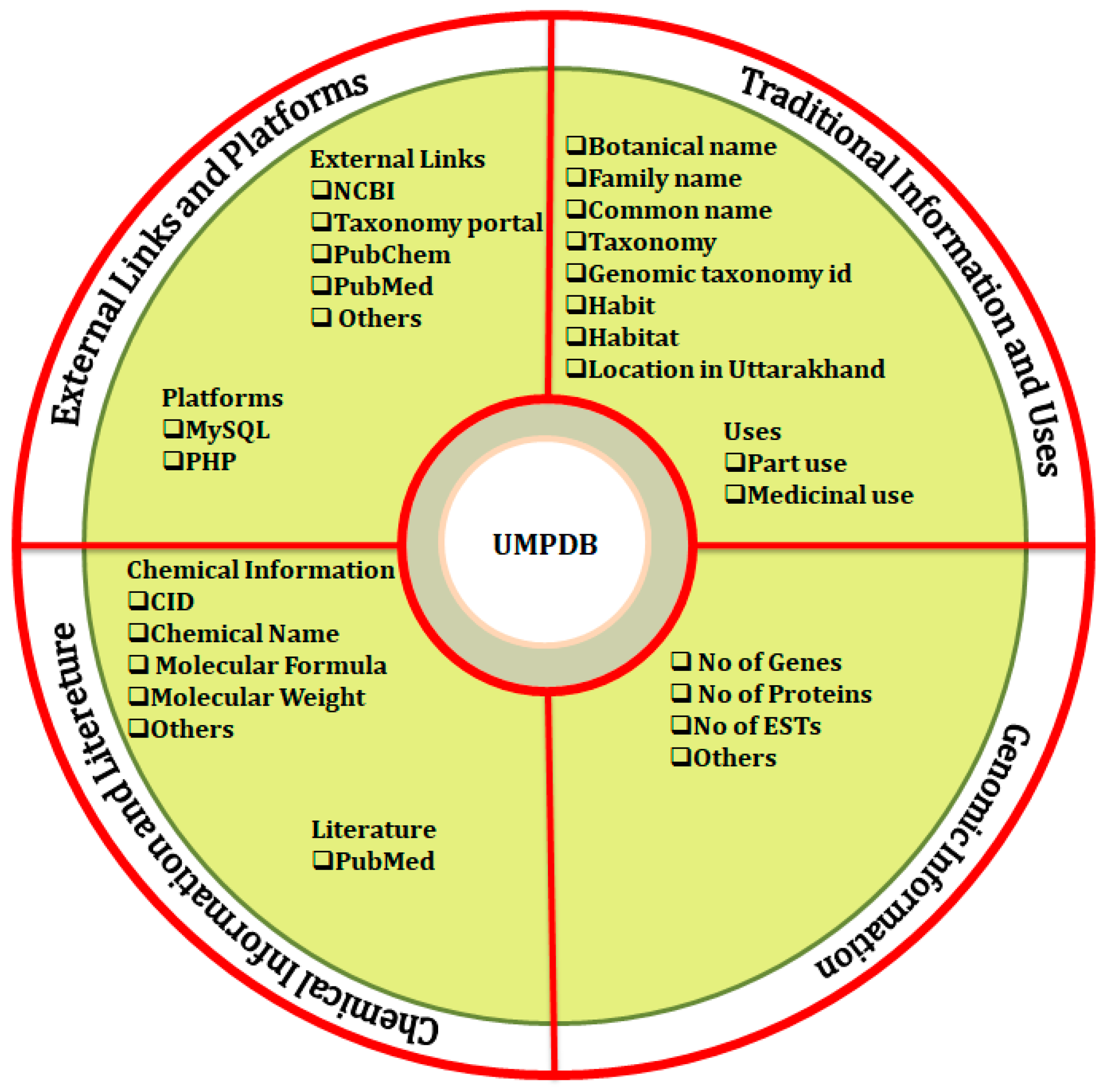
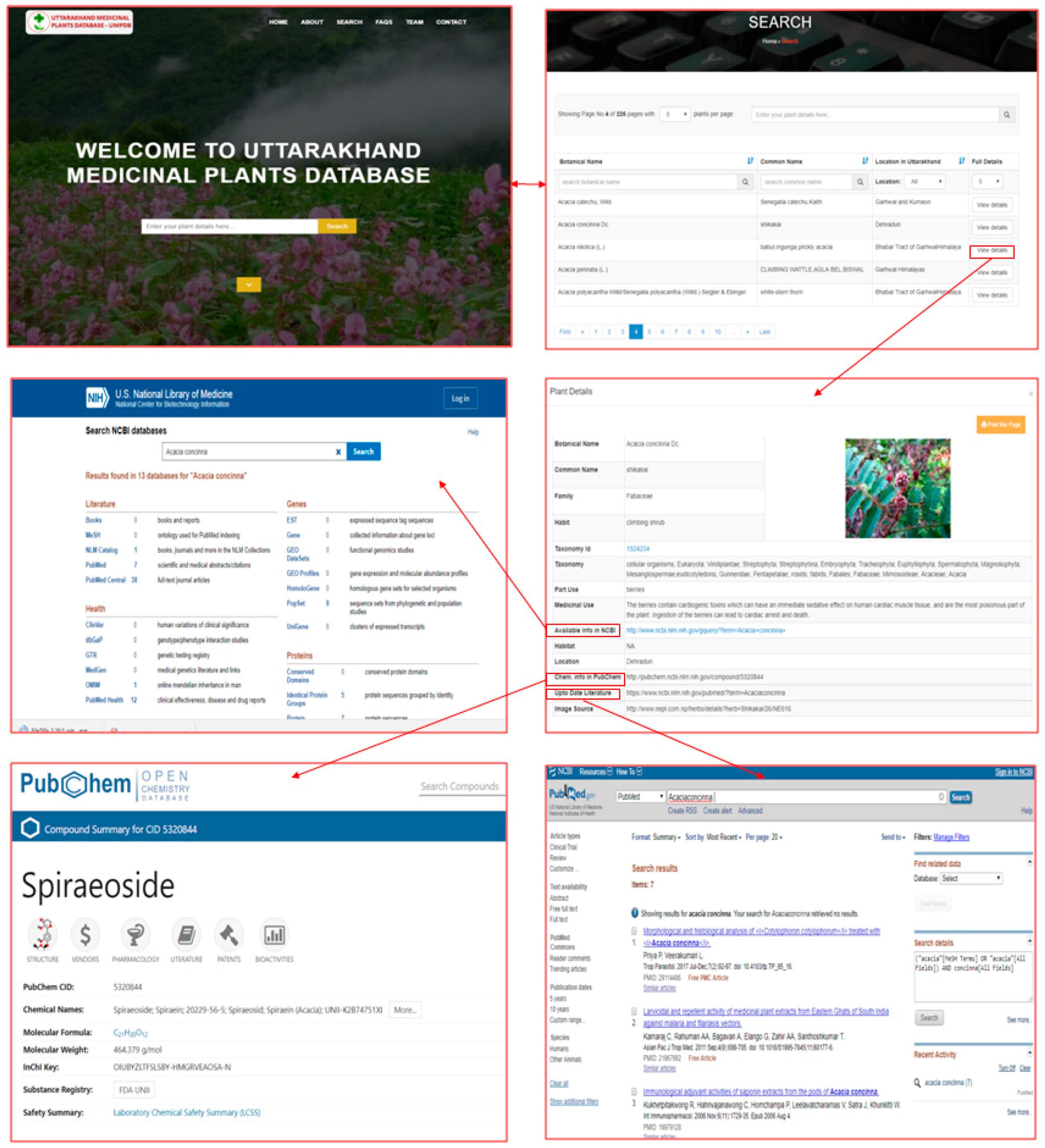
2.1. PlantCentric Portal
2.2. Data Access
2.3. Comparison of UMPDB with ENVIS Medicinal Plants: Uttarakh and Database
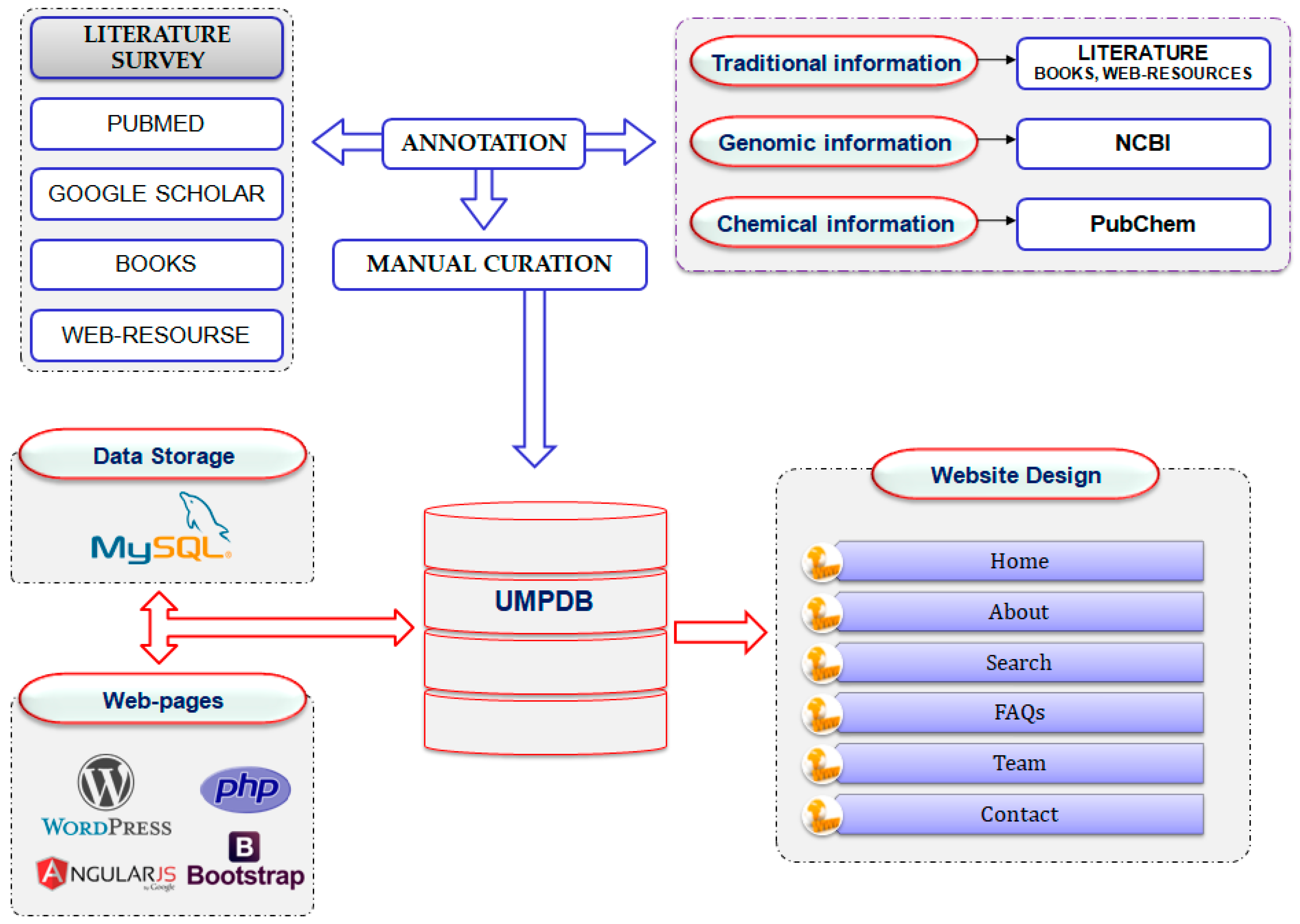
3. Materials and Methods
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mani, M.S. Biogeography of the Himalaya. In Ecology and Biogeography of India; Springer: The Hague, The Netherlands, 1978; pp. 664–681. [Google Scholar]
- Samant, S.S.; Dhar, U.; Palni, L.M.S. Medicinal Plants of Himalaya, Diversity, Distribution and Potential Values; Gyanodaya Prakashan: Nainital, India, 1998. [Google Scholar]
- Singh, K.P.; Kumar, A.; Kumar, U. Medicinal Plants of Uttarakhand; Astral International: New Delhi, India, 2017; Volume 1–3. [Google Scholar]
- Singh, J.S.; Singh, S.P. Forest vegetation of the Himalaya. Bot. Rev. 1987, 53, 80–192. [Google Scholar] [CrossRef]
- Gangwar, R.S.; Joshi, B.D. Some medicinal flora in the riparian zone of river Ganga at Saptrishi, Haridwar, Uttaranchal. Himal. J. Environ. Zool. 2006, 20, 237–241. [Google Scholar]
- Nautiyal, S.; Rajan, K.S.; Shibasaki, R. Environmental conservation vs compensation: Explorations from the Uttaranchal Himalaya. Environ. Inform. Arch. 2004, 2, 24–35. [Google Scholar]
- Sharma, J.; Gaur, R.D.; Paiuli, R.M. Conservation status and diversity of some important plant in the Shiwalik Himalaya of Uttarakhand, India. Int. J. Med. Aromat. Plants 2011, 1, 75–82. [Google Scholar]
- Gangwar, K.K.; Deepali, G.R.; Gangwar, R.S. Ethnomedicinal plant diversity in Kmaun Himalaya of uttarakhand, India. Nat. Sci. 2010, 8, 66–78. [Google Scholar]
- Mukhergee, T.K. Protection of Indian traditional knowledge: In Ethno Medicinal Plants; Trivedi, P.C., Sharma, N.K., Eds.; Poiner Publishers: Jaipur, India, 2004; pp. 18–33. [Google Scholar]
- Naini, V.; Mamidala, E. An ethnobotanical study of plants used for the treatment of diabetes in the Warangal district, Andhra Pradesh, India. Biolife 2013, 1, 24–28. [Google Scholar]
- Panchangam, S.S.; Vahedi, M.; Megha, M.J.; Kumar, A.; Raithatha, K.; Suravajhala, P.; Reddy, P. Saffron’omics’: The challenges of integrating omic technologies. Avicenna J. Phytomed. 2016, 6, 604–620. [Google Scholar] [PubMed]
- Jee, B.; Kumar, S.; Yadav, R.; Singh, Y.; Kumar, A.; Sharma, N. Ursolic acid and carvacrol may be potential inhibitors of dormancy protein small heat shock protein16.3 of Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, J.; Meinel, T.; Dunkel, M.; Murqueitio, M.S.; Adams, R.; Blasse, C.; Eckert, A.; Preissner, S.; Preissner, R. CancerResource: A comprehensive database of cancer-relevant proteins and compound interactions supported by experimental knowledge. Nucleic Acids Res. 2011, 39, D960–D967. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, C.; Gupta, A.; Kanjilal, S.; Katiyar, S. Drug discovery from plant sources: An integrated approach. Ayu 2012, 33, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Modak, M.; Dixit, P.; Londhe, J.; Ghaskadbi, S.; Devasagayam, T.P.A. Indian herbs and herbal drugs used for the treatment of diabetes. J. Clin. Boichem. Nutr. 2007, 40, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochem. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar]
- Sen, S.; Chakraborty, R. Revival, modernization and integration of India traditional herbal medicine in clinical practice: Importance, challenges and future. J. Tradit. Complement. Med. 2017, 7, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.G.; Wu, J.Y. Development and application of medicinal plant tissue cultures for production of drugs and herbal medicinals in China. Nat. Prod. Rep. 2006, 23, 789–810. [Google Scholar] [CrossRef] [PubMed]
- Thatoi, H.; Patra, J.K. Biotechnology and pharmacological evaluation of medicinal plants: An overview. J. Herbs Species Med. Plants 2011, 17, 214–218. [Google Scholar] [CrossRef]
- Shakya, A.K. Medicinal plants: Future source of new drugs. Int. J. Herb. Med. 2016, 4, 59–64. [Google Scholar]
- Mumtaz, A.; Ashfaq, U.A.; ul Qamar, M.T.; Anwar, F.; Gulzar, F.; Ali, M.A.; Saari, N.; Pervez, M.T. MPD3: A useful medicinal plants database for drug designing. Nat. Prod. Res. 2017, 31, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Ramakrishnan, S.M.; Bagler, G. Phytochemica: A platform to explore phytochemicals of medicinal plants. Database 2015. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Ramakrishnan, S.M.; Randhawa, V.; Bagler, G. SerpentinaDB: A database of plant-derived molecules of Rauvolfia serpentina. BMC Complement. Altern. Med. 2015, 15, 262. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.B.; Ghorpade, P.N.; Kale, M.V.; Sonawane, K.D. FERN ethnomedicinal plant database: Exploring fern ethnomedicinal plants knowledge for computational drug discovery. Curr. Comput. Adied Drug Res. 2015, 11, 266–271. [Google Scholar] [CrossRef]
- Mangal, M.; Sagar, P.; Singh, H.; Raghava, G.P.; Agarwal, S.M. NPACT: Naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Res. 2013, 41, D1124–D1129. [Google Scholar] [CrossRef] [PubMed]
- Dunkel, M.; Fullbeck, M.; Neumann, S.; Preissner, R. SuperNatural: A searchable database of available natural compounds. Nucleic Acids Res. 2006, 34, D678–D683. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Ye, L.; Kang, H.; Zhang, D.; Tao, L.; Tang, K.; Liu, X.; Zhu, R.; Liu, Q.; Chen, Y.Z.; et al. HIT: Linking herbal active ingredients to targets. Nucleic Acids Res. 2011, 39, D1055–D1059. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhou, J.; Xu, Z. Concept design of computer-aided study on traditional Chinese drugs. J. Chem. Inf. Comput. Sci. 1999, 39, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xu, X.; Cheng, F.; Liu, H.; Luo, X.; Shen, J.; Chen, K.; Zhao, W.; Shen, X.; Jiang, H. Virtual screening on natural products for discovering active compounds and target information. Curr. Med. Chem. 2003, 10, 2327–2342. [Google Scholar] [CrossRef] [PubMed]
- Ehrman, T.M.; Barlow, D.J.; Hylands, P.J. Phytochemical databases of Chinese herbal constituents and bioactive plant compounds with known target specificities. J. Chem. Inf. Model. 2007, 47, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Valli, M.; dos Santos, R.N.; Figueira, L.D.; Nakajima, C.H.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. Development of a natural products database from the biodiversity of Brazil. J. Nat. Prod. 2013, 76, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Valli, M.; Dametto, A.C.; Pinto, M.E.F.; Freire, R.T.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. NuBBEDB: An updated database to uncover chemical and biological information from Brazilian biodiversity. Sci. Rep. 2017, 7, 7215. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Telukunta, K.K.; Doring, K.; Simoben, C.V.; A Moumbock, A.F.; Malange, Y.I.; Njuma, L.E.; Young, J.N.; Sippl, W.; Gunther, S. NANPDB: A resource for natural products from northern African sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Zofou, D.; Babiaka, S.B.; Meudom, R.; Scharfe, M.; Lifongo, L.L.; Mbah, J.A.; Mbaze, L.M.; Sippl, W.; Efange, S.M. AfroDb: A select highly potent and diverse natural product library from African medicinal plants. PLoS ONE 2013, 8, e78085. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Nwodo, J.N.; Ibezim, M.; Simoben, C.V.; Karman, B.; Ngwa, V.F.; Sippl, W.; Adikwu, M.U.; Mbaze, L.M. Molecular modeling of potential anticancer agents from African medicinal plants. J. Chem. Inf. Model. 2014, 54, 2433–2450. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.M.; Sharma, M.; Fatima, S. VOCC: A database of volatile organic compounds in cancer. RSC Adv. 2016, 6, 114783–114789. [Google Scholar] [CrossRef]
- Kumar, A.; Pandeya, A.; Malik, G.; Kumari, P.H.; Kumar, S.A.; Sharma, M.; Gahlaut, V.; Gajula, M.N.V.P.; Suravajhala, P.; Singh, K.P.; et al. A web-resource for nutrient use efficiency related genes, QTLSs, and microRNA in important cereals and model plants. bioRxiv 2017. [Google Scholar] [CrossRef]
- Sood, S.K.; Kumari, P.; Thakur, R.; Bassi, S.K.; Thakur, A. Herbal Medicine; Pointer Publishers: Jaipur, India, 2015. [Google Scholar]
- Shah, R. Edible Plants of North West Himalaya (Uttarakhand); Bishen Singh Mahinder Pal Singh: Dehradun, India, 2015. [Google Scholar]
- Chaduvula, P.K.; Bonthala, V.S.; Manjusha, V.; Siddiq, E.A.; Polumetla, A.K.; Prasad, G.M.N.V. CmMDb: A versatile database for Cucumis melo microsatellite markers and other horticulture crop research. PLoS ONE 2015, 10, e0118630. [Google Scholar]
- Bhawna, V.S.; Gajula, M.N.V. PvTFDB: A Phaseolus vulgaris transcription factors database for expediting functional genomics in legumes. Database 2016. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, A.; Kumar, R.; Sharma, M.; Kumar, U.; Gajula, M.N.V.P.; Singh, K.P. Uttarakhand Medicinal Plants Database (UMPDB): A Platform for Exploring Genomic, Chemical, and Traditional Knowledge. Data 2018, 3, 7. https://doi.org/10.3390/data3010007
Kumar A, Kumar R, Sharma M, Kumar U, Gajula MNVP, Singh KP. Uttarakhand Medicinal Plants Database (UMPDB): A Platform for Exploring Genomic, Chemical, and Traditional Knowledge. Data. 2018; 3(1):7. https://doi.org/10.3390/data3010007
Chicago/Turabian StyleKumar, Anuj, Rohit Kumar, Mansi Sharma, Upendra Kumar, M. N. V. Prasad Gajula, and Krishna Pal Singh. 2018. "Uttarakhand Medicinal Plants Database (UMPDB): A Platform for Exploring Genomic, Chemical, and Traditional Knowledge" Data 3, no. 1: 7. https://doi.org/10.3390/data3010007