Isolation, Characterization and Whole Genome Analysis of an Avian Pathogenic Escherichia coli Phage vB_EcoS_GN06
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
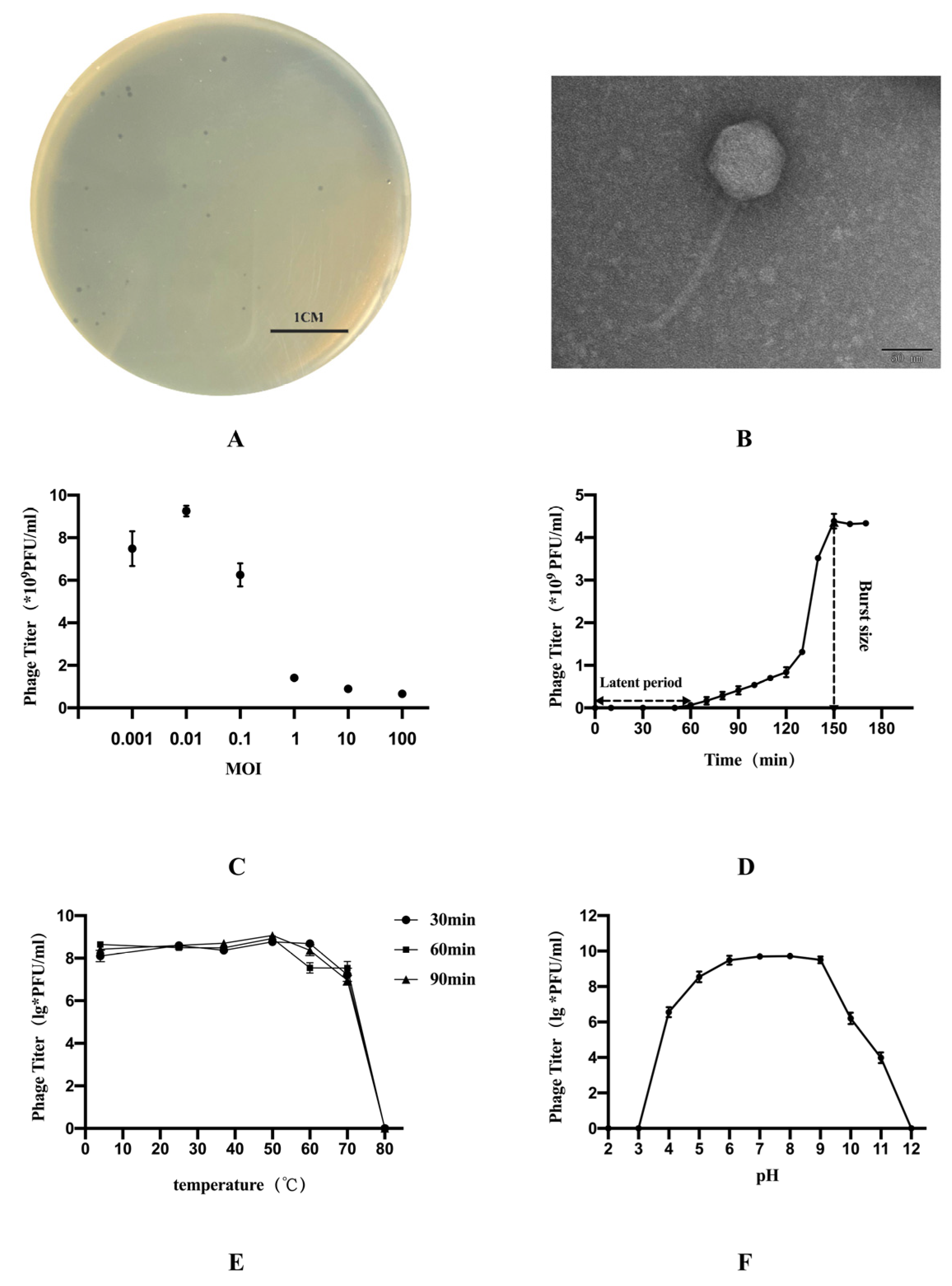
2.1. Isolation and Purification of Phage and Electron Microscopic Observation
2.2. Determination of Phage Lysis Profile
2.3. Optimal Multiplicity of Infection and One-Step Growth Curve
2.4. Thermolability and pH Sensitivity
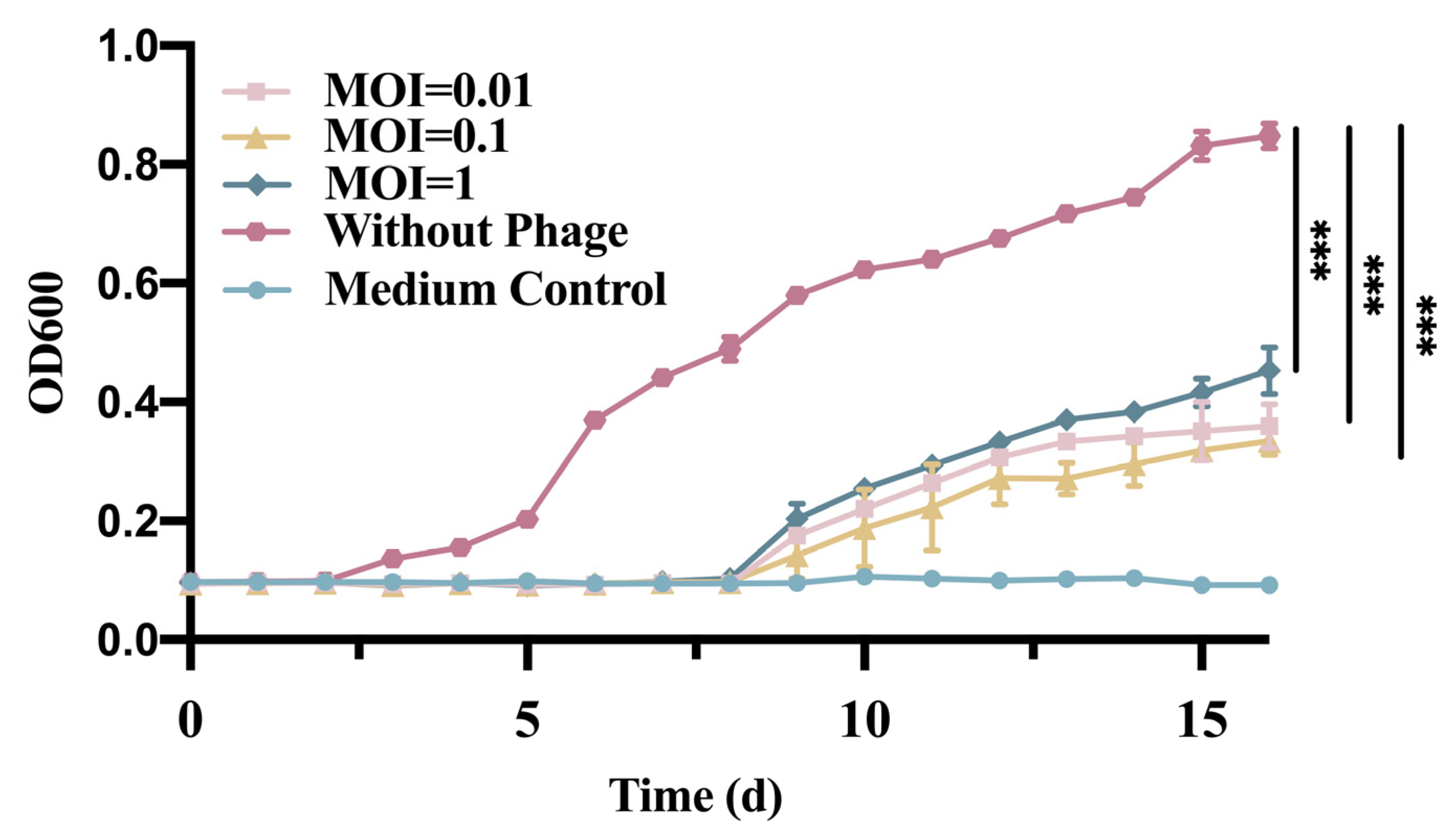
2.5. In Vitro Inhibition Test of Phage
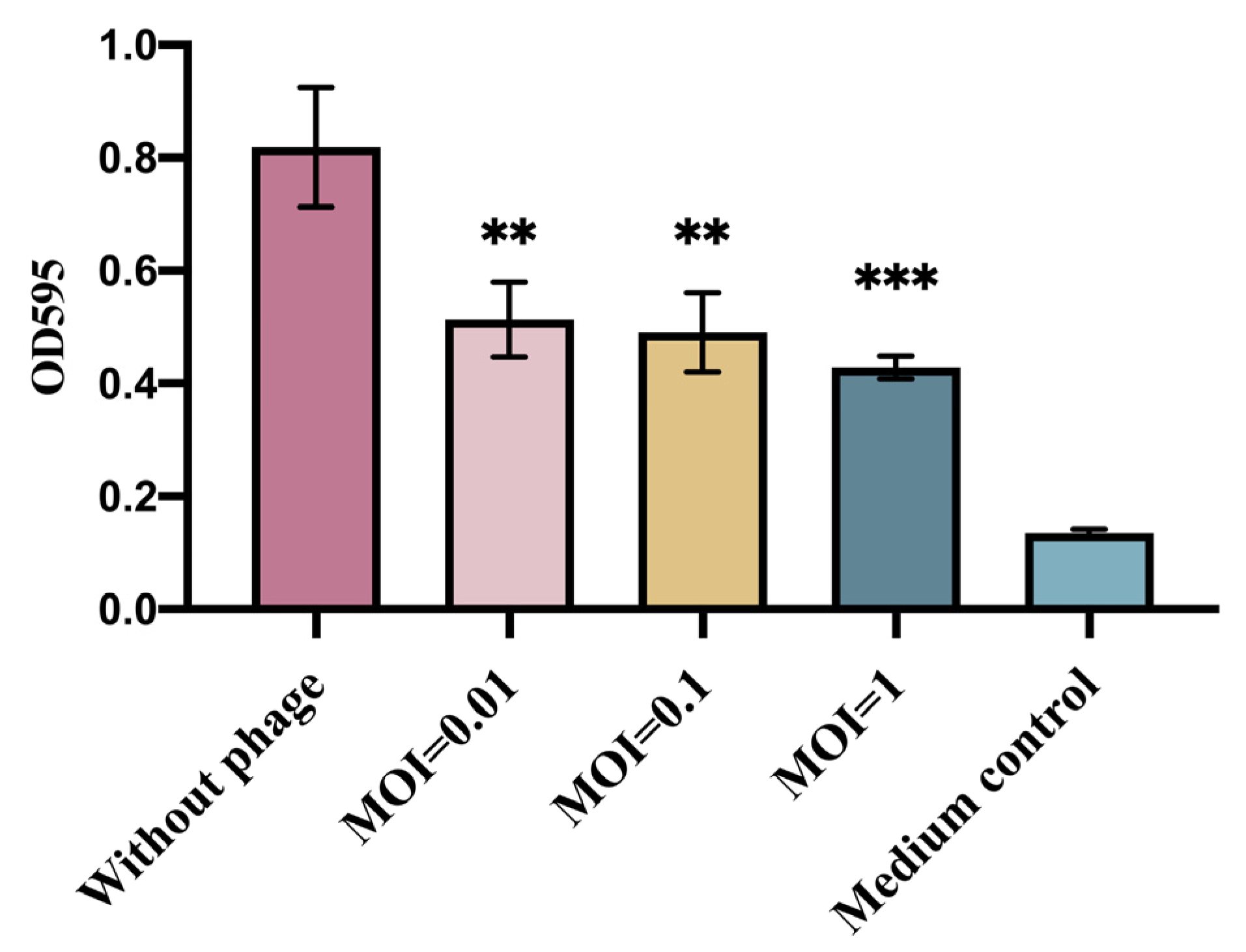
2.6. Determination of the Ability of Phages to Inhibit the Biofilm of Escherichia coli
2.7. Extraction and Analysis of Phage vB_EcoS_GN06 Genome
2.8. Statistical Analysis
3. Results
3.1. Characteristics of Phage vB_EcoS_GN06
3.2. In Vitro Phage vB_EcoS_GN06 Inhibition Test
3.3. Determination of the Ability of Phage vB_EcoS_GN06 to Inhibit the Biofilm of Escherichia coli
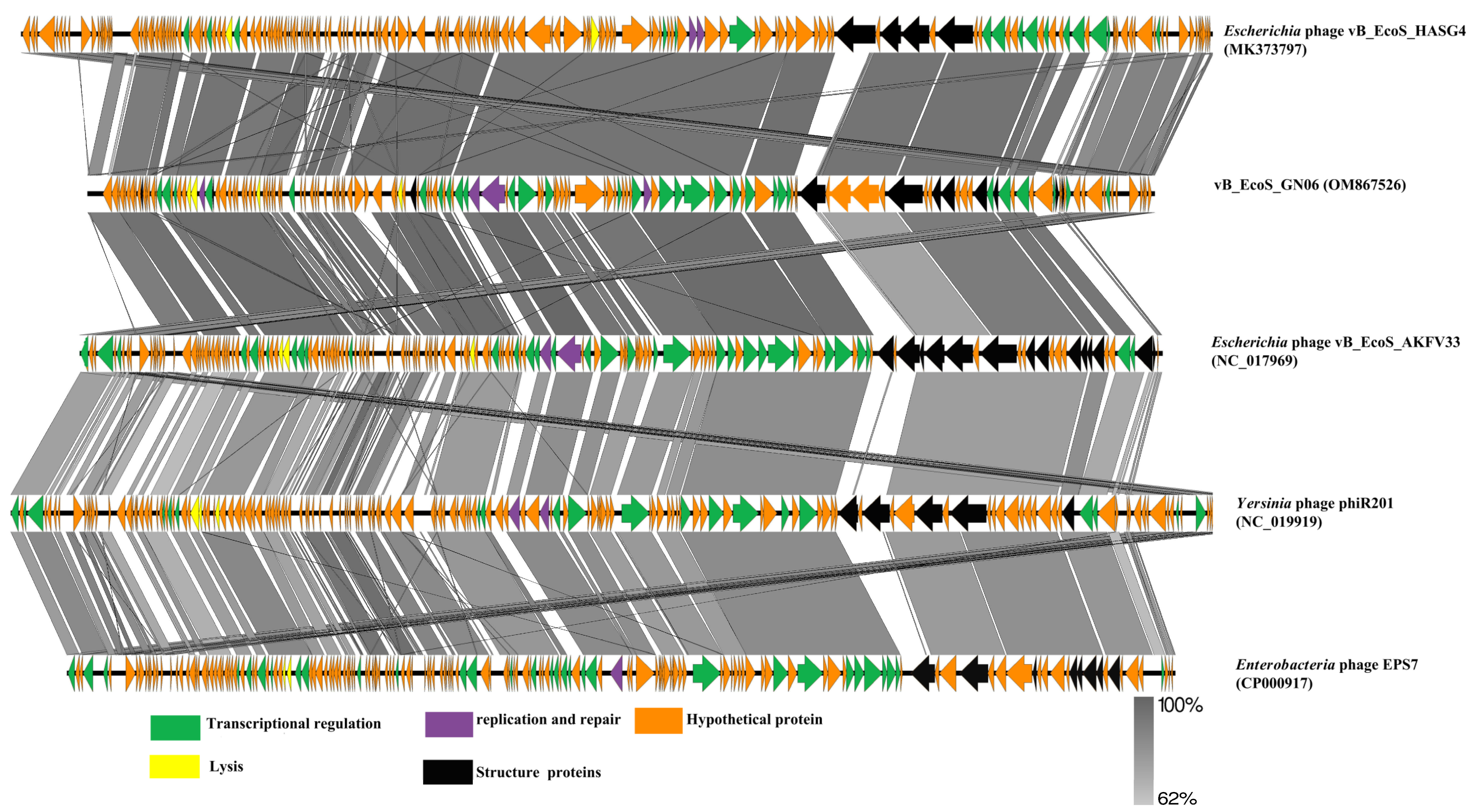
3.4. The Whole Genomic Analysis of the Phage vB_EcoS_GN06
3.5. Comparative Genomics and Phylogenetic Analysis of Phage vB_EcoS_GN06
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guabiraba, R.; Schouler, C. Avian colibacillosis: Still many black holes. FEMS Microbiol. Lett. 2015, 362, fnv118. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Mills, A.; Rushton, J.; Yeung, S. How antibiotics are used in pig farming: A mixed-methods study of pig farmers, feed mills and veterinarians in Thailand. BMJ Glob. Health 2020, 5, e001918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa, S.A.; Cruz-Cordova, A.; Luna-Pineda, V.M.; Reyes-Grajeda, J.P.; Cazares-Dominguez, V.; Escalona, G.; Sepulveda-Gonzalez, M.E.; Lopez-Montiel, F.; Arellano-Galindo, J.; Lopez-Martinez, B.; et al. Multidrug- and Extensively Drug-Resistant Uropathogenic Escherichia coli Clinical Strains: Phylogenetic Groups Widely Associated with Integrons Maintain High Genetic Diversity. Front. Microbiol. 2016, 7, 2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuniati, Y.; Hasanah, N.; Ismail, S.; Anitasari, S.; Paramita, S. Antibacterial Activity of Dracontomelon dao Extracts on Methicillin-Resistant S. aureus (Mrsa) and E. coli Multiple Drug Resistance (MDR). Afr. J. Infect. Dis. 2018, 12, 62–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Jamal, M.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Ur Rahman, S.; Das, C.R. Isolation, characterization and efficacy of phage MJ2 against biofilm forming multi-drug resistant Enterobacter cloacae. Folia Microbiol. 2019, 64, 101–111. [Google Scholar] [CrossRef]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef]
- Ripp, S. Bacteriophage-based pathogen detection. Adv. Biochem. Eng. Biotechnol. 2010, 118, 65–83. [Google Scholar] [CrossRef]
- Tan, D.; Zhang, Y.; Cheng, M.; Le, S.; Gu, J.; Bao, J.; Qin, J.; Guo, X.; Zhu, T. Characterization of Klebsiella pneumoniae ST11 Isolates and Their Interactions with Lytic Phages. Viruses 2019, 11, 1080. [Google Scholar] [CrossRef] [Green Version]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef] [Green Version]
- Seo, B.J.; Song, E.T.; Lee, K.; Kim, J.W.; Jeong, C.G.; Moon, S.H.; Son, J.S.; Kang, S.H.; Cho, H.S.; Jung, B.Y.; et al. Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella Typhimurium. J. Vet. Med. Sci. 2018, 80, 851–860. [Google Scholar] [CrossRef] [Green Version]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dabrowska, K.; Krylov, V.N.; Balcao, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Costa, A.R.; Pinto, G.; Meneses, L.; Azeredo, J. Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 2020, 44, 684–700. [Google Scholar] [CrossRef]
- Han, J.E.; Kim, J.H.; Hwang, S.Y.; Choresca, C.H., Jr.; Shin, S.P.; Jun, J.W.; Chai, J.Y.; Park, Y.H.; Park, S.C. Isolation and characterization of a Myoviridae bacteriophage against Staphylococcus aureus isolated from dairy cows with mastitis. Res. Vet. Sci. 2013, 95, 758–763. [Google Scholar] [CrossRef]
- Shende, R.K.; Hirpurkar, S.D.; Sannat, C.; Rawat, N.; Pandey, V. Isolation and characterization of bacteriophages with lytic activity against common bacterial pathogens. Vet. World 2017, 10, 973–978. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Bao, H.; Zhang, H.; Wang, R. Isolation and Characterization of Lytic Phage vB_EcoM_JS09 against Clinically Isolated Antibiotic-Resistant Avian Pathogenic Escherichia coli and Enterotoxigenic Escherichia coli. Intervirology 2015, 58, 218–231. [Google Scholar] [CrossRef]
- Holguin, A.V.; Rangel, G.; Clavijo, V.; Prada, C.; Mantilla, M.; Gomez, M.C.; Kutter, E.; Taylor, C.; Fineran, P.C.; Barrios, A.F.; et al. Phage PhiPan70, a Putative Temperate Phage, Controls Pseudomonas aeruginosa in Planktonic, Biofilm and Burn Mouse Model Assays. Viruses 2015, 7, 4602–4623. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef]
- Merril, C.R.; Scholl, D.; Adhya, S.L. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2003, 2, 489–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauden, M.I.; Martin-Benito, J.; Sanchez-Ferrero, J.C.; Pulido-Cid, M.; Valpuesta, J.M.; Carrascosa, J.L. Large terminase conformational change induced by connector binding in bacteriophage T7. J. Biol. Chem. 2013, 288, 16998–17007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afayibo, D.J.A.; Zhu, H.; Zhang, B.; Yao, L.; Abdelgawad, H.A.; Tian, M.; Qi, J.; Liu, Y.; Wang, S. Isolation, Molecular Characterization, and Antibiotic Resistance of Avian Pathogenic Escherichia coli in Eastern China. Vet. Sci. 2022, 9, 319. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Q.; Cheng, Y.; Liu, R.; Zhao, R.; Wang, J.; Wang, Y.; Yang, S.; Chen, A. Effect of Bacterial Resistance of Escherichia coli From Swine in Large-Scale Pig Farms in Beijing. Front. Microbiol. 2022, 13, 820833. [Google Scholar] [CrossRef]
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Li, L.; Wu, Y.; Ma, D.; Zhou, Y.; Wang, L.; Han, K.; Cao, Y.; Wang, X. Isolation and characterization of a novel Escherichia coli phage Kayfunavirus ZH4. Virus Genes 2022, 58, 448–457. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, J.; Guo, C.; Ge, C.; Wang, X.; Wei, D.; Li, X.; Si, H.; Hu, C. Identification and complete genome of lytic “Kp34likevirus” phage vB_KpnP_Bp5 and therapeutic potency in the treatment of lethal Klebsiella pneumoniae infections in mice. Virus Res. 2021, 297, 198348. [Google Scholar] [CrossRef]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Aríñez-Soriano, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Microencapsulation with alginate/CaCO3: A strategy for improved phage therapy. Sci. Rep. 2017, 7, 41441. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Ma, W.; Li, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 2020, 132, 109011. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Casjens, S.R.; Thuman-Commike, P.A. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 2011, 411, 393–415. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, A.N.; Woudstra, C.; Sorensen, M.C.H.; Brondsted, L. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput. Struct. Biotechnol. J. 2021, 19, 4854–4867. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, L.; Han, K.; Wang, L.; Cao, Y.; Ma, D.; Wang, X. A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods. Viruses 2022, 14, 286. [Google Scholar] [CrossRef]
- Casey, E.; van Sinderen, D.; Mahony, J. In Vitro Characteristics of Phages to Guide ‘Real Life’ Phage Therapy Suitability. Viruses 2018, 10, 163. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Mei, S.; Zhu, H. Multi-label multi-instance transfer learning for simultaneous reconstruction and cross-talk modeling of multiple human signaling pathways. BMC Bioinform. 2015, 16, 417. [Google Scholar] [CrossRef] [Green Version]
- Albers, S.; Czech, A. Exploiting tRNAs to Boost Virulence. Life 2016, 6, 4. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef] [PubMed]


| Strains | Genus | Place | Source | Serotype/Capsular Type | Sensitivity |
|---|---|---|---|---|---|
| GXEC-N01 | Escherichia coli | Guangxi | Avian | O157: H7 | - |
| GXEC-N02 | Escherichia coli | Guangxi | Human | Undetected | - |
| GXEC-N03 | Escherichia coli | Guangxi | Human | Undetected | - |
| GXEC-N04 | Escherichia coli | Guangxi | Avian | O126: K71(B16) | - |
| GXEC-N05 | Escherichia coli | Guangxi | Avian | O26: K60(B6) | - |
| GXEC-N06 | Escherichia coli | Guangxi | Avian | O126: K71(B16) | - |
| GXEC-N07 | Escherichia coli | Guangxi | Avian | O157: H7 | - |
| GXEC-N08 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N09 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N10 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N11 | Escherichia coli | Guangxi | Avian | O142: K86(B) | - |
| GXEC-N12 | Escherichia coli | Guangxi | Human | O127a: K63(B8) | - |
| GXEC-N13 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N14 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N15 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N16 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N17 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N18 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N19 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N20 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N21 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N22 | Escherichia coli | Guangxi | Avian | O78 | + |
| GXEC-N23 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N24 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N25 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N26 | Escherichia coli | Guangxi | Avian | Undetected | - |
| GXEC-N27 | Escherichia coli | Guangxi | Human | Undetected | - |
| GXEC-B01 | Escherichia coli | Guangxi | Pet | Undetected | - |
| GXEC-B02 | Escherichia coli | Guangxi | Pet | Undetected | - |
| GXEC-C01 | Escherichia coli | Guangxi | Bovine | O127a: K63(B8) | - |
| GDEC-F01 | Escherichia coli | Guangdong | Pig | Undetected | - |
| GDEC-F02 | Escherichia coli | Guangdong | Pig | Undetected | - |
| GDEC-F03 | Escherichia coli | Guangdong | Environment | Undetected | - |
| GDEC-F04 | Escherichia coli | Guangdong | Environment | O127a: K63(B8) | - |
| GDEC-F05 | Escherichia coli | Guangdong | Environment | O127a: K63(B8) | - |
| GDEC-F06 | Escherichia coli | Guangdong | Environment | O127a: K63(B8) | - |
| GDEC-F07 | Escherichia coli | Guangdong | Environment | O111: K58(B4) | - |
| GDEC-F08 | Escherichia coli | Guangdong | Environment | Undetected | - |
| GDEC-F09 | Escherichia coli | Guangdong | Environment | Undetected | - |
| GDEC-F10 | Escherichia coli | Guangdong | Environment | Undetected | - |
| GDEC-F11 | Escherichia coli | Guangdong | Avian | Undetected | - |
| GDEC-F12 | Escherichia coli | Guangdong | Avian | Undetected | - |
| GDEC-F13 | Escherichia coli | Guangdong | Avian | Undetected | - |
| GDEC-F14 | Escherichia coli | Guangdong | Avian | Undetected | - |
| GDEC-F15 | Escherichia coli | Guangdong | Environment | Undetected | - |
| SCEC-Z01 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z02 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z03 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z04 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z05 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z06 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z07 | Escherichia coli | Sichuan | Avian | O157 | - |
| SCEC-Z08 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z09 | Escherichia coli | Sichuan | Avian | Undetected | - |
| SCEC-Z10 | Escherichia coli | Sichuan | Avian | Undetected | - |
| CVCC1527 | Escherichia coli | China Veterinary Culture Collection Center (CVCC) | Pig | O8: K88 | - |
| CVCC4050 | Escherichia coli | China Veterinary Culture Collection Center (CVCC) | O157: H7 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Tan, Y.; Liao, Y.; Li, L.; Han, K.; Bai, H.; Cao, Y.; Li, J.; Gong, Y.; Wang, X.; et al. Isolation, Characterization and Whole Genome Analysis of an Avian Pathogenic Escherichia coli Phage vB_EcoS_GN06. Vet. Sci. 2022, 9, 675. https://doi.org/10.3390/vetsci9120675
Wang L, Tan Y, Liao Y, Li L, Han K, Bai H, Cao Y, Li J, Gong Y, Wang X, et al. Isolation, Characterization and Whole Genome Analysis of an Avian Pathogenic Escherichia coli Phage vB_EcoS_GN06. Veterinary Sciences. 2022; 9(12):675. https://doi.org/10.3390/vetsci9120675
Chicago/Turabian StyleWang, Leping, Yizhou Tan, Yuying Liao, Lei Li, Kaiou Han, Huili Bai, Yajie Cao, Jun Li, Yu Gong, Xiaoye Wang, and et al. 2022. "Isolation, Characterization and Whole Genome Analysis of an Avian Pathogenic Escherichia coli Phage vB_EcoS_GN06" Veterinary Sciences 9, no. 12: 675. https://doi.org/10.3390/vetsci9120675





